Background: mTOR integrates various intracellular and extracellular signals to regulate cell growth, proliferation, and survival.
Results: PDK4 promotes tumorigenesis through activation of the CREB-RHEB-mTORC1 signaling cascade.
Conclusion: PDK4 is a novel activator of mTOR signaling.
Significance: PDK4 activation of mTORC1 may be targeted for the treatment of mTOR-mediated cancer and other diseases.
Keywords: cAMP-response Element-binding Protein (CREB), Cellular Regulation, DNA-Protein Interaction, Mechanistic Target of Rapamycin (mTOR), Warburg Effect, Ras Homolog Enriched in Brain, Pyruvate Dehydrogenase Kinase 4, Signaling, Tumorigenesis
Abstract
Mechanistic target of rapamycin (mTOR) integrates multiple extracellular and intracellular signals to regulate cell growth and survival. Hyperactivation of mTOR has been observed in various cancers. Regulation of mTOR activity is thus of importance in physiological processes and tumor development. Here, we present pyruvate dehydrogenase kinase 4 (PDK4) as a novel regulator of mTORC1 signaling. mTORC1 activity was augmented with PDK4 overexpression and reduced by PDK4 suppression in various cell lines. Furthermore, PDK4 bound to cAMP-response element-binding protein (CREB) and prevented its degradation. The enhanced CREB consequently transactivated the expression of Ras homolog enriched in brain (RHEB), a direct key activator of mTORC1, independent of AMP-activated protein kinase or tuberous sclerosis complex protein 2. PDK4 potentiated the mTORC1 effectors hypoxia-inducible factor 1α and pyruvate kinase isozymes M2 and promoted aerobic glycolysis (Warburg effect). Knockdown of PDK4 suppressed the tumor development of cancer cells with activated mTORC1. The abundance of PDK4 dictated the responsiveness of cells to the mTOR inhibitor, rapamycin. Combinatory suppression of mTOR and PDK4 exerted synergistic inhibition on cancer cell proliferation. Therefore, PDK4 promotes tumorigenesis through activation of the CREB-RHEB-mTORC1 signaling cascade.
Introduction
The mechanistic target of rapamycin (mTOR)2 is an evolutionarily conserved serine/threonine kinase that integrates various intracellular and extracellular signals and regulates cell survival, growth, proliferation, differentiation, and metabolism (1–4). The receptor tyrosine kinase/PI3K/AKT pathway relays signaling from growth factors to mTOR by first suppressing the tuberous sclerosis complex 1 and 2 (TSC1 and TSC2) protein complex. The TSC1/2 protein complex is a negative regulator of the small GTPase Ras homolog enriched in brain (RHEB) as TSC2 exerts its GTPase-activating activity toward RHEB. The unleashed RHEB then activates mTOR by binding to its catalytic domain and stimulating mTOR phosphorylation (5, 6). mTOR exists in rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-insensitive mTOR complex 2 (mTORC2). Activated mTORC1 exercises its cellular functions by modulating downstream effectors such as S6 kinase and 4E-binding protein 1 (4E-BP1), whereas mTORC2 phosphorylates AKT at Ser-473 (4). Conversely, hyperactive mTORC1 also induces feedback inhibition of AKT through up-regulation of S6 kinase, growth factor receptor-bound protein 10 (GRB10), and store-operated Ca2+ entry and down-regulation of platelet-derived growth factor receptors and insulin receptor substrate (5–8). Genetic mutations in human cancers that cause gain of function for proto-oncogenes or loss of function for tumor suppressors in this pathway lead to hyperactivation of mTORC1. Mutations of tumor suppressor TSC1 or TSC2 cause TSC, a multiorgan tumor syndrome, through activation of mTOR (9–12). mTOR is also a sensor of energy status downstream of the liver kinase B1 (LKB1)/AMP-activated protein kinase (AMPK)-TSC1/2 signaling pathway. Deletion of either LKB1 or AMPK also activates mTOR (13). An important role of mTOR has been established in the regulation of cellular processes such as metabolism, autophagy, and protein synthesis (14–17). Hyperactive mTOR augments angiogenesis, aerobic glycolysis (Warburg effect), and cell proliferation in tumor development (18–20). Therefore, the regulation of mTOR activity is of significance in normal cellular processes as well as tumor development.
Pyruvate is an intermediate pivot in glucose metabolism and can be decarboxylated to acetyl-CoA by pyruvate dehydrogenase (PDH) (21). PDH links glycolysis to the citric acid cycle and releases energy via NADH. Pyruvate dehydrogenase kinases (PDKs) inactivate PDH through phosphorylation of its E1a subunit. PDKs are mitochondrial proteins encoded by genomic genes, and their expressions vary in diverse tissues, indicating that they may have different functions. PDK4 is one of four isozymes of PDKs and is highly expressed in heart and skeletal muscle (22). The role of PDK4 in tumorigenesis is unclear. As an inhibitor of PDK4 and its isoforms, dichloroacetic acid (DCA) has been used to treat some mitochondrial diseases (23). DCA has antiproliferative and pro-apoptotic effects on cancer cells due to unknown mechanisms. Furthermore, DCA can suppress hypoxia-inducible factor 1α (HIF-1α) (24), a downstream effector of mTOR signaling (25).
Here, we show that PDK4 activates mTORC1 through up-regulation of the CREB-RHEB cascade. Overexpression of PDK4 sensitizes cells to mTOR inhibition, and suppression of PDK4 blunts tumorigenesis. Therefore, PDK4 may be targeted for the treatment of certain cancers.
EXPERIMENTAL PROCEDURES
Reagents
Rapamycin and DCA were obtained from Sigma. MG-132 was obtained from Beyotime (Shanghai, China). Cycloheximide (CHX) was obtained from Genview (Galveston, TX). Torin-1 was obtained from Selleckchem (Houston, TX). Fetal bovine serum, Dulbecco's modified Eagle's medium/F-12 (1:1), Dulbecco's modified Eagle's medium, Lipofectamine 2000, and 4–12% BisTris NuPAGE gels were purchased from Invitrogen. Restriction enzymes were from Takara (Otsu Shiga, Japan).
Antibodies
The antibodies against total S6 and phospho-S6 (Ser-235/236) have been previously described (6). PDK1, PDK2, PDK3, PDK4, and phospho-PDH (Ser-293) antibodies were from Abgent (San Diego). Antibodies for α-tubulin, β-actin, ERK, phospho-ERK (Tyr-204), GRB10, AKT, normal IgG antibody, and all HRP-labeled secondary antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). TSC2, PTEN, phospho-AKT (Ser-473), RHEB, CREB, and phospho-CREB (Ser-133) antibodies were from Cell Signaling Technology (Beverly, MA). Phospho-AMPK (Thr-183/172) antibody was from Bioworld (Atlanta, GA), and phospho-GRB10 (Ser-501/503) antibody was from Millipore (Bedford, MA). PDH antibody was from Invitrogen.
Preparation of Plasmids and Viruses
The pLL3.7-shRNA vector was used for suppression of PDK4 expression. The target sequence for human PDK4 was 5′-GGACGTAAGAGATTCTCAT-3′. The target of mouse Pdk4 was 5′-GGATTTGGTGGAGTTCCAT-3′. Lentiviruses were generated by co-transfecting pLL3.7 and the packaging vectors (VSVG, REV, and pMDL) into HEK293T cells. After 48 h of transfection, viruses were collected by filtering through a 0.45-μm filter. PC3 cells were infected by incubation with viruses for 2 days, and the expression of GFP protein was used to evaluate the infection rate.
To generate a Pdk4 overexpression vector, Pdk4 was amplified from cDNA of wild-type mouse embryonic fibroblasts (MEFs) as a template using the following primers: 5′-TAAGCGGCCGCACCATGAAGGCAGCCCGCTT-3′ and 5′-CCCCAGGATCCTCACACTGCCAGCTTCTCCTTCG-3′. The PCR fragment was then cloned into the pQCXIP-puro vector at NotI/BamHI sites to generate pQCXIP-puro-PDK4.
Cell Culture
Pten−/−, Tsc2−/−, myr-Akt, Ampk−/− MEF cells and ELT-3 cells have been described previously (2, 7, 26, 27). A549, PC3, MDA-MB-468, PANC-1, MCF7, NIH/3T3, and HEK293T were obtained from ATCC. ELT-3 cells were cultured in Dulbecco's modified Eagle's medium/F-12 (1:1) with 10% fetal bovine serum and 1% antibiotics in 5% CO2 at 37 °C. The other cell lines used in this study were maintained and propagated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% antibiotics in 5% CO2 at 37 °C. The dosage of DCA treatment of cells was chosen according to McFate et al. (28).
Immunoblotting
Immunoblotting was conducted as described previously (2, 9). Cells were lysed in lysis/loading buffer (10 mm Tris, pH 6.8, 10% glycerol, 2% SDS, and 100 mm DTT) and boiled for 10 min. The cell lysates were then subjected to immunoblotting.
Cell Proliferation Assay (MTT)
Cell proliferation was measured using an MTT assay kit (BioDev-Tech, Beijing, China). Cells were plated at 4 × 103 cells per well, seeded in quintuplet in 96-well plates for 24 h, and then treated with rapamycin at various concentrations. Cells were incubated with 200 μl of medium containing 20 μl of MTT reagent at 37 °C. After 2 h, the supernatant was removed, and 150 μl of dimethyl sulfoxide was added. The plates were shaken under protection from light for 10 min, and the spectrometric absorbance at 490 nm was then recorded. Cell proliferation assays were replicated in at least two independent experiments.
RNA Interference
A total of 5–8 × 104 cells were seeded in a 12-well plate and transfected with synthesized siRNA by Lipofectamine 2000 following the manufacturer's instructions. Cell lysates were harvested for immunoblotting after 48 h of transfection. The target sequences for RNAi were as follows: pdk4-target 1, 5′-GGACGTAAGAGATTCTCAT-3′; pdk4-target 2, 5′-GGATTTGGTGGAGTTCCAT-3′; rheb-target 1, 5′-GGAGCAGCTTCACAAGGAA-3′; creb-target 1, 5′-GGAGTCTGTGGATAGTGTA-3′; creb-target 2, 5′-CAATACAGCTGGCTAACAA-3′; pdh-target 1, 5′-CCTATCGAGCACATGGCTT-3′; pdh-target 2, 5′-CCGAGAGGCAACAAAGTTT-3′; pdk1, 5′-GCAAAGTTGGTATATCCAA-3′; pdk2, 5′-CCTGGCCAACATCATGAAA-3′; pdk3, 5′-GGATAACTTTCTCAACGTT-3′; and for negative control, 5′-TTCTCCGAACGTGTCACGT-3′.
Reverse Transcription of mRNA and Quantitative Real Time-PCR (qRT-PCR)
A total of 1 μg of RNA was reverse-transcribed using the PrimeScriptTM RT reagent kit (Takara). cDNA was used as template using TransStart Green quantitative PCR SuperMix (TransGen Biotech, Beijing, China) in a 20-μl qRT-PCR. Amplifications were performed for 40 cycles using CFX on an iCycler (Bio-Rad). β-Actin served as the internal control. The number of samples from an individual experiment is three or 5, and the qRT-PCR assays data were replicated in at least two independent experiments. The qRT-PCR primer sequences were as follows: BACT forward, AGAGGGAAATCGTGCGTGAC and reverse, CAATAGTGATGACCTGGCCGT; pdk1 forward, AGGATCAGAAACCGGCACAAT, and reverse, GTGCTGGTTGAGTAGCATTCTAA; pdk2 forward, AACCTGCTTCCTGACCGAGT, and reverse GAACTGGCTTAGAGTCCGGTG; pdk3 forward, CCAGAAGACCCACGAGTTTTG, and reverse, GGCCATTGTAGGAACAACATCA; pdk4 forward, CCGCTTAGTGAACACTCCTTC, and reverse, TCTACAAACTCTGACAGGGCTTT; rheb forward, AAGTCCCGGAAGATCGCCA, and reverse GGTTGGATCGTAGGAATCAACAA; c-fos forward, ATCGGCAGAAGGGGCAAAGTAG and reverse, GCAACGCAGACTTCTCATCTTCAAG; sst-1 forward, CCACCGGGAAACAGGAACTG, and reverse, TTGCTGGGTTCGAGTTGGC; nurr-1 forward, GTGTTCAGGCGCAGTATGG, and reverse TGGCAGTAATTTCAGTGTTGGT.
Immunoprecipitation
The co-immunoprecipitation of CREB and PDK4 was performed as described previously (29). NIH/3T3 or HEK293T cells were cultured in 15-cm plates until they reached 80–90% confluence and were then lysed in 1 ml of lysis buffer. The samples were centrifuged to remove insoluble debris, and the supernatant was split into 2 equal aliquots. Anti-phospho-CREB antibody and control IgG antibody were added separately to each aliquot, and samples were incubated at 4 °C overnight. After incubation, 100 μl of a 50% slurry of protein G-agarose beads (Millipore) was added, and samples were rotated for 2 h. Immunoprecipitates were spun down and washed three times with lysis buffer. Immunocomplexes were then subjected to immunoblotting.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (19). NIH/3T3 cells were cultured in 15-cm plates until they reached 80–90% confluence. Cross-linking was achieved by incubation with 1% formaldehyde for 10 min and then stopped by the addition of glycine to a 0.125 m final concentration. Chromatin was sheared by sonication to fragment sizes between ∼500 and 1000 bp and then immunoprecipitated with either anti-phospho-CREB antibody or normal IgG antibody at 4 °C overnight. Sixty microliters of salmon sperm DNA/protein G-agarose (Cell Signaling Technology) were added to immunocomplexes, and samples were incubated at 4 °C for 2 h. The immunocomplexes were sequentially washed once for 10 min in ChIP low salt wash buffer and once in ChIP high salt wash buffer and twice in ChIP LiCl buffer and TE buffer. DNA-protein complexes were eluted twice with elution buffer. Finally, the released DNA was extracted using phenol/chloroform followed by ethanol precipitation. DNA was resuspended in 30 μl of MilliQ water and amplified by real time-PCR. The RHEB primers were designed by the computer software program PRIMER 6 within the predicted binding region (PBR) and 2 kb upstream of the transcription start site as no-binding control (NBR). The primer sequences were as follows: PBR forward, ACCTCTCCTTGGCTCCACCCTT, and PBR reverse, TCCCACCTACTTCCGCCGCTTT; NBR forward, AAGCTCCTCAAGGGACAATGGT, and NBR reverse, TGGCTCTCTCCAATGAGCATCC.
Measurements of Glucose and Lactate
Glucose and lactate in medium were measured as described previously (19). A total of 5 × 104 cells per well were seeded in 12-well plates (n = 3) for 12 h, and then cells were incubated with fresh medium with or without rapamycin for 48 h. Cell numbers were counted before measurement. The medium was collected, and the glucose and lactate concentrations were examined immediately using a glucose and lactate measuring instrument (EKF-Diagnostic GmbH, Magdeburg, Germany). The consumption of glucose and production of lactate were normalized by cell number. The assay data were replicated in three independent experiments.
Induction of Subcutaneous Tumors in Nude Mice
Subcutaneous tumors were established as described previously (2). Immunodeficient nude mice (BALB/c, 4–6 weeks old) were obtained from the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences. Eight mice were used in each cohort.
Tumor growth and mouse survival were assessed following subcutaneous inoculation of 1 × 106 PC3 cells in 100 μl of DMEM into the flank region near the axilla. Mice were sacrificed when tumor size was 1000 mm3. The animal study protocol was approved by the Animal Center of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences, and was in compliance with the regulations of the Beijing Administration Office of Laboratory Animals on the care of experimental animals. The data were replicated in two independent experiments.
Histology
Nude mouse tumor tissues were fixed in 4% buffered paraformaldehyde at room temperature and embedded in paraffin. Immunohistochemistry for detection of Ki67 and CD31 was conducted using a standard protocol. Anti-Ki67 and anti-CD31 antibodies were diluted at 1:500 and incubated in a humidified chamber overnight at 4 °C. The secondary antibody used was horseradish peroxidase-goat anti-rabbit IgG (Zhongshan Jinqiao, Beijing, China). Diaminobenzidine was used to develop the signal. Sections were lightly counterstained with hematoxylin.
Statistics
The Kaplan-Meier log-rank test was used for analysis of mouse tumor development with GraphPad Prism software (GraphPad Software). Comparisons between two groups of data were conducted using the Student's t test. Drug combination effect was evaluated by calculating the combination index as described previously (19). Differences were considered significant when p < 0.05.
RESULTS
PDK4 Activates mTORC1
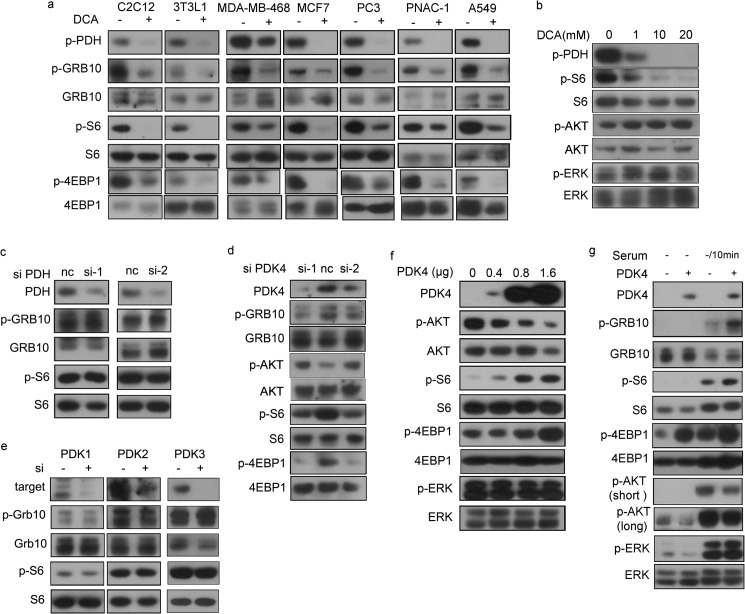
DCA, a PDK inhibitor, was previously shown to suppress HIF-1α expression. Because mTORC1 is a critical regulator of HIF-1α, we treated cells with DCA and examined mTOR activity. As a substrate of PDKs, phosphorylated PDH (p-PDH) was used to indicate the action of DCA. DCA reduced the phosphorylation of S6, 4EBP1, and GRB10, three mTORC1 effectors, in mouse myoblast and adipoblast cell lines and various human cancer cells (Fig. 1a), and the inhibition was dose-dependent (Fig. 1b). However, we did not observe any difference in p-S6 and p-Grb10 between pdh knockdown cells and control cells (Fig. 1c), suggesting that DCA suppression of mTOR activity likely does not occur through PDH. To investigate the specificity of PDK4 regulation on mTORC1, we knocked down pdk4 and the other three pdk isozymes in NIH/3T3 cells. Knockdown of pdk4, but not pdk1, pdk2, or pdk3, reduced mTORC1 activity (Fig. 1, d and e). Taken together, our data demonstrate that DCA suppresses mTORC1 specifically through Pdk4 and independent of Pdh.
FIGURE 1.
PDK4 is a positive regulator of mTOR. Cell lysates were analyzed by immunoblotting for the indicated proteins. a, MEFs and cancer cell lines treated with or without 20 mm DCA for 24 h. b, protein lysates from NIH/3T3 cells treated with varying doses of DCA were analyzed by immunoblotting. NIH/3T3 cells were transfected with control siRNA and two independent siRNAs for PDH (c) or PDK4 (d). e, NIH/3T3 cells were transfected with siRNA for PDK1–3 or control siRNA, and protein expressions were evaluated by immunoblotting. f, PDK4-expressing plasmids (0, 0.4, 0.8, and 1.6 μg) were transfected into NIH/3T3 cells. g, NIH/3T3 cells transfected with PDK4 expression plasmids or control plasmids were starved for 24 h and then stimulated with or without 10% serum for 10 min. Short and long indicate short and long exposure, respectively, of immunoblotting reaction. nc indicates negative control.
To further investigate PDK4 regulation of mTOR, we overexpressed PDK4 in NIH/3T3 cells. p-S6 and p-4EBP1 were elevated, and p-Akt was decreased in response to Pdk4 expression in a dose-dependent manner (Fig. 1f). In contrast, Pdk4 overexpression had no impact on p-Erk, a marker of the MAPK signaling pathway (Fig. 1f). Similarly, overexpression of Pdk4 increased phosphorylation of S6, 4EBP1, and Grb10 but not Erk upon serum stimulation after 24 h of starvation of NIH/3T3 cells (Fig. 1g), with a decrease of p-Akt. Consistently, inhibition of Pdk4 did not affect p-Erk (Fig. 1b). We also observed an increase of p-Akt in DCA-treated cells in a dose-dependent manner (Fig. 1b) and an increase of p-Akt in PDK4 knocked down cells (Fig. 1d), which attributed to the feedback inhibition of mTORC1.
DCA-induced Inactivation of mTORC1 Is Independent of AMPK and PTEN-AKT-TSC2
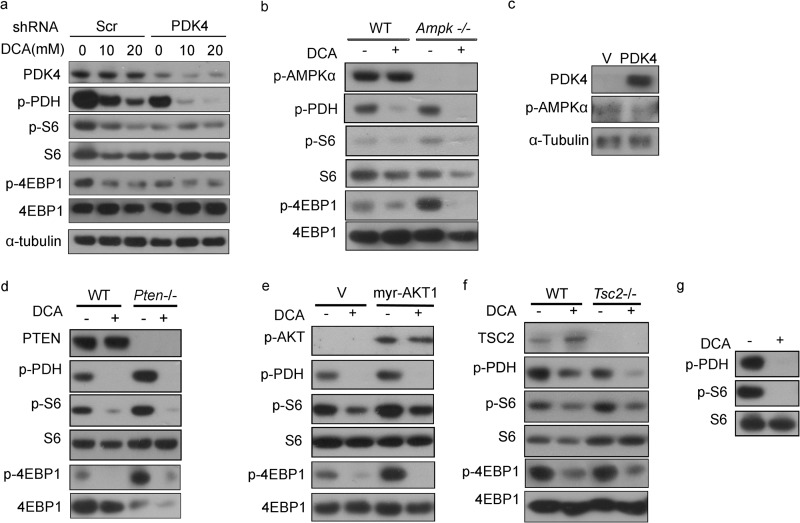
After knocking down PDK4, suppression of mTOR activity by DCA was compromised (Fig. 2a), consistent with PDK4 regulation on mTOR. Both LKB1-AMPK and PTEN/AKT/TSC1/2 pathways are major mTOR regulatory networks. To examine the potential involvement of AMPK in PDK4 regulation of mTOR, AMPK α1/α2 double knock-out MEFs were used. DCA-mediated p-S6 and p-4EBP1 reductions were not affected in Ampk−/− MEFs compared with wild-type MEFs. Furthermore, DCA did not alter p-Ampk levels in wild-type MEFs (Fig. 2b). Finally, overexpression of pdk4 had no effect on p-Ampk levels in NIH/3T3 cells (Fig. 2c). Taken together, these data indicate that PDK4 is unlikely upstream of AMPK.
FIGURE 2.
DCA inhibition of mTOR is independent of AMPK and PTEN-AKT-TSC2. Cell lysates were analyzed by immunoblotting for the indicated proteins. a, Scr and shPDK4 PC3 cells treated with or without 10 and 20 mm DCA for 24 h. b, wild-type and Ampk−/− MEFs treated with or without 20 mm DCA for 24 h. c, NIH/3T3 cells transfected with control and PDK4 overexpression plasmids. d, wild-type and Pten−/− MEFs treated with or without 20 mm DCA for 24 h. e, wild-type and myr-AKT1 MEFs treated with or without 20 mm DCA for 24 h. f, wild-type and Tsc2−/− MEFs treated with or without 20 mm DCA for 24 h. g, ELT-3 (TSC2-null) cells were treated with or without 20 mm DCA for 24 h. V indicates control plasmids.
DCA down-regulated S6 and 4EBP1 phosphorylation in both wild-type and Pten−/− MEFs (Fig. 2d). Neither constitutive AKT1 activation (myr-AKT1) (Fig. 2e) nor TSC2 deletion (Fig. 2f) compromised DCA-induced mTORC1 suppression. DCA also decreased p-S6 in ELT-3 cells, a Tsc2-null cell line derived from an Eker rat uterine leiomyoma (Fig. 2g). Collectively, these data demonstrate that PDK4 regulation of mTORC1 is in parallel with PTEN/AKT/TSC1/2 and LKB1/AMPK/TSC1/2 pathways.
PDK4 Up-regulates RHEB Transcription
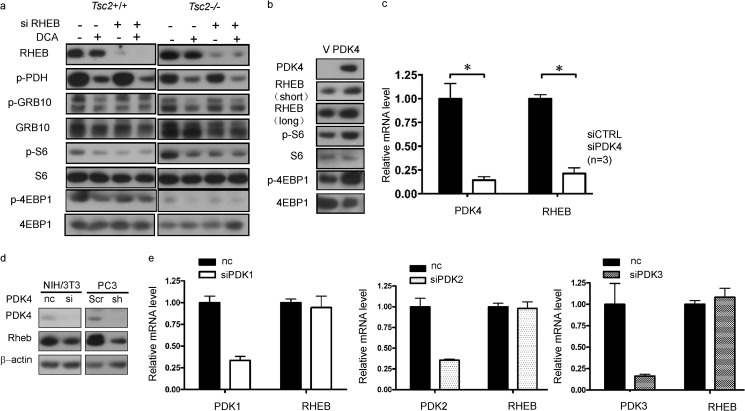
Through its GTPase-activating protein (GAP) activity, TSC2 negatively regulates the small GTPase RHEB, which functions as an activator of mTOR (30). Because PDK4 up-regulated mTOR independently of TSC2, we explored the possibility of RHEB serving as a link between PDK4 and mTOR. We knocked down RHEB in both Tsc2+/+ and Tsc2−/− MEFs and then treated these cells with DCA. DCA-induced depression of p-S6, p-4EBP1, and p-Grb10 was abolished in rheb knockdown cells (Fig. 3a). Furthermore, overexpressed Pdk4 stimulated Rheb expression in NIH/3T3 cells (Fig. 3b). Taken together, RHEB is involved in PDK4 regulation of mTORC1.
FIGURE 3.
PDK4 activates mTOR through transcriptional up-regulation of RHEB. Lysates of wild-type and Tsc2−/− MEFs transfected with one of the siRNAs for RHEB or control siRNA treated with or without 20 mm DCA for 24 h (a) and NIH/3T3 cells transfected with PDK4 expression plasmids or control plasmids (b) were subjected to immunoblotting. c, total RNA was extracted from NIH/3T3 cells transfected with PDK4 siRNA or negative control siRNA for qRT-PCR. d, protein lysates were extracted from NIH/3T3 cells transfected with PDK4 siRNA or negative control siRNA and PC3 cells with shScr or shPDK4 for immunoblotting, respectively. e, relative expression of RHEB mRNA from NIH/3T3 cells transfected with siRNA for PDK1–3 or negative control siRNA was determined by qRT-PCR (n = 3). nc indicates negative control. V indicates control plasmids. *, p < 0.05.
In addition to TSC2 suppression of RHEB, there are other distinct mechanisms that regulate RHEB (27, 29, 31). pdk4 knockdown reduced both mRNA (Fig. 3c) and protein levels (Fig. 3d) of Rheb, suggesting that Pdk4 regulated Rheb at the transcription level. Knockdown of pdk1, pdk2, or pdk3 had no effect on rheb mRNA levels (Fig. 3e).
PDK4 Stimulates RHEB Expression through CREB
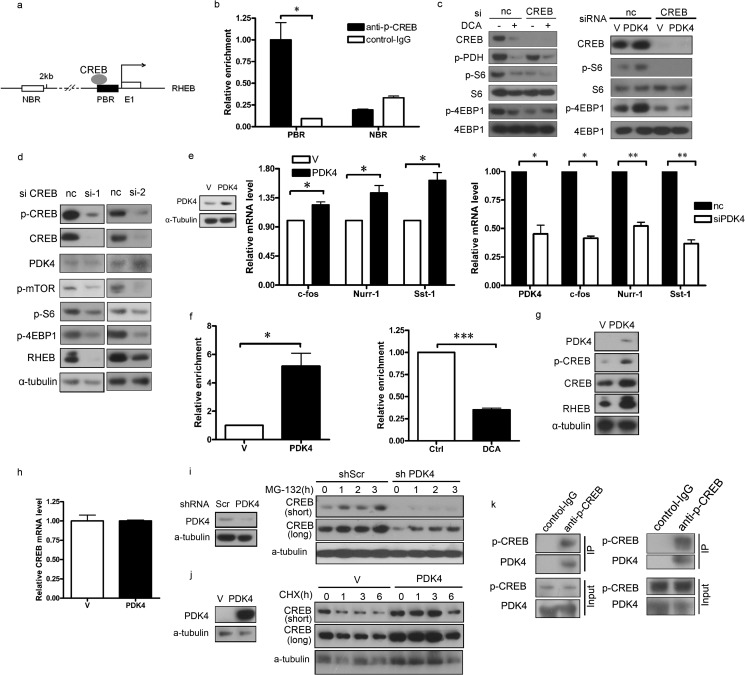
To identify the link between PDK4 and RHEB, we analyzed the promoter region of rheb for potential protein-binding sites and identified binding sites for the transcription factor CREB (Fig. 4a). CREB is a cellular transcription factor. After phosphorylation at Ser-133, CREB binds to certain DNA sequences and increases or decreases the transcription of targeted genes (32, 33). Chromatin immunoprecipitation assay was performed with cross-linked chromatin from NIH/3T3 cells treated with p-CREB antibody or control IgG. CREB indeed bound to the predicted CREB-binding regions of the RHEB gene (Fig. 4b). To provide more direct evidence of CREB involvement in PDK4-regulated mTORC1 activity, we treated CREB-knocked down NIH/3T3 cells with DCA. DCA-induced depression of mTOR activity was abolished in rheb knockdown cells (Fig. 4c, left panel). Similarly, Pdk4 no longer activated mTORC1 in creb knockdown cells (Fig. 4c, right panel), which indicates that CREB is involved in PDK4 regulation of mTOR. A marked reduction of Rheb was observed in CREB knockdown cells compared with control cells (Fig. 4d), implicating PDK4 in the upstream of CREB and CREB in the stimulation of RHEB expression. Furthermore, Creb downstream target genes, such as c-fos, nurr-1, sst1, were enhanced in pdk4 overexpression cells (Fig. 4e, left and middle panels) and suppressed in pdk4 knockdown cells (Fig. 4e, right panel). The interaction between CREB protein and the RHEB promoter was significantly higher in pdk4-overexpressing cells than in control cells (Fig. 4f, left panel) and was suppressed in NIH/3T3 cells treated with DCA (Fig. 4f, right panel), indicating that CREB is the potential bridge between PDK4 and RHEB.
FIGURE 4.
PDK4 stimulates RHEB expression through stabilization of CREB. a, schematic illustration of the promoter regions of the mouse RHEB gene. E1 represents RHEB exon 1; the dark rectangle denotes a PBR, and the open rectangle indicates an NBR in the RHEB promoter. The arrow above the gene indicates the transcription start site. b, NIH/3T3 cells were subjected to ChIP assay using an anti-phospho-CREB antibody; normal rabbit IgG antibody served as the negative control. c, lysates of NIH/3T3 cells transfected with CREB siRNA or negative control siRNA treated with or without 20 mm DCA for 24 h and transfected with PDK4 or control plasmids were subjected to immunoblotting. d, lysates of NIH/3T3 cells transfected with two independent siRNAs for CREB or negative control siRNA were subjected to immunoblotting. e, total RNAs were extracted from NIH/3T3 cells transfected with PDK4 or control plasmids and NIH/3T3 cells transfected with siRNA for PDK4 or control siRNA for qRT-PCR. f, CREB antibody-immunoprecipitated DNA from PDK4-overexpressing and control NIH/3T3 cells and NIH/3T3 cells treated with 20 mm DCA for 24 h were PCR-amplified for regions indicated in a. g and h, total protein lysates and RNA were extracted from NIH/3T3 cells transfected with PDK4 or control plasmids for immunoblotting and qRT-PCR, respectively. i, immunoblotting of CREB in PDK4-overexpressing or control NIH/3T3 treated with MG-132 (10 μm). j, immunoblotting of CREB in NIH/3T3 with shPDK4 or shScr treated with CHX (25 μm). k, co-immunoprecipitation of endogenous proteins from NIH/3T3 cells and HEK293T cells. Immunoprecipitation (IP) was performed with anti-phospho-CREB or control IgG, and complexes were examined by immunoblotting with either phospho-CREB or PDK4 antibody. Co-immunoprecipitation and ChIP assay data were replicated in two independent experiments. Data represent mean ± S.E. of replicate real time PCR. nc indicates negative control. V indicates control plasmids. *, p < 0.05. **, p < 0.01; p < 0.001.
Next, we examined the regulatory mechanism of PDK4 on CREB. PDK4 overexpression increased both total Creb and p-Creb protein levels in NIH/3T3 cells (Fig. 4g). However, PDK4 did not affect the mRNA levels of Creb (Fig. 4h), suggesting that PDK4 regulates CREB post-transcriptionally. To test whether PDK4 stabilizes CREB, we established pdk4 knockdown NIH/3T3 cells (Fig. 4i, left panel). The synthesis rate of Creb protein, measured by inhibiting degradation with the proteasome inhibitor MG-132, was similar between shPDK4 NIH/3T3 cells and shScr control cells (Fig. 4i, right panel). To test whether Creb degradation was inhibited in pdk4-overexpressing cells (Fig. 4j, left panel), we treated cells with protein biosynthesis inhibitor cycloheximide (CHX). We found apparent suppression of Creb degradation in PDK4-overexpressing cells compared with control cells (Fig. 4j, right panel). In addition, endogenous PDK4 co-immunoprecipitated CREB (Fig. 4k), indicating that PDK4 physically interacts with CREB. Taken together, we speculated that PDK4 interacts with CREB and prevents CREB degradation. Therefore, PDK4 enhances the transactivation of CREB on RHEB.
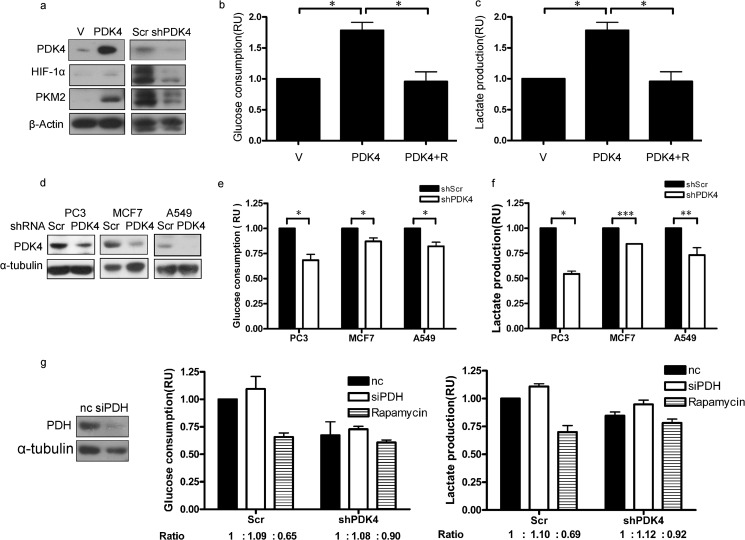
PDK4 Potentiates HIF-1α and Its Target Pyruvate Kinase Isozymes M2 and Promotes Aerobic Glycolysis
Among the mTOR effectors, HIF-1α is one of the critical regulators in tumorigenesis. Elevated/attenuated HIF-1α and its downstream target pyruvate kinase isozymes M2 (PKM2) were observed in PDK4-overexpressing/knockdown cells (Fig. 5a). Increased glucose consumption and lactate production are manifestations of aerobic glycolysis in mTOR-hyperactivated cells (19). The potentiated Warburg effect was reversed by the mTOR inhibitor rapamycin in PDK4-overexpressing NIH/3T3 cells (Fig. 5, b and c). Glucose consumption (Fig. 5e) and lactate production (Fig. 5f) were reduced by PDK4 depletion in three human cancer cell lines, PC3, A549, and MCF7 (Fig. 5d). To clarify PDK4-induced metabolic changes whether through mTORC1 or PDH, we knocked down PDH in shPDK4 and control cells. Glucose consumption and lactate production showed a slight increase in both cells (Fig. 5g). On the contrary, after knocking down PDK4, the inhibition of the Warburg effect by mTOR inhibitor rapamycin was significantly compromised (Fig. 5g). Thus mTOR indeed plays an important role in PDK4-related metabolic changes. These data also add additional lines of evidence to the PDK4 activation of mTORC1.
FIGURE 5.
PDK4 potentiates HIF-1α and its target PKM2 and promotes aerobic glycolysis. a, cell lysates from NIH/3T3 transfected with PDK4 expression plasmids or control plasmids and PC3 with shScr or shPDK4 were collected and analyzed by immunoblotting. b and c, glucose consumption (b) and lactate production (c) of control or PDK4-overexpressing NIH/3T3 cells treated with or without 10 nm rapamycin (R) for 48 h. d, immunoblotting of indicated proteins in control and PDK4 knockdown human cancer cells (PC3, A549, and MCF7). Glucose consumption (e) and lactate production (f) in control or PDK4-knockdown human cancer cells (n = 3) are shown. g, glucose consumption and lactate production of shScr or shPDK4 NIH/3T3 cells transfected with siRNA for PDH or negative control siRNA treated with or without 10 nm rapamycin for 48 h. Data are presented as the mean ± S.E. nc indicates negative control. V indicates control plasmids. *, p < 0.05. **, p < 0.01; ***, p < 0.001. RU indicates relative units.
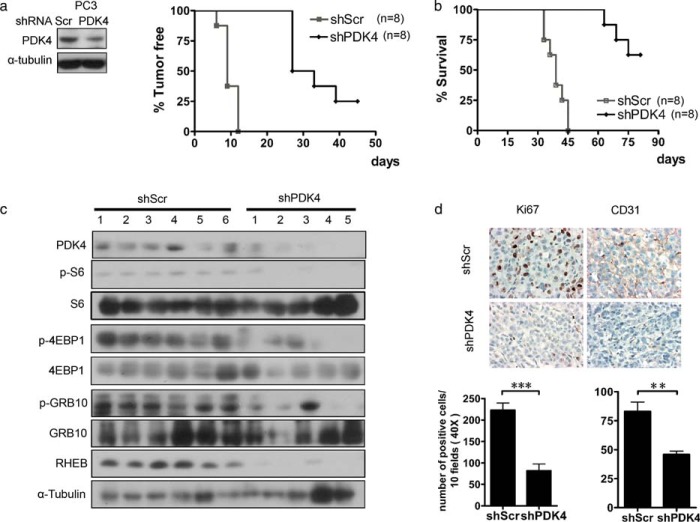
Inhibition of PDK4 Suppresses Cell Proliferation and Tumorigenesis
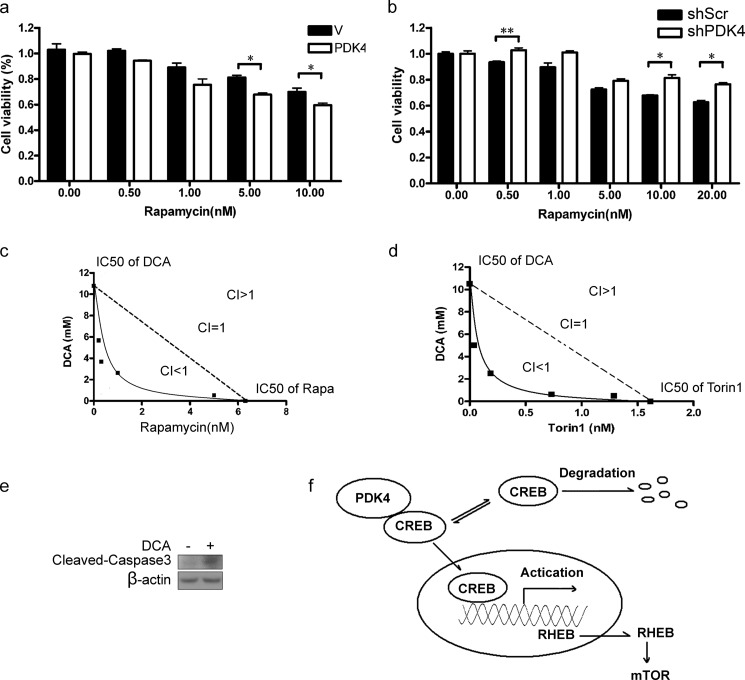
To investigate the role of PDK4 in tumor development of mTOR-activated cancer cells, we injected PTEN-mutated PC3 cells expressing control shRNA (shScr) or shPDK4 subcutaneously into nude mice. Reduction of PDK4 in PC3 cells dramatically retarded tumor initiation and development and extended the survival of tumor-bearing mice (Fig. 6, a and b). Additionally, the attenuated PDK4 expression with the marked decrease of RHEB expression and lower mTOR activity were detected in tumor tissues dissected from nude mice (Fig. 6c). Reduced Ki67 staining and microvessel density (CD31 staining) illustrated suppressed proliferation and angiogenesis, manifestations of blunted mTOR activity, in PC3 shPDK4 tumors (Fig. 6d). In addition, PDK4 highly expressing cells displayed greater sensitivity to rapamycin treatment (Fig. 7a). Consistently, knockdown of PDK4 rendered NIH/3T3 cells less sensitive to rapamycin (Fig. 7b) Furthermore, enhanced inhibition of proliferation was observed in PC3 cells treated with DCA and rapamycin (Fig. 7c). Consistently, the catalytic mTOR inhibitor Torin-1 also enhanced the inhibition of PC3 cell proliferation by DCA (Fig. 7d). DCA induced cleavage of caspase3 (Fig. 7e), a manifestation of cellular apoptosis. The MTT readout correlated with caspase activation, indicating a connection between MTT and viability.
FIGURE 6.
Depletion of PDK4 suppresses tumorigenesis. PC3 cells with shScr or shPDK4 were injected subcutaneously into nude mice (n = 8), and tumor formation (a) and survival of the mice (b) were analyzed. The Kaplan-Meier log-rank test was used, p < 0.001. c, tumor lysates were examined for indicated protein expression by immunoblotting. d, Ki67 and CD31 immunostaining of PC3 tumors with shScr or shPDK4 (left panel). The graphs represent the number of Ki67 and CD31 positive cells/field; 10 fields per tumor were counted. Magnification, ×40. Data are calculated as the mean ± S.E. **, p < 0.01; ***, p < 0.001.
FIGURE 7.
Combinatory suppression of mTOR and PDK4 synergistically inhibits cell proliferation. a, NIH/3T3 cells transfected with PDK4 expression plasmids or control plasmids were treated with rapamycin for 48 h at the indicated concentrations. b, NIH/3T3 cells with shPDK4 or shScr were treated with rapamycin for 48 h at the indicated concentrations. c, PC3 cells were treated with various doses of rapamycin and DCA in a 96-well plate for 48 h. Isoboles for the combination of rapamycin with DCA were isoeffective (IC50) for inhibition of proliferation of PC3. The dashed line indicates the zero interaction of the isobole. d, PC3 cells were treated with varying doses of Torin-1 and DCA in a 96-well plate for 48 h. e, cell lysates from NIH/3T3 treated with or without 10 mm DCA for 24 h were collected and analyzed by immunoblotting. f, schematic illustration of PDK4-CREB-RHEB-mTOR signaling cascade. PDK4 binds to CREB and prevents its degradation. The enhanced CREB consequently transactivates the expression RHEB and potentiates mTOR. Cell viability was assessed with MTT assay. Data are calculated as the mean ± S.E. *, p < 0.05; **, p < 0.01.
DISCUSSION
mTOR is a central controller of multiple cellular processes. Here, we have identified PDK4 as a positive regulator of mTORC1 and demonstrated that PDK4 activates mTORC1 through CREB-mediated transcriptional regulation of RHEB (Fig. 7f). PDK4-CREB-RHEB-mTOR signaling cascade is critical for tumorigenesis.
mTOR integrates intracellular and extracellular signals and modulates cell growth and proliferation. Environmental cues that affect mTOR include growth factors, stress, energy status, and amino acids (14). By coordinating these upstream inputs, mTORC1 controls a variety of cellular processes. Gain of function mutations of proto-oncogenes or loss of function mutations of tumor suppressors can augment mTORC1 signaling and result in tumorigenesis. Therefore, the regulation on mTOR is of considerable importance both in physiological and pathological states. Guo et al. (34) observed increased PDK4 and reduced mTOR signaling in H2O2-induced senescent cells in comparison with control cells. The authors proposed that PDK4 negatively regulated mTOR. In contrast, our findings from MEFs, human cancer cells, and a xenograft tumor model demonstrated that PDK4 is an activator of mTORC1 in various tissues (Figs. 1 and 6).
Both AMPK and TSC2 are negative regulators of mTOR (13, 35–37). Our data suggest that PDK4 regulation of mTOR is independent of AMPK or TSC2 (Fig. 2). We identified RHEB, an immediate activator of mTOR, as the connector between PDK4 and mTOR. TSC1/2, Bnip3, or p38-PRAK cascade inactivates mTORC1 through suppression of RHEB (27, 29, 38, 39). The transcriptional regulation of Rheb was discovered in ATF6-Rheb-mTOR signaling (31). We found that the transcriptional activity of RHEB is significantly higher in PDK4-overexpressing cells than in control cells, demonstrating that PDK4 activation of mTORC1 occurs through up-regulation of RHEB transcription. We then demonstrated that CREB serves as the transcriptional activator of RHEB. Furthermore, PDK4 post-transcriptionally regulates CREB by enhancing CREB accumulation (Fig. 4). Previous studies have shown that CREB is degraded by phosphatases or the ubiquitin-proteasome pathway (40, 41). Because PDK4 physically interacts with CREB, PDK4 may bind to CREB and prevent its degradation. Taken together, we speculate that PDK4 stabilizes CREB and potentiates CREB activation of RHEB transcription.
Hyperactive mTOR promotes angiogenesis, potentiates aerobic glycolysis, and accelerates tumor growth (19, 20). In this study, we identified PDK4-CREB-RHEB as the activating axis upstream of mTOR. This cascade may augment mTOR downstream events in tumorigenesis. Indeed, overexpression of PDK4 increased expression of mTOR effectors HIF-1α and PKM2 and promoted aerobic glycolysis. This critical role of PDK4 in aerobic glycolysis is mTOR-dependent as inhibition of mTOR with rapamycin reversed the glycolytic phenotype caused by PDK4 overexpression (Fig. 5). Moreover, the attenuated PDK4 expression suppressed cell proliferation and tumor angiogenesis (Fig. 6). The Warburg effect is characterized by an increase of lactate production with relative reduction of acetyl-CoA. This effect is partially achieved through mTOR-mediated HIF-1α up-regulation of glycolytic enzymes and PDK. Alternatively, the putative PDK4 suppression of PDH may prevent transformation of pyruvate to acetyl-CoA and consequently promote aerobic glycolysis.
As PDK4-CREB-RHEB signaling cascade activates mTORC1 and promotes tumorigenesis, components in this cascade may be potential targets for cancer therapy. Because PDK4-overexpressed cells displayed higher sensitivity to rapamycin, the level of PDK4 may be a sensitivity biomarker of cancer cells to mTOR inhibitors. In addition, combination of mTOR inhibitors and the PDK4 inhibitor exerted synergistic suppression on the proliferation of cancer cells with activated mTOR signaling (Fig. 7). This result is a proof of principle that combination therapy may be a superior approach for the management of cancers caused by the hyperactivated mTOR signaling cascade with fewer side effects. As the pan-inhibitor of PDKs, DCA has antiproliferative and pro-apoptotic effects on cancer cells, possibly through depolarization of mitochondria and induction of apoptosis of cancer cells (24). However, its potential to be translated into clinical treatment is debatable because of its broad side effects (42, 43). Our data indicate that DCA inhibits mTOR activity, and this effect is specifically through PDK4, independent of PDH and the other three PDK isoforms (Fig. 1). Our study reveals an additional mechanism of DCA inhibition of cancer. To avoid the toxicity of DCA, it will be worthwhile to screen specific inhibitors of PDK4.
In summary, we have identified PDK4 regulation of mTOR through a newly identified PDK4-CREB-RHEB-mTOR signaling cascade. Activated PDK4 may potentiate mTOR and promote cell growth and tumorigenesis. Our study uncovers a novel therapeutic target that controls mTOR signaling, and the development of PDK4 inhibitors is thus warranted. In addition, the interplay between PDK4 and mTOR may display wider significance in physiological and pathological states, such as metabolic disorders, beyond the potential connection in cancers.
Acknowledgments
We are grateful to Dr. Jiahuai Han (Xiamen University) for providing Ampk−/− cells, Dr. Huihua Li (Capital Medical University) for instruction, and Yunzhou Gao (Chinese Academy of Medical Sciences) for technical assistance in histology experiments.
This work was supported by the National Science and Technology Major Project (2013ZX10002008), the National Basic Research Program of China 973 Program Grant (2011CB965002), the National Natural Science Foundation of China (Grants 81101516 and 81130085), and the Ministry of Education of China 211 Project (B08007).
This article was selected as a Paper of the Week.
- AMPK
- AMP-activated protein kinase
- CREB
- cAMP-response element-binding protein
- DCA
- dichloroacetic acid
- HIF-1α
- hypoxia-inducible factor 1α
- mTOR
- mechanistic target of rapamycin
- PDK4
- pyruvate dehydrogenase kinase 4
- PDH
- pyruvate dehydrogenase
- RHEB
- Ras homolog enriched in brain
- TSC1 and TSC2
- tuberous sclerosis complex 1 and 2
- MEF
- mouse embryonic fibroblast
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDK
- pyruvate dehydrogenase kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- qRT-PCR
- quantitative real time-PCR
- CHX
- cycloheximide
- PBR
- predicted binding region
- NBR
- nonspecific binding region.
REFERENCES
- 1. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma J., Meng Y., Kwiatkowski D. J., Chen X., Peng H., Sun Q., Zha X., Wang F., Wang Y., Jing Y., Zhang S., Chen R., Wang L., Wu E., Cai G., Malinowska-Kolodziej I., Liao Q., Liu Y., Zhao Y., Sun Q., Xu K., Dai J., Han J., Wu L., Zhao R. C., Shen H., Zhang H. (2010) Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J. Clin. Invest. 120, 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 4. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shah O. J., Wang Z., Hunter T. (2004) Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 6. Zhang H., Bajraszewski N., Wu E., Wang H., Moseman A. P., Dabora S. L., Griffin J. D., Kwiatkowski D. J. (2007) PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J. Clin. Invest. 117, 730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peng H., Liu J., Sun Q., Chen R., Wang Y., Duan J., Li C., Li B., Jing Y., Chen X., Mao Q., Xu K. F., Walker C. L., Li J., Wang J., Zhang H. (2013) mTORC1 enhancement of STIM1-mediated store-operated Ca2+ entry constrains tuberous sclerosis complex-related tumor development. Oncogene 32, 4702–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villén J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwiatkowski D. J., Zhang H., Bandura J. L., Heiberger K. M., Glogauer M., el-Hashemite N., Onda H. (2002) A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 [DOI] [PubMed] [Google Scholar]
- 10. Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Potter C. J., Pedraza L. G., Xu T. (2002) Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 4, 658–665 [DOI] [PubMed] [Google Scholar]
- 12. Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 13. Inoki K., Zhu T., Guan K. L. (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 14. Hay N., Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 15. Kim J., Kundu M., Viollet B., Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yecies J. L., Manning B. D. (2011) Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 71, 2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma J., Sun Q., Mi R., Zhang H. (2011) Avian influenza A virus H5N1 causes autophagy-mediated cell death through suppression of mTOR signaling. J. Genet. Genomics 38, 533–537 [DOI] [PubMed] [Google Scholar]
- 18. Hudson C. C., Liu M., Chiang G. G., Otterness D. M., Loomis D. C., Kaper F., Giaccia A. J., Abraham R. T. (2002) Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22, 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Q., Chen X., Ma J., Peng H., Wang F., Zha X., Wang Y., Jing Y., Yang H., Chen R., Chang L., Zhang Y., Goto J., Onda H., Chen T., Wang M. R., Lu Y., You H., Kwiatkowski D., Zhang H. (2011) Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4129–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guertin D. A., Sabatini D. M. (2007) Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 21. Holness M. J., Sugden M. C. (2003) Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 31, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 22. Bowker-Kinley M. M., Davis W. I., Wu P., Harris R. A., Popov K. M. (1998) Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 329, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stacpoole P. W., Kurtz T. L., Han Z., Langaee T. (2008) Role of dichloroacetate in the treatment of genetic mitochondrial diseases. Adv. Drug Deliv. Rev. 60, 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michelakis E. D., Sutendra G., Dromparis P., Webster L., Haromy A., Niven E., Maguire C., Gammer T. L., Mackey J. R., Fulton D., Abdulkarim B., McMurtry M. S., Petruk K. C. (2010) Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2, 31ra34. [DOI] [PubMed] [Google Scholar]
- 25. Meric-Bernstam F., Gonzalez-Angulo A. M. (2009) Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 27, 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi T., Hirayama Y., Kobayashi E., Kubo Y., Hino O. (1995) A germline insertion in the tuberous sclerosis (Tsc2) gene gives rise to the Eker rat model of dominantly inherited cancer. Nat. Genet. 9, 70–74 [DOI] [PubMed] [Google Scholar]
- 27. Zheng M., Wang Y. H., Wu X. N., Wu S. Q., Lu B. J., Dong M. Q., Zhang H., Sun P., Lin S. C., Guan K. L., Han J. (2011) Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat. Cell Biol. 13, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McFate T., Mohyeldin A., Lu H., Thakar J., Henriques J., Halim N. D., Wu H., Schell M. J., Tsang T. M., Teahan O., Zhou S., Califano J. A., Jeoung N. H., Harris R. A., Verma A. (2008) Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J. Biol. Chem. 283, 22700–22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Wang Y., Kim E., Beemiller P., Wang C. Y., Swanson J., You M., Guan K. L. (2007) Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J. Biol. Chem. 282, 35803–35813 [DOI] [PubMed] [Google Scholar]
- 30. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 31. Schewe D. M., Aguirre-Ghiso J. A. (2008) ATF6α-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 10519–10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montminy M. R., Bilezikjian L. M. (1987) Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328, 175–178 [DOI] [PubMed] [Google Scholar]
- 33. Mayr B., Montminy M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 34. Guo L., Xie B., Mao Z. (2012) Autophagy in premature senescent cells is activated via AMPK pathway. Int. J. Mol. Sci. 13, 3563–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Downward J. (1998) Mechanisms and consequences of activation of protein kinase B/Akt. Curr. Opin. Cell Biol. 10, 262–267 [DOI] [PubMed] [Google Scholar]
- 36. Shackelford D. B., Shaw R. J. (2009) The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tee A. R., Anjum R., Blenis J. (2003) Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 278, 37288–37296 [DOI] [PubMed] [Google Scholar]
- 38. Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 39. Zhang H., Cicchetti G., Onda H., Koon H. B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C. L., Kwiatkowski D. J. (2003) Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Invest. 112, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Comerford K. M., Leonard M. O., Karhausen J., Carey R., Colgan S. P., Taylor C. T. (2003) Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 100, 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mu J., Ostrowski R. P., Soejima Y., Rolland W. B., Krafft P. R., Tang J., Zhang J. H. (2013) Delayed hyperbaric oxygen therapy induces cell proliferation through stabilization of cAMP responsive element binding protein in the rat model of MCAo-induced ischemic brain injury. Neurobiol. Dis. 51, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeAngelo A. B., Daniel F. B., Stober J. A., Olson G. R. (1991) The carcinogenicity of dichloroacetic acid in the male B6C3F1 mouse. Fundam. Appl. Toxicol. 16, 337–347 [DOI] [PubMed] [Google Scholar]
- 43. Stacpoole P. W., Henderson G. N., Yan Z., James M. O. (1998) Clinical pharmacology and toxicology of dichloroacetate. Environ. Health Perspect. 106, 989–994 [DOI] [PMC free article] [PubMed] [Google Scholar]