Background: Accumulation of the plant hormone jasmonoyl-l-isoleucine (JA-Ile) is tightly controlled to prevent overactivation of defense responses.
Results: Cytochrome P450 94s (CYP94s) with distinct tissue expression patterns localize to ER and oxidize JA-Ile to a dicarboxy derivative that fails to assemble JA-Ile co-receptor complexes.
Conclusion: Sequential CYP94-catalyzed oxidations block receptor activation and signaling.
Significance: P450s inactivate fatty acid-derived signals in both plants and animals.
Keywords: Cytochrome P450, Fatty Acid Oxidation, Plant Biochemistry, Plant Hormone, Stress Response, Jasmonate
Abstract
The plant hormone jasmonate (JA) controls diverse aspects of plant immunity, growth, and development. The amplitude and duration of JA responses are controlled in large part by the intracellular level of jasmonoyl-l-isoleucine (JA-Ile). In contrast to detailed knowledge of the JA-Ile biosynthetic pathway, little is known about enzymes involved in JA-Ile metabolism and turnover. Cytochromes P450 (CYP) 94B3 and 94C1 were recently shown to sequentially oxidize JA-Ile to hydroxy (12OH-JA-Ile) and dicarboxy (12COOH-JA-Ile) derivatives. Here, we report that a third member (CYP94B1) of the CYP94 family also participates in oxidative turnover of JA-Ile in Arabidopsis. In vitro studies showed that recombinant CYP94B1 converts JA-Ile to 12OH-JA-Ile and lesser amounts of 12COOH-JA-Ile. Consistent with this finding, metabolic and physiological characterization of CYP94B1 loss-of-function and overexpressing plants demonstrated that CYP94B1 and CYP94B3 coordinately govern the majority (>95%) of 12-hydroxylation of JA-Ile in wounded leaves. Analysis of CYP94-promoter-GUS reporter lines indicated that CYP94B1 and CYP94B3 serve unique and overlapping spatio-temporal roles in JA-Ile homeostasis. Subcellular localization studies showed that CYP94s involved in conversion of JA-Ile to 12COOH-JA-Ile reside on endoplasmic reticulum (ER). In vitro studies further showed that 12COOH-JA-Ile, unlike JA-Ile, fails to promote assembly of COI1-JAZ co-receptor complexes. The double loss-of-function mutant of CYP94B3 and ILL6, a JA-Ile amidohydrolase, displayed a JA profile consistent with the collaborative action of the oxidative and the hydrolytic pathways in JA-Ile turnover. Collectively, our results provide an integrated view of how multiple ER-localized CYP94 and JA amidohydrolase enzymes attenuate JA signaling during stress responses.
Introduction
The fatty-acid derived plant hormone jasmonate (JA)4 controls a wide range of chemical and morphological defense responses to arthropod herbivores, microbial pathogens and other forms of biotic stress (1, 2). JA also plays essential roles in plant growth and development, including sexual reproduction and growth regulation (3–5). Components of the core JA signaling module interact directly with many other phytohormone response pathways to mediate adaptive responses to changing environments (6, 7). Most JA responses are regulated at the transcriptional level by fluctuations in the intracellular level of the bioactive form of JA, jasmonoyl-l-isoleucine (8–10). Despite detailed knowledge of the enzymatic steps involved in the production of JA-Ile (2, 11, 12), relatively little is known about the enzymatic and cellular processes underlying other aspects of JA-Ile homeostasis, including JA transport and catabolism.
JA-Ile exerts its effects on gene expression by promoting degradation of Jasmonate ZIM-domain (JAZ) repressor proteins via the ubiquitin-proteasome system. In cells containing low levels of JA-Ile, the activity of transcription factors such as MYC2 that positively regulate JA-responsive genes is repressed by direct binding of JAZ proteins and associated co-repressors (13–15). Elevated JA-Ile levels promote binding of JAZ proteins to Coronatine Insensitive1 (COI1), which is the F-box protein component of the E3 ubiquitin ligase SCFCOI1 (14, 16). Proteolytic degradation of JAZ substrates via the ubiquitin-proteasome system releases MYC2 and other transcription factors from repression, thereby leading to expression of JA-response genes.
The JA signaling module is part of a complex phytohormone network that exerts exquisite control over the allocation of metabolic resources between the competing processes of growth and defense (17–19). Given the fitness costs associated with expression of JA-regulated defenses, which are energetically demanding and potentially toxic, plants appear to have evolved various strategies to attenuate the strength of these responses (20, 21). Several mechanisms to reign in JA responses have recently been elucidated (22). The rapid JA-dependent transcriptional induction of JAZ genes implies that de novo synthesized JAZ proteins exert negative feedback control (23). Alternative splicing events that remove or weaken the COI1-interacting JAZ degron provide a mechanism to produce JAZ variants that are more resistant to ubiquitin-mediated degradation, thus repressing JA responses in cells containing high JA-Ile levels (24–26). Similarly, some full-length JAZ proteins (e.g. JAZ8) have a non-canonical degron that does not interact with COI1 in the presence of JA-Ile (27). These JAZs therefore accumulate and repress their target transcription factors in the presence of JA-Ile. Another mechanism of negative feedback involves JA-dependent expression of negative-acting bHLH transcription factors that compete with MYC2 for cis-regulatory elements in the promoters of JA-responsive genes (28–31).
Superimposed upon these transcription-based mechanisms for signal attenuation is an independent negative feedback loop involving JA-induced catabolism of JA-Ile. Two major pathways for catabolic inactivation of JA-Ile have recently been described. One pathway is catalyzed by amidohydrolases that cleave JA-Ile to jasmonic acid and Ile (32–34). A second pathway involves oxidation of the C12 position JA-Ile to generate 12OH-JA-Ile, which is further oxidized to the carboxyl derivative 12COOH-JA-Ile (35–37). Elucidation of this pathway was advanced by the discovery that two members (CYP94B3 and CYP94C1) of the CYP94 family of cytochromes P450 in Arabidopsis catalyze the sequential oxidation reactions (38–40). 12OH-JA-Ile exhibits reduced capacity to promote formation of COI1-JAZ co-receptor complexes (38) but the activity of other oxidized derivatives of the hormone remains unknown.
Here, we report the identification and characterization of CYP94B1 as a third enzyme in the ω-oxidation pathway for JA-Ile catabolism. Our results indicate that CYP94B1 and CYP94B3 work coordinately to catalyze the initial oxidation of JA-Ile in overlapping and distinct tissues of Arabidopsis. We also show that sequential oxidation of JA-Ile by CYP94s occurs on endoplasmic reticulum (ER), and that the 12COOH-JA-Ile product of this pathway fails to promote the formation of COI1-JAZ co-receptor complexes. Collectively, our results provide an integrated view of how ER-localized CYP94 enzymes attenuate JA signaling during stress responses.
EXPERIMENTAL PROCEDURES
Plant Material, Growth, and Treatments
Arabidopsis thaliana ecotype Col-0 was used as the wild-type (WT) for all experiments, and was grown as previously described (41). T-DNA insertion lines cyp94b1-1 (SALK_129672) and cyp94b1-2 (SALK_132621 were obtained from the Arabidopsis Biological Resource Center. Previously described (33) ill6-1 (SALK_024894C) seed was provided by Dr. John Browse at Washington State University. A complete list of primers used in this study is provided in supplemental Table S1. Nicotiana tabacum (cv Petit Havana) plants used to infiltrate Agrobacterium tumefaciens harboring constructs for subcellular localization studies were grown in a growth chamber maintained under 18 h of light (40 μE m−2 s−1) at 23 °C and 6 h of dark at 18 °C.
Transgenic lines overexpressing CYP94B3 (At3g48520) were as described previously (38). Binary vector constructs for generating CYP94B1-OE lines were made by PCR-amplifying the full-length CYP94B1 (At5g63450) open reading frame (ORF) from WT genomic DNA template using the primer pair JH3_XbaI F and JH3_XhoI R1 (supplemental Table S1) and ligating the resulting PCR fragment into the XbaI and XhoI sites of a modified pBI121 vector (41), which places the gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter.
Transgenic plants expressing β-glucuronidase (GUS) under the control of the respective promoters were generated for the tissue-specific expression study of CYP94B1 and CYP94B3. Promoter regions 1.8 kb and 2.0 kb upstream of CYP94B1 and CYP94B3 translation initiation sites were PCR amplified from Arabidopsis (Col-0) genomic DNA using primers shown in supplemental Table S1 and were cloned in front of uidA gene (GUS) of pBI121 using ClaI and XmaI restriction sites.
The resulting 35S:CYP94B1, CYP94B1promoter:GUS and CYP94B3promoter:GUS constructs were transformed first into A. tumefaciens strain C58C1 then to Arabidopsis by the floral dip method. T1 seeds were first screened for their antibiotic resistance on Murashige and Skoog (MS) medium containing kanamycin (50 μg ml−1) and vancomycin (100 μg ml−1), and upon transfer to soil, 20 lines harboring 35S:CYP94B1 (CYP94B1-OE) were analyzed for mRNA expression of the transgene, jasmonate profile, and fertility. Among these, three CYP94B1-OE lines were propagated to obtain homozygous T3 lines. CYP94B1promoter:GUS (B1pro:GUS) and CYP94B3promoter:GUS (B3pro:GUS)lines were selected by staining 20 ∼ 24-d old rosette leaves for GUS activity after either treating with methyl jasmonate (MeJA) or mechanically wounding for 24 h. For MeJA treatment, ten microliter of 50 μm MeJA was spotted onto a leaf surface divided into 4∼6 droplets. For wounding, one leaf per plant was crushed twice across the midrib with a hemostat. MeJA treated or wounded leaves were collected after 24 h of treatment for histochemical GUS staining. Control untreated leaves were collected prior to MeJA or wounding treatment. Wounding treatment for JA measurements was conducted on fully expanded rosette leaves of 4∼5-week-old plants just before bolting by wounding twice across the midrib with a hemostat (42). At indicated times after wounding (Fig. 2), damaged leaves were harvested, weighed, flash frozen in liquid nitrogen, and stored at −80 °C until processed for JA extraction.
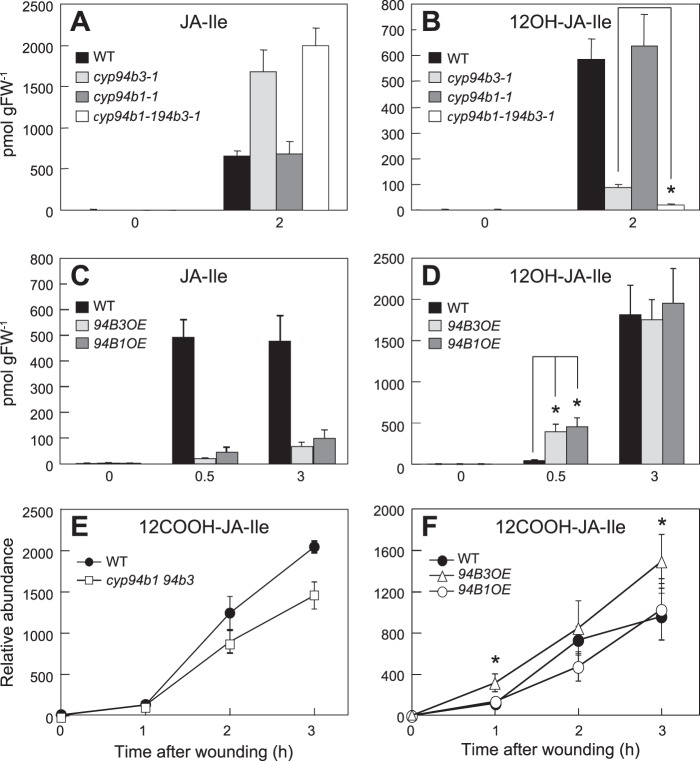
FIGURE 2.
Jasmonate profile in wounded rosette leaves of CYP94 T-DNA insertion mutants and overexpressing lines. Rosette leaves were wounded twice across the midrib with a hemostat. Damaged leaves were harvested for JA extraction at indicated times after wounding and analyzed by LC-MS/MS. A and B, JA-Ile and 12OH-JA-Ile levels in wounded (2 h) and unwounded (0 h) leaves of WT, cyp94b3-1, cyp94b1-1, and cyp94b1-1cyp94b3-1 plants. *, p < 0.001; Student's t test. C and D, time course of JA-Ile and 12OH-JA-Ile accumulation in wounded leaves of plants overexpressing CYP94B3 (94B3OE) or CYP94B1 (94B1OE). *, p < 0.01; Student's t test. E and F, relative abundance of 12COOH-JA-Ile in wounded leaves of WT, cyp94b1-1cyp94b3-1, 94B3OE, and 94B1OE at various times after wounding. *, p < 0.05 between WT and 94B3OE; Student's t test. Each data point represents the mean ± S.D. of three biological replicates.
Root growth inhibition assays were conducted with seedlings grown vertically on MS agar medium (0.8% sucrose) in square Petri plates supplemented with JA (10 μm) or coronatine (0.1 μm). Seedlings were grown for 8∼10 days under continuous light (100 μE m−2 s−1) in a growth chamber maintained at 21 °C.
Histochemical Staining and Microscopy
Histochemical detection of GUS activity was as described previously (43). Images of flowers were taken with a Leica MZ16 dissecting microscope. Pollen viability was determined with the fluorescein diacetate (FDA)/propidium iodide (PI) double staining procedure (3). Between 250 and 500 pollen grains were assessed in four independent trials for each genotype with an epifluorescence microscope (Zeiss Axiophot) (excitation at 365 nm, emission at 450 nm longpass). Green fluorescence under UV excitation emitted by de-esterified product of FDA indicates live cells whereas nonviable cells incorporate only PI, which fluoresce red orange under UV light.
For subcellular localization studies, ORFs of CYP94B3 and CYP94C1 were PCR amplified from WT genomic DNA using primer sets indicated in supplemental Table S1 and ligated into the XbaI and SalI restriction sites of the binary vector pVKH18En6-mRFP. An internal XbaI restriction site within the CYP94B3 gene was eliminated by using overlapping PCR to introduce a nonsense mutation (primers FBP-3873 and FBP-3874). Similarly, a SalI restriction site within CYP94C1 was removed using FBP-3877 and FBP-3878 primer set. The two gene constructs were transiently expressed in N. tabacum leaves by syringe-infiltrating the culture suspension of Agrobacterium strains harboring each construct. A second strain of Agrobacterium contained a construct encoding a soluble ER marker, GFP-HDEL (44), and was co-infiltrated to test co-localization. GFP and mRFP fluorescence was analyzed with an inverted Zeiss LSM 510 Meta Confocal Microscope using microscope settings described earlier (45).
Heterologous Expression in Yeast and In Vitro Assays
The CYP94B1 ORF was PCR amplified from WT Arabidopsis genomic DNA template using primers JH3_XmaI_F and JH3_XmaI_R (supplemental Table S1). Subsequent ligation into the XmaI site of yeast expression vector pYeDP60 was followed by transformation into Saccharomyces cerevisiae WAT11 strain (46). Preparation of microsomes and in vitro hydroxylation assays were described previously (38). Reactions were terminated by adding methanol (to 50% final concentration) containing internal standards for LC-MS/MS analysis.
In Vitro COI1-JAZ Pull-down Assays
Constructs and expression of JAZ proteins in E. coli were as previously described (25, 27). In vitro COI1-JAZ pull-down assays were performed as previously described (25). Ligands used in the COI1-JAZ pull-down assays were enantiomeric mixtures of chemically synthesized JA-Ile and 12COOH-JA-Ile (37).
Jasmonate Metabolite Analysis
Jasmonate extraction and analysis by LC-MS/MS was according to previously established methods (47). Transitions from deprotonated molecules to characteristic product ions were monitored in electrospray negative mode for JA-Ile (322→130), [13C6]JA-Ile (328→136), 12OH-JA-Ile (338→130), and 12COOH-JA-Ile (352→130). [13C6]JA-Ile was used as an internal standard for the quantification of JA-Ile and its derivatives. Peak area was integrated using Masslynx software (Waters), and the analytes were quantified based on standard curves generated by comparing analyte responses to the corresponding internal standard. Peak area normalized by the internal standard [13C6]JA-Ile and tissue weight was used to determine the relative abundance of 12COOH-JA-Ile. The 12OH-JA-Ile was a gift from Paul Staswick (University of Nebraska, Lincoln, NE) and [13C6]-JA-Ile was synthesized by conjugation of (±)-JA (Sigma) to [13C6]-l-Ile (Cambridge Isotope Laboratories) (48).
RNA Analysis
Total RNA from leaf tissue was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA blot analysis was as described previously (41). Gene-specific probes for CYP94B1, CYP94B3, and Actin-8 (ACT8) (At1g49240) were prepared by PCR amplification from genomic DNA template using primers shown in supplemental Table S1. Reverse transcription-PCR was used to detect CYP94B1 and CYP94B3 transcripts in cyp94b1-2cyp94b3-1 double mutant using primers indicated in supplemental Table S1.
RESULTS
CYP94B1 Encodes a JA-Ile 12-Hydroxylase
Previous analyses of wound responses in cyp94b3 single and cyp94b3cyp94c1 double mutants suggested that additional enzymes are involved in oxidation of JA-Ile to 12OH-JA-Ile (38–40). Candidates for this activity include uncharacterized members of the CYP94 family, which in Arabidopsis consists of six enzymes (20). The deduced amino acid sequence of CYP94B3 is most similar (77% identity) to that of CYP94B1, with both enzymes predicted to have the hallmark features of cytochromes P450 (49). Consistent with the hypothesis that CYP94B1 is a JA-Ile 12-hydroxylase, the wound-inducible expression pattern of the gene (At5g63450) encoding CYP94B1 was similar to that of the CYP94B3-encoding gene (At3g48520) (38, 40). To directly test this hypothesis, we expressed recombinant CYP94B1 in yeast (S. cerevisiae) and assayed the capacity of microsomal fractions to produce 12OH-JA-Ile from JA-Ile in vitro. Microsomes prepared from yeast cells expressing CYP94B1 produced a compound whose retention time and mass spectrum was identical to an authentic 12OH-JA-Ile standard (Fig. 1B, left panel). Low but detectable levels of a compound corresponding to 12COOH-JA-Ile (m/z 352 > 130) were also observed in the same reaction mixture (Fig. 1B, right panel). Control assays performed with microsomes isolated from cells expressing the pYeDP60 empty vector lacked JA-Ile hydroxylase activity (Fig. 1B). These results demonstrate that CYP94B1 converts JA-Ile to 12OH-JA-Ile and, to a lesser extent, 12COOH-JA-Ile in vitro.
FIGURE 1.
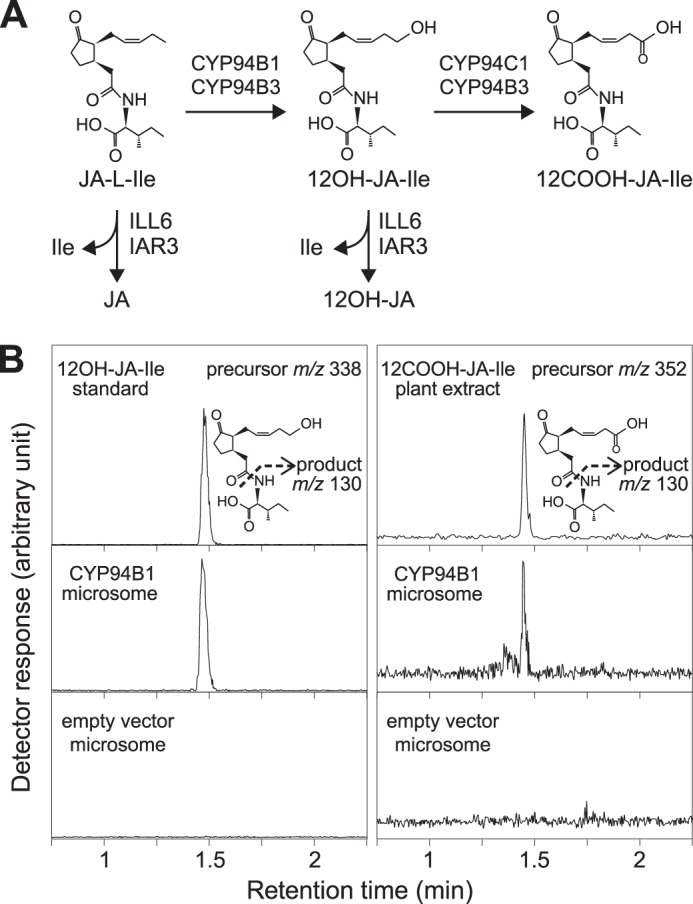
CYP94B1 expressed in yeast catalyzes ω-oxidation of JA-Ile. A, schematic summary of reactions catalyzed by CYP94s and JA amidohydrolases, ILL6 and IAR3. Assignment of enzymes for each step is based on in vitro and in vivo data described in the text and literatures. B, LC chromatogram of products obtained in an in vitro JA-Ile hydroxylation assay. Microsomal preparations from yeast transformed with either CYP94B1 or empty vector control were incubated with 50 μm JA-Ile for 2 h. Reaction products were analyzed by LC-MS/MS to detect 12OH-JA-Ile and 12COOH-JA-Ile. Precursor and product ions used for MS/MS detection are shown next to the chromatograms.
To investigate the biochemical function of CYP94B1 in planta, we used LC-MS/MS to measure the wound-induced production of JA-Ile and its oxidized derivatives in a T-DNA insertion mutant (cyp94b1-1) that fails to express CYP94B1 transcripts (data not shown). Consistent with our previous observation (38), the wound-induced accumulation of JA-Ile and 12OH-JA-Ile in wild-type (WT) and cyp94b1-1 plants was not significantly different (Fig. 2, A and B). To determine whether 12-hydroxylation activity of CYP94B3 compensates for the loss of CYP94B1, we also analyzed a double mutant that lacks both cyp94b1-1 and cyp94b3-1. Wounded leaves of a cyp94b3-1 single mutant exhibited a significant reduction in 12OH-JA-Ile levels (15% of WT) and also hyper-accumulated JA-Ile (Fig. 2, A and B), similar to previous reports (38–40). In the cyp94b1-1cyp94b3-1 double mutant, however, the level of 12OH-JA-Ile was further reduced to ∼3% of WT levels (Fig. 2B). We also observed that JA-Ile levels in wounded leaves of the double mutant were consistently higher than that in the cyp94b3-1 single mutant, although this increase was not statistically significant. These hormone profiles were reproduced in a cyp94b1-2cyp94b3-1 double mutant constructed with an independent mutant allele of cyp94b1, cyp94b1-2 (data not shown).
We also assessed the in vivo enzymatic function of CYP94B1 in transgenic lines (CYP94B1-OE) that overexpress CYP94B1 from the constitutive Cauliflower Mosaic Virus (CaMV) 35S promoter. Of 20 independent lines (T1 generation) tested, five showed high constitutive accumulation of CYP94B1 transcripts and strongly reduced levels of JA-Ile in wounded leaves (data not shown). Side-by-side comparison of selected CYP94B1-OE lines with previously characterized CYP94B3-OE lines showed that both CYP94Bs have similar effect on wound-induced accumulation of JA-Ile and 12OH-JA-Ile. For example, the level of JA-Ile was only 10 to 20% of that in WT plants at 0.5 and 3 h post wounding (Fig. 2, C and D). Likewise, wounded leaves of CYP94B1-OE and CYP94B3-OE plants hyperaccumulated 12OH-JA-Ile (relative to WT) at the 0.5 h time point. The levels of 12OH-JA-Ile in both lines were comparable to that in the WT leaves at the 3 h time point (Fig. 2D), which may reflect depletion of JA-Ile substrate (38). These collective findings show that CYP94B1 has JA-Ile 12-hydroxylase activity in vivo.
Given that recombinant CYP94B1 converts JA-Ile to 12COOH-JA-Ile in vitro, we measured 12COOH-JA-Ile levels in wounded leaves of the mutant (cyp94b1cyp94b3) and the overexpressing (CYP94B1-OE and CYP94B3-OE) lines (Fig. 2, E and F). The 12COOH-JA-Ile content in wounded leaves of the cyp94b1cyp94b3 line was only marginally affected, accumulating to ∼70% of that in WT leaves 2 h and 3 h after wounding. This finding reflects the major role of CYP94C1 in formation of 12COOH-JA-Ile (40). However, overexpression of CYP94B3 resulted in 1.5 ∼ 2.5-fold increase in 12COOH-JA-Ile compared with the WT, indicating the catalytic capacity of CYP94B3 to further oxidize 12OH-JA-Ile to 12COOH-JA-Ile (Fig. 2F). Even though recombinant CYP94B1 converted JA-Ile to 12COOH-JA-Ile in vitro (Fig. 1B), increased 12COOH-JA-Ile level was not observed in the CYP94B1-OE plants (Fig. 2F).
The ω-Oxidation and the Hydrolytic Pathways Cooperatively Contribute to JA Homeostasis in Vivo
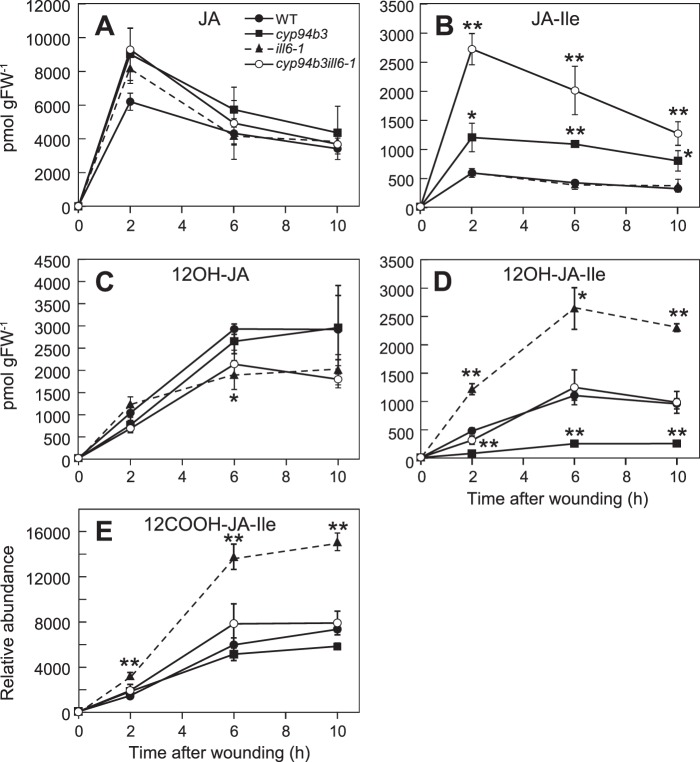
In addition to CYP94s, amidohydrolases previously studied for their function in auxin metabolism have been shown to metabolize JA substrates (Fig. 1A) (32–34). To test how these two classes of enzymes contribute to JA metabolism, JA profiles in the single and double T-DNA insertion mutants of cyp94b3-1 and ill6-1 were compared (Fig. 3). JA-Ile levels were increased in wounded leaves of the cyp94b3 but not the ill6-1 single mutant (Fig. 3B). Notably, however, JA-Ile levels in the cyp94b3ill6-1 double mutant were significantly greater than that in the cyp94b3 single mutant, indicating that both enzymes contribute to JA-Ile catabolism. We also found that the cyp94b3 and ill6-1 single mutations decreased and increased, respectively, wound-induced formation of 12OH-JA-Ile, consistent with previous reports (34, 38). Combining the two mutations appeared to cancel these effects, such that the level of 12OH-JA-Ile in the double mutant was comparable to that in WT plants (Fig. 3D). Similar to previous observations (34, 38, 40), the wound-induced JA content was not affected by either mutation (Fig. 3A). 12OH-JA levels, however, were reduced by the ill6-1 mutation, indicating that hydrolysis of 12OH-JA-Ile leads to 12OH-JA formation. 12COOH-JA-Ile levels were also significantly increased in wounded leaves of ill6-1 plants (Fig. 3E), presumably due to increased production of its precursor, 12OH-JA-Ile.
FIGURE 3.
Interaction between two major JA-Ile turnover pathways mediated by CYP94B3 and ILL6. A–E, time course of jasmonate accumulation in wounded rosette leaves of WT, cyp94b3-1, ill6-1, and cyp94b3-1ill6-1 plants. Asterisks denote significant difference between WT and mutant at p < 0.01 (*) or p < 0.001 (**); Student's t test. Each data point represents the mean ± S.D. of three biological replicates.
Overexpression of CYP94B1 Results in JA-deficient Phenotypes
We next studied the physiological effects of CYP94B1 overexpression in CYP94B1-OE lines in two well-established JA-mediated physiological processes, fertility and JA-induced root growth inhibition. Five independent T1 lines expressing high levels of CYP94B1 transcripts displayed defects in fertility (Fig. 4, panels A–C). Unlike WT flowers, the stigma of CYP94B1-OE flowers protruded through closed petals at developmental stages prior to pollination (Fig. 4B, bottom panel). Elongated stigma papillae were also visible in the infertile flowers (Fig. 4C, right panel). Anther dehiscence and release of pollen grains occurred without noticeable defects but shorter anther filaments relative to the taller pistils on CYP94B1-OE flowers prevented successful pollination (Fig. 4C). Pollen collected from CYP94B1-OE flowers was also less viable than WT pollen (40∼60% WT; Fig. 4D).
FIGURE 4.
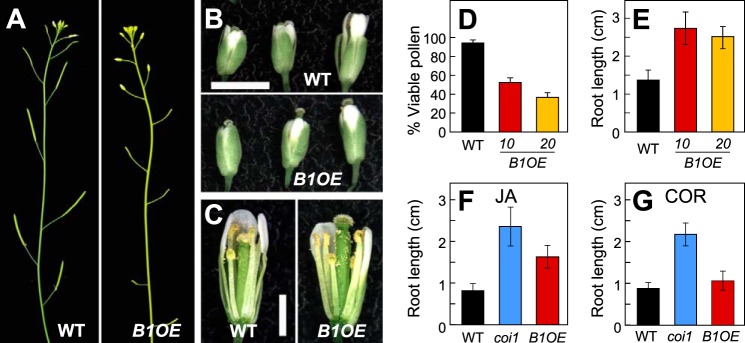
CYP94B1-OE plants display JA-resistant phenotypes in roots and flowers. A, photographs showing silique development in WT (left panel) and CYP94B1-OE (right). B, photographs of representative WT (top panel) and CYP94B1-OE (bottom) flowers at three developmental stages. Scale bar, 2 mm. C, developing stamens and pistils of WT (left) and CYP94B1-OE (right) at time of pollination. Petals and sepals were removed to expose the interior parts of flowers. Scale bar, 1 mm. D, pollen viability of WT and two independent homozygous lines of CYP94B1-OE (B1OE-10 and B1OE-20). Pollen viability was assessed in four independent trials. E - G, root growth inhibition assays. WT, CYP94B1-OE (B1OE), and coi1-1 were grown on MS medium containing either 10 μm jasmonic acid (E, F) or 0.1 μm coronatine (COR) (G). Root length was determined on 10-d (E, F) or 8-d (G) old plants. Data show the mean ± S.D. (n > 20).
In addition to reproductive phenotypes, roots of CYP94B1-OE seedlings were highly resistant to the growth inhibitory effects of exogenous JA (Fig. 4, E and F). The extent of insensitivity to JA was not as strong in CYP94B1-OE as in coi1-1, which have completely lost responsiveness to JA (Fig. 4F). To examine if the reduced sensitivity to JA-induced growth inhibitory effects is due to a defect in JA perception or downstream signaling events, root growth inhibition assays were performed with the bacterial toxin coronatine, which is a structural mimic of JA-Ile and potent agonist of the COI1-JAZ receptor (16, 50). Unlike control coi1-1 seedlings that are insensitive to coronatine, growth of CYP94B1-OE roots was as sensitive to coronatine as WT roots, indicating that CYP94B1-OE seedlings possess an intact COI1-JAZ signaling system (Fig. 4G). This also suggests that coronatine is not a substrate for CYP94B1. Together, these findings demonstrate that overexpression of CYP94B1 depletes JA-Ile to attenuate JA signaling in vivo.
Tissue-specific Expression of CYP94B1 and CYP94B3
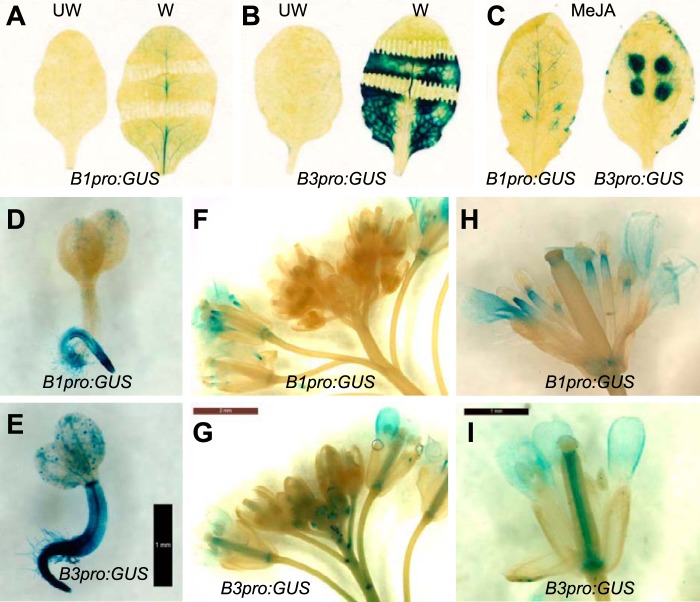
Given the finding that CYP94B1 and CYP94B3 serve a similar biochemical role in oxidation and inactivation of JA-Ile, we next addressed the question of whether the corresponding genes have similar tissue specific expression patterns. The promoter regions of CYP94B1 (1.8 kb) and CYP94B3 (2.0 kb) were fused to the β-glucuronidase (GUS)-encoding uidA reporter gene and the resulting constructs, designated as B1pro:GUS and B3pro:GUS, were transformed into Arabidopsis. At least 25 independent T1 lines were subject to histochemical staining with 5-bromo-4-chloro-3-indolyl β-glucuronic acid (X-gluc) to determine the range of expression patterns; subsequent experiments were performed with lines exhibiting a representative staining pattern (Fig. 5). Mature rosette leaves of B1pro:GUS and B3pro:GUS lines displayed a very low background of GUS staining prior to elicitation (Fig. 5, A and B labeled UW). Mechanical wounding and MeJA treatment, however, induced GUS expression patterns that were spatially distinct between the two lines (Fig. 5, A–C). Wounding activated B1pro:GUS expression mainly in the vascular regions, whereas it elicited very strong expression of the B3pro:GUS reporter throughout the leaf lamina (Fig. 5B). MeJA treatment induced GUS expression in both reporter lines, with staining in B3pro:GUS being more intense (Fig. 5C). GUS staining in the B3pro:GUS was more spatially restricted to the site of MeJA application, producing dark blue circles with sharply defined borders (Fig. 5C, right panel). More prominent staining in the vascular system was observed in MeJA-treated B3pro:GUS leaves (Fig. 5C).
FIGURE 5.
Tissue-specific expression of CYP94B1 and CYP94B3. A and B, X-gluc staining of control unwounded (UW) or wounded (W) (for 24 h) leaves of CYP94B1promoter:GUS (B1pro:GUS) and CYP94B3promoter:GUS (B3pro:GUS) plants. C, B1pro:GUS (left panel) and B3pro:GUS (right) leaves treated with MeJA (for 24 h). Several droplets (10 μl) of a solution containing 50 μm MeJA were spotted onto the leaf surface. D and E, X-gluc staining of B1pro:GUS and B3pror:GUS seedlings 2∼3-days after germination. Bar, 1 mm. F-I, influorescence and flowers of B1pro:GUS and B3pro:GUS. Bars, 2 mm (G) and 1 mm (I).
GUS staining of whole seedlings and reproductive tissue also revealed differences in the spatial expression pattern of CYP94B1 and CYP94B3. In seedlings, expression of B1pro:GUS was largely restricted to the roots (Fig. 5D), whereas B3pro:GUS was highly expressed both in the shoot and root (Fig. 5E). In the influorescence, B3pro:GUS was expressed in younger unopened buds as well as in opened mature flowers (Fig. 5, G and I), whereas B1pro:GUS was expressed predominantly in opened mature flowers (Fig. 5, F and H). Within the flower, CYP94B1 was expressed in the stamen at the joint between anther and filament. CYP94B3 expression was more localized to the center of pistil. Additionally, CYP94B3 was expressed strongly at the base of the initiating lateral peduncles. Both genes were expressed in petals (Fig. 5, F-I) but neither gene appeared to be expressed in mature pollen, where some JA biosynthetic genes are expressed (3, 43). In summary, results from these promoter-GUS studies show that CYP94B1 and CYP94B3 have both overlapping and distinct tissue-specific expression patterns.
Subcellular Localization of JA-Ile-oxidizing CYP94 Enzymes
The cytosolic location of JA-Ile-conjugation (51), together with the site of JA-Ile perception in the nucleus (52), raises the question about the site of JA-Ile inactivation. Although many P450s reside in the ER facing the cytosolic side (49, 53), there are examples of plant P450s that are localized to other subcellular compartments, including mitochondria (54, 55) and chloroplasts (56, 57). We therefore determined the subcellular location of CYP94B3 and CYP94C1 by generating constructs in which the monomeric red fluorescent protein (mRFP) was fused to the C terminus of CYP94B3 and CYP94C1, designated CYP94B3-mRFP and CYP94C1-mRFP, respectively. An Agrobacterium tumefaciens co-infiltration assay (44) was used to transiently co-express these constructs in tobacco (N. tabacum) leaves along with a soluble ER localization marker fused to the green fluorescent protein (GFP-HDEL) (44). Laser scanning confocal microscopy analyses of the CYP94B3-mRFP and CYP94C1-mRFP infiltrated leaves produced images typical of the ER network. The ER localization pattern was confirmed by the overlap of the ER marker GFP-HDEL with that of CYP94B3-mRFP and CYP94C1-mRFP (Fig. 6). These findings indicate that the ER is the site of JA-Ile oxidative turnover by CYP94 P450s.
FIGURE 6.
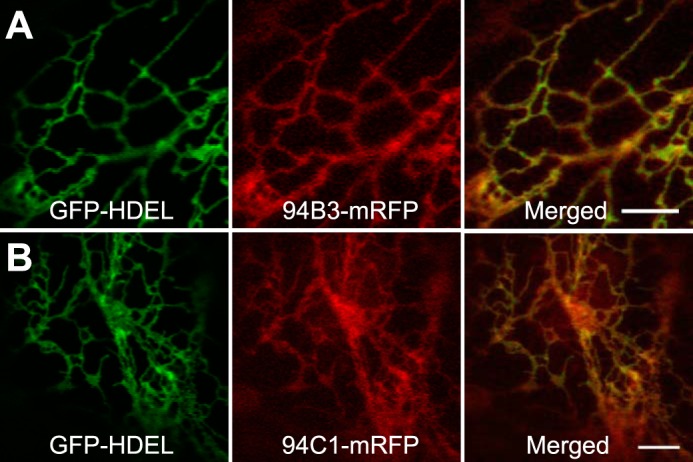
Subcellular localization of CYP94B3 and CYP94C1. Confocal images of tobacco leaf epidermal cells co-expressing the GFP-HDEL ER marker (left panel of A and B), and either CYP94B3 (94B3-mRFP) or CYP94C1 (94C1-mRFP) fused to the monomeric red fluorescent protein (middle panel of A and B). Merge between GFP and mRFP indicates co-localization (right panel of A and B). Bars, 5 μm.
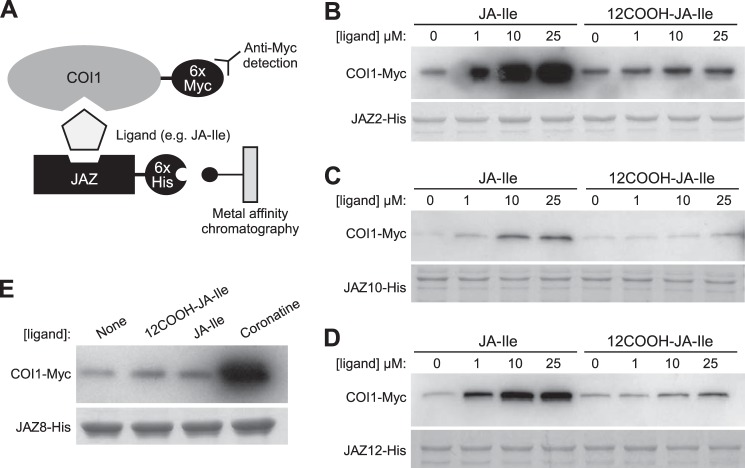
12COOH-JA-Ile Is Inactive in Promoting COI1-JAZ Interaction
It was previously observed that although 12OH-JA-Ile is less effective than JA-Ile in promoting COI1-JAZ interaction, this oxidized derivative maintains some activity as a receptor ligand (38). To test whether further oxidation to 12COOH-JA-Ile has a more pronounced effect on receptor activation, we used an in vitro pull-down assay (Fig. 7A) (14, 16, 27) to compare the ability of JA-Ile and 12COOH-JA-Ile to promote the COI1 interaction with four representative JAZ substrates. As shown in Fig. 7, JA-Ile stimulated COI1 binding to JAZ2, JAZ10, and JAZ12 in a dose-dependent manner, as previously reported (25, 27). 12COOH-JA-Ile, however, was largely inactive in promoting COI1-JAZ interaction with all JAZs tested (Fig. 7). We also found that 12COOH-JA-Ile fails to promote interaction of COI1 with JAZ8 (Fig. 7E), which contains a non-canonical degron that binds COI1 in the presence of coronatine but not JA-Ile (27).
FIGURE 7.
12COOH-JA-Ile does not promote assembly of COI1-JAZ receptor complexes. A, schematic of in vitro pull-down assays. Ligand-dependent interaction between COI1 and (B) JAZ2, (C) JAZ10, (D) JAZ12, and (E) JAZ8. Pull-down assays were performed using recombinant JAZ-His protein and crude leaf extract from Arabidopsis plants expressing 35S:COI1-Myc in the presence of the indicated concentrations of JA-Ile, 12COOH-JA-Ile or coronatine. Reactions with JAZ8 contained 25 μm of 12COOH-JA-Ile and JA-Ile or 2.5 μm coronatine. Equal recovery and loading of JAZ-His protein is indicated by a Coomassie Blue-stained acrylamide gel.
DISCUSSION
Hydroxylation is commonly used in detoxification pathways to convert hydrophobic toxic chemicals into more water-soluble products to be stored away from active sites or to be excreted (58, 59). Overactivation of JA pathway can have “toxic” effects on plants, for instance, by causing growth arrest. Multiple lines of evidence presented in this paper demonstrate that CYP94B1 is a JA-Ile-12-hydroxylase that participates in metabolic inactivation of the JA-Ile signal.
Recombinant CYP94B1 expressed in yeast converted JA-Ile substrate into 12OH-JA-Ile and also to lesser amounts of 12COOH-JA-Ile. Two consecutive oxidations of JA-Ile to 12COOH-JA-Ile were previously observed with CYP94B3 and CYP94C1, with major products being 12OH-JA-Ile and 12COOH-JA-Ile, respectively (38, 40). Our results indicate that CYP94B1, like B3, acts primarily in catalysis of the first hydroxylation step. That wound-induced formation of 12OH-JA-Ile in the leaves of the cyp94b1cyp94b3 double mutant was reduced to only 3% of that in WT indicates that CYP94B1 and CYP94B3 account for ∼ 97% of the 12-hydroxylation activity in vivo. The remainder may result from combined contributions by other CYP94 subfamily enzymes such as CYP94B2 or CYP94C1. Considering the near complete depletion of 12OH-JA-Ile in wounded cyp94b1cyp94b3 leaves (Fig. 2B), it was unexpected that the production of 12COOH-JA-Ile in this mutant was maintained at near (70%) WT levels (Fig. 2E). This implies that CYP94C1 can catalyze two consecutive oxidation steps using JA-Ile as substrate. On the other hand, the fact that loss of CYP94C1, which is the major enzyme for 12COOH-JA-Ile formation, results in only a ∼50% reduction in 12COOH-JA-Ile, implies the existence of another enzyme(s) that can catalyze 12COOH-JA-Ile formation. A near-to-complete loss of 12COOH-JA-Ile in cyp94b3cyp94c1 (40) indicates that CYP94B3 is directly involved in 12COOH-JA-Ile formation in vivo, especially in the absence of CYP94C1. Increased accumulation of 12COOH-JA-Ile in CYP94B3-OE (Fig. 2F) also supports a role for CYP94B3 in the formation of 12COOH-JA-Ile. Overexpression of CYP94B1 did not increase the level of 12COOH-JA-Ile over WT, despite its ability to phenocopy the effects of CYP94B3-OE on JA-Ile and 12OH-JA-Ile levels (Fig. 2, D and F). Therefore, our collective results indicate that CYP94B1 is mainly involved in the initial oxidation of JA-Ile to 12OH-JA-Ile, but that a role for this CYP94 member in metabolism of other substrates cannot be excluded.
The decline in 12OH-JA-Ile levels in the cyp94b3cyp94c1 double mutant, in which 12OH-JA-Ile conversion to 12COOH-JA-Ile is blocked (40), indicates pathways for 12OH-JA-Ile metabolism other than oxidization to 12COOH-JA-Ile. A recently reported O-linked glucosyl derivative of 12OH-JA-Ile (37) could potentially serve as a sink for 12OH-JA-Ile. Additionally, 12OH-JA-Ile is hydrolyzed to 12OH-JA and Ile by the IAR3 and ILL6 members of the amidohydrolase superfamily in Arabidopsis (33, 34) and homologous enzymes (JIH1) in tobacco (32). Even though the effect of ILL6 mutation on JA-Ile level was minimal in the ill6-1 single mutant (Fig. 3B) (34), additional loss of CYP94B3 function created a major blockage in JA-Ile turnover and, consequently, a massive accumulation of JA-Ile in the cyp94b3ill6-1 double mutant. Together with the reported in vitro hydrolytic activity (33, 34), this genetic evidence confirms the function of ILL6 in JA-Ile turnover in vivo. It remains to be determined whether the 'ω-oxidation' and the hydrolysis pathways catalyzed by CYP94s and amidohydrolases, respectively, are solely responsible for JA-Ile catabolism, or whether additional pathways are involved (e.g. methylation). Higher order knock-out mutants defective in multiple CYP94s and amidohydrolases will help answer this question.
Strong JA-deficient phenotypes created by overexpressing CYP94B1 indicates that increased flux through ω-oxidation leads to deactivation of JA signaling, implying that the oxidized products are inactive in inducing JA responses. Our previous study showed that 12OH-JA-Ile is significantly reduced in its ability to promote interaction between COI1 and JAZs but retains some activity (38). Therefore, we could not exclude the possibility that 12OH-JA-Ile associates with the receptor in vivo. We show here that 12COOH-JA-Ile is essentially inactive in promoting COI1 interaction with a broad range of JAZ substrates, indicating that consecutive oxidation of JA-Ile to 12COOH-JA-Ile is an effective means to terminate signaling (Fig. 7). Additional modifications to the C12 hydroxy group, such as sulfation or glucosylation, may also inhibit COI1-JAZ complex-promoting activity by restricting entry of the ligand into the binding pocket of COI1 (50). Additional structural studies are needed to better understand how modification of JA-Ile by CYP94 P450s and other enzymes affect ligand interaction with the COI1-JAZ co-receptor system.
In addition to the compromised receptor binding capacity, altered physical properties (e.g. polarity) may play role in the loss of signaling activity by oxidized JA-Ile derivatives, for example by changing their subcellular partitioning. The subcellular distribution of various JA derivatives is not well understood. Our subcellular localization of CYP94B3 and CYP94C1 demonstrates the ER as the major site for JA-Ile oxidation. Given the established topology of many P450s in ER membranes (49, 53), we presume that the CYP94 N terminus is imbedded in the ER with the enzyme's active site facing the cytosol. Even though CYP94B1 was not tested for its subcellular location, the similarities of this isoform with CYP94B3 suggest that it also localizes to the ER. It is plausible that JA metabolites are rapidly exchanged between the nucleus and the ER through lateral diffusion in the ER membrane, which forms a continuum with the outer membrane of the nuclear envelope (60). The molecular organization of JA-metabolizing enzymes and metabolites in multiple subcellular compartments needs further investigation. Given the known biological activities of some oxidized- and glucosylated JAs (61, 62), it is also possible that these JA-Ile derivatives may have molecular targets other than the COI1.
The promoter-GUS reporters of CYP94B1 and CYP94B3 revealed their tissue specific expression patterns. Notable differences in the expression pattern of these two genes include: i) in reproductive organs where CYP94B1 was strongly expressed (e.g. upper anther filament), CYP94B3 expression was not detected; ii) in wounded leaves where CYP94B1 expression was strongest (e.g. in vascular tissues), expression of CYP94B3 was more evenly dispersed in all cell types; and iii) in seedlings where CYP94B1 expression was largely restricted to the roots. In the inflorescence stems, CYP94B3 was also strongly expressed at the base of the initiating lateral peduncles, reminiscent of the expression of lateral organ boundaries (LOB) gene, recently shown to function in defining organ boundaries through interplay with brassinosteroids (63).
The combined effects of the hydrolysis pathway catalyzed by amidohydrolases and the ω-oxidation pathway catalyzed by CYP94s represent an evolutionarily conserved mechanism to maintain optimal levels of the potent JA-Ile signal. At least 70 CYP94-like sequences have been reported in sequenced plant genomes (20). The vast majority of these have not been characterized (59, 64) but based on function of Arabidopsis CYP94s, many of them are expected to be involved in metabolism of JAs. Likewise, although auxin amidohydrolases have been characterized in only a handful of plant species (65, 66), their related sequences have been reported from many plant species (67). In addition to their conservation across higher plants, tight co-regulation of these pathways (33, 34, 38, 40) implies selection pressure to limit the production of JA-Ile and thereby attenuate the strength of JA responses. Reverse genetic approaches aimed at blocking all catabolic steps may be useful to elucidate those essential functions of JA-Ile catabolism. Attenuation of the JA signal depends also on other mechanisms, including negative transcriptional control of JA responses that are also tightly co-regulated with JA-regulated genes (24, 25, 27, 28). Additional studies are needed to understand how the concerted action of these various negative feedback loops restrains JA-responses and impacts plant growth under various stress conditions.
Supplementary Material
Acknowledgments
We thank Nizam Zubir Mohamad, Noor Kamila Ahmad Shafiai, Li Zhang, Thomas Marcink, Priya Bariya, and Hannah Wahl for technical assistance. We are grateful to Paul Staswick for providing 12OH-JA-Ile, Joe Chappell for providing yeast strain WAT11, John Browse for ill6-1 seeds, Sheng Yang He for providing the Arabidopsis 35S:COI1-Myc line, and the Arabidopsis Biological Resource Center for providing T-DNA insertion lines.
This work was supported in part by National Institutes of Health Grant No. GM57795 (to G. A. H.) and the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U. S. Department of Energy (Grant No. DE-FG02-91ER20021) for partial support of A. J. K. and C. T.

This article contains supplemental Table S1.
- JA
- jasmonate
- JAZ
- Jasmonate ZIM-domain
- CYP
- cytochrome P450
- JA-Ile
- jasmonoyl-l-isoleucine
- MeJA
- methyl-jasmonic acid
- 12OH-JA-Ile
- 12-hydroxy-jasmonoyl-l-isoleucine
- 12COOH-JA-Ile
- 12-carboxy-jasmonoyl-isoleucine
- GUS
- β-glucuronidase
- X-glu
- β-glucuronic acid.
REFERENCES
- 1. Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 [DOI] [PubMed] [Google Scholar]
- 2. Wasternack C., Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 111, 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McConn M., Browse J. (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li L., Zhao Y., McCaig B. C., Wingerd B. A., Wang J., Whalon M. E., Pichersky E., Howe G. A. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y., Turner J. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3, e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song S., Qi T., Wasternack C., Xie D. (2014) Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21, 112–119 [DOI] [PubMed] [Google Scholar]
- 7. Vanstraelen M., Benková E. (2012) Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487 [DOI] [PubMed] [Google Scholar]
- 8. Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 [DOI] [PubMed] [Google Scholar]
- 9. Koo A. J., Howe G. A. (2009) The wound hormone jasmonate. Phytochemistry 70, 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 [DOI] [PubMed] [Google Scholar]
- 11. Schaller A., Stintzi A. (2009) Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry 70, 1532–1538 [DOI] [PubMed] [Google Scholar]
- 12. Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., Jez J. M. (2012) Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 13. Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E. E. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S. Y., Howe G. A., Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 [DOI] [PubMed] [Google Scholar]
- 15. Chini A., Fonseca S., Fernández G., Adie B., Chico J. M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F. M., Ponce M. R., Micol J. L., Solano R. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 [DOI] [PubMed] [Google Scholar]
- 16. Katsir L., Schilmiller A. L., Staswick P. E., He S. Y., Howe G. A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang D. L., Yao J., Mei C. S., Tong X. H., Zeng L. J., Li Q., Xiao L. T., Sun T. P., Li J., Deng X. W., Lee C. M., Thomashow M. F., Yang Y., He Z., He S. Y. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noir S., Bömer M., Takahashi N., Ishida T., Tsui T. L., Balbi V., Shanahan H., Sugimoto K., Devoto A. (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161, 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ullmann-Zeunert L., Stanton M. A., Wielsch N., Bartram S., Hummert C., Svatoš A., Baldwin I. T., Groten K. (2013) Quantification of growth–defense trade-offs in a common currency: nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant J. 75, 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koo A. J., Howe G. A. (2012) Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huot B., Yao J., Montgomery B. L., He S. Y. (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campos M. L., Kang J. H., Howe G. A. (2014) Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung H. S., Koo A. J., Gao X., Jayanty S., Thines B., Jones A. D., Howe G. A. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moreno J. E., Shyu C., Campos M. L., Patel L. C., Chung H. S., Yao J., He S. Y., Howe G. A. (2013) Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 162, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chung H. S., Howe G. A. (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chung H. S., Niu Y., Browse J., Howe G. A. (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70, 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shyu C., Figueroa P., Depew C. L., Cooke T. F., Sheard L. B., Moreno J. E., Katsir L., Zheng N., Browse J., Howe G. A. (2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakata M., Mitsuda N., Herde M., Koo A. J., Moreno J. E., Suzuki K., Howe G. A., Ohme-Takagi M. (2013) A bHLH-type transcription factor, ABA-inducible BHLH-type transcription factor/JA-associated MYC2-like1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25, 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sasaki-Sekimoto Y., Jikumaru Y., Obayashi T., Saito H., Masuda S., Kamiya Y., Ohta H., Shirasu K. (2013) Basic helix-loop-helix transcription factors Jasmonate-associated MYC2-like1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 163, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song S., Qi T., Fan M., Zhang X., Gao H., Huang H., Wu D., Guo H., Xie D. (2013) The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9, e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fonseca S., Fernández-Calvo P., Fernández G. M., Díez-Díaz M., Gimenez-Ibanez S., Lopez-Vidriero I., Godoy M., Fernández-Barbero G., Van Leene J., De Jaeger G., Franco-Zorrilla J. M., Solano R. (2014) bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS ONE 9, e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woldemariam M. G., Onkokesung N., Baldwin I. T., Galis I. (2012) Jasmonoyl-l-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-l-isoleucine levels and attenuates plant defenses against herbivores. Plant J. 72, 758–767 [DOI] [PubMed] [Google Scholar]
- 33. Bhosale R., Jewell J. B., Hollunder J., Koo A. J., Vuylsteke M., Michoel T., Hilson P., Goossens A., Howe G. A., Browse J., Maere S. (2013) Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell 25, 2865–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Widemann E., Miesch L., Lugan R., Holder E., Heinrich C., Aubert Y., Miesch M., Pinot F., Heitz T. (2013) The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J. Biol. Chem. 288, 31701–31714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glauser G., Grata E., Dubugnon L., Rudaz S., Farmer E. E., Wolfender J. L. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 283, 16400–16407 [DOI] [PubMed] [Google Scholar]
- 36. Guranowski A., Miersch O., Staswick P. E., Suza W., Wasternack C. (2007) Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 581, 815–820 [DOI] [PubMed] [Google Scholar]
- 37. Kitaoka N., Kawaide H., Amano N., Matsubara T., Nabeta K., Takahashi K., Matsuura H. (2014) CYP94B3 activity against jasmonic acid amino acid conjugates and the elucidation of 12-O-β-glucopyranosyl-jasmonoyl-l-isoleucine as an additional metabolite. Phytochemistry 99, 6–13 [DOI] [PubMed] [Google Scholar]
- 38. Koo A. J., Cooke T. F., Howe G. A. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 40. Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., Pinot F. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287, 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koo A. J. K., Chung H. S., Kobayashi Y., Howe G. A. (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520 [DOI] [PubMed] [Google Scholar]
- 42. Herde M., Koo A. K., Howe G. (2013) Elicitation of jasmonate-mediated defense responses by mechanical wounding and insect herbivory in Jasmonate Signaling (Goossens A., Pauwels L. eds), pp. 51–61, Humana Press; [DOI] [PubMed] [Google Scholar]
- 43. Schilmiller A. L., Koo A. J., Howe G. A. (2007) Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 143, 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brandizzi F., Hanton S., DaSilva L. L., Boevink P., Evans D., Oparka K., Denecke J., Hawes C. (2003) ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34, 269–281 [DOI] [PubMed] [Google Scholar]
- 45. Brandizzi F., Fricker M., Hawes C. (2002) A greener world: The revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 3, 520–530 [DOI] [PubMed] [Google Scholar]
- 46. Pompon D., Louerat B., Bronine A., Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 47. Koo A. J., Gao X., Jones A. D., Howe G. A. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986 [DOI] [PubMed] [Google Scholar]
- 48. Kramell R., Schmidt J., Schneider G., Sembdner G., Schreiber K. (1988) Synthesis of N-(jasmonoyl) amino acid conjugates. Tetrahedron 44, 5791–5807 [Google Scholar]
- 49. Werck-Reichhart D. l., Bak S. r., Paquette S. (2009) Cytochromes P450 in The Arabidopsis Book, The American Society of Plant Biologists, pp. 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sheard L. B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T. R., Kobayashi Y., Hsu F. F., Sharon M., Browse J., He S. Y., Rizo J., Howe G. A., Zheng N. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsieh H. L., Okamoto H., Wang M., Ang L. H., Matsui M., Goodman H., Deng X. W. (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- 52. Withers J., Yao J., Mecey C., Howe G. A., Melotto M., He S. Y. (2012) Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 20148–20153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamamoto K., Gildenberg M., Ahuja S., Im S.-C., Pearcy P., Waskell L., Ramamoorthy A. (2013) Probing the transmembrane structure and topology of cicrosomal cytochrome-P450 by solid-state NMR on temperature-resistant bicelles. Sci. Rep. 3, 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benveniste I., Salaün J.-P., Durst F. (1978) Phytochrome-mediated regulation of a monooxygenase hydroxylating cinnamic acid in etiolated pea seedlings. Phytochemistry 17, 359–363 [Google Scholar]
- 55. Okuda K. I. (1994) Liver mitochondrial P450 involved in cholesterol catabolism and vitamin D activation. J. Lipid Res. 35, 361–372 [PubMed] [Google Scholar]
- 56. Froehlich J. E., Itoh A., Howe G. A. (2001) Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Helliwell C. A., Sullivan J. A., Mould R. M., Gray J. C., Peacock W. J., Dennis E. S. (2001) A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 28, 201–208 [DOI] [PubMed] [Google Scholar]
- 58. Hardwick J. P. (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 75, 2263–2275 [DOI] [PubMed] [Google Scholar]
- 59. Pinot F., Beisson F. (2011) Cytochrome P450 metabolizing fatty acids in plants: characterization and physiological roles. FEBS J. 278, 195–205 [DOI] [PubMed] [Google Scholar]
- 60. Goldberg M. W., Allen T. D. (1995) Structural and functional organization of the nuclear envelope. Curr. Opin. Cell Biol. 7, 301–309 [DOI] [PubMed] [Google Scholar]
- 61. Yoshihara T., Omer E.-L., Koshino H., Sakamura S., Kikuta Y., Koda Y. (1989) Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.). Agric. Biol. Chem. 53, 2835–2837 [Google Scholar]
- 62. Nakamura Y., Mithöfer A., Kombrink E., Boland W., Hamamoto S., Uozumi N., Tohma K., Ueda M. (2011) 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in samanea saman. Plant Physiol. 155, 1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bell E. M., Lin W. C., Husbands A. Y., Yu L., Jaganatha V., Jablonska B., Mangeon A., Neff M. M., Girke T., Springer P. S. (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. U.S.A. 109, 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mizutani M., Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315 [DOI] [PubMed] [Google Scholar]
- 65. Bartel B., Fink G. R. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748 [DOI] [PubMed] [Google Scholar]
- 66. Ludwig-Müller J. (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 [DOI] [PubMed] [Google Scholar]
- 67. Campanella J. J., Larko D., Smalley J. (2003) A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp. Funct. Genomics 4, 584–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.