Background: SREBP-1 plays a critical role in maintaining lipid homeostasis by activating lipogenic gene transcription.
Results: LSD1 is required for SREBP1-mediated gene expression through multiple mechanisms in mammalian cells.
Conclusion: LSD1 is a novel regulator of lipid metabolism.
Significance: LSD1 is a potential target for treating diseases with aberrant lipid homeostasis.
Keywords: Lipid Metabolism, Lipogenesis, Sirtuin 1 (SIRT1), Transcription, Triglyceride, LSD1, SREBP1
Abstract
Dysregulation of lipid homeostasis is a common feature of several major human diseases, including type 2 diabetes and cardiovascular disease. However, because of the complex nature of lipid metabolism, the regulatory mechanisms remain poorly defined at the molecular level. As the key transcriptional activators of lipogenic genes, such as fatty acid synthase (FAS), sterol regulatory element-binding proteins (SREBPs) play a pivotal role in stimulating lipid biosynthesis. Several studies have shown that SREBPs are regulated by the NAD+-dependent histone deacetylase SIRT1, which forms a complex with the lysine-specific histone demethylase LSD1. Here, we show that LSD1 plays a role in regulating SREBP1-mediated gene expression. Multiple lines of evidence suggest that LSD1 is required for SREBP1-dependent activation of the FAS promoter in mammalian cells. LSD1 knockdown decreases SREBP-1a at the transcription level. Although LSD1 affects nuclear SREBP-1 abundance indirectly through SIRT1, it is also required for SREBP1 binding to the FAS promoter. As a result, LSD1 knockdown decreases triglyceride levels in hepatocytes. Taken together, these results show that LSD1 plays a role in regulating lipogenic gene expression, suggesting LSD1 as a potential target for treating dysregulation of lipid metabolism.
Introduction
Among the known regulators of lipid metabolism, SREBPs2 are conserved transcription factors in maintaining lipid homeostasis (1, 2). In invertebrates, such as Caenorhabditis elegans and Drosophila, there is only one SREBP transcription factor that critically controls fatty acid biosynthesis, whereas in mammals there are three SREBP isoforms (SREBP-1a, -1c, and -2) encoded by two different genes, SREBF1 and SREBF2 (3). Two distinct promoters generate SREBP-1a and -1c isoforms from the SREBF1 gene with several amino acids longer at the N terminus of SREBP-1a proteins (3). SREBP-1a is expressed at a higher level in comparison with SREBP-1c in proliferating cells, such as cancer cells, but SREBP-1c is the predominant form of the SREBF1 gene in vivo, particularly in hepatocytes (4). Interestingly, a recent study has shown that SREBP-1a is also highly expressed in macrophages (5). All SREBPs are synthesized as inactive precursors that are tethered to the endoplasmic reticulum membrane (6). For SREBP-2, reduction of intracellular sterols induces the precursor transportation to the Golgi apparatus, where it is processed by proteolytic cleavage, and the resulting N-terminal fragment of SREBP-2 is then translocated into the nucleus and activates transcription of target genes as homodimers (7). Similar to SREBP-2, the two SREBP-1 isoforms are also processed in the Golgi apparatus to generate the mature/nuclear SREBP-1 proteins (8, 9). However, SREBP-1c is primarily activated by insulin at several steps, including transcription, proteolytic maturation from its precursor, and nuclear protein stability (3). In addition, SREBP-1c and SREBP-2 target different genes. SREBP-1c activates the expression of enzymes in the fatty acid and triglyceride biosynthesis pathway, including fatty acid synthase (FAS), whereas SREBP-2 preferentially stimulates the expression of genes responsible for cholesterol biosynthesis and uptake, including HMG-CoA reductase (HMGCR) and the LDL receptor (LDLR) (2). In general, SREBP-1a can activate both SREBP-1c and SREBP-2 target genes (10).
Although the proteolytic processing of SREBP precursors is a key step of controlling SREBP activation, nuclear SREBP proteins are further regulated (2, 3, 11). Like many other transcription factors, the transcriptional activity of SREBPs can be stimulated or repressed by transcriptional cofactors, including CBP/p300 (12) and the Mediator complex (13, 14). In addition to the maturation process of SREBP precursors, protein stability also critically contributes to the control of nuclear SREBP protein abundance. Phosphorylation of nuclear SREBPs within a conserved domain facilitates the binding to the E3 ligase, SCFFbw7b, and thus controls their ubiquitination and subsequent proteasome-mediated degradation (15, 16). For SREBP-1c, phosphorylation at the conserved threonine 402 residue by either GSK-3β (15) or CDK8 (17) is key to promote its degradation. Both kinases can be inactivated or down-regulated by insulin (17, 18), consistent with the positive role for insulin on nuclear SREBP-1c protein stability (15, 17).
In addition to phosphorylation, the stability of nuclear SREBP proteins is also regulated by acetylation and deacetylation (3, 11). Interaction of SREBPs with CBP/p300 not only results in acetylation of histone tails in the promoters of SREBP target genes but also causes acetylation at the conserved lysine residues of SREBPs (12). For human nuclear SREBP-1a, acetylation at the lysine 324 and lysine 333 inhibits its ubiquitination and degradation (12). It is known that the NAD+-dependent histone deacetylase SIRT1 can not only remove acetyl groups from histone tails but can also deacetylate several transcription factors, including p53, FOXO, and NF-κB/p65 (19). Recently, SIRT1 has been demonstrated as an important regulator of SREBPs by deacetylation, thus promoting nuclear SREBP protein degradation and repression of SREBP target genes (20–22). Consistent with the inverse correlation of SREBP and SIRT1 levels in response to food intake (20, 23), during fasting SIRT1 or its orthologs down-regulate SREBPs in mouse (20, 22), Drosophila (20), and C. elegans (20), thereby inhibiting lipid biosynthesis. Moreover, genetic manipulation of SIRT1 levels in mice results in altered triglyceride and cholesterol levels in serum and liver (23–27). These studies show that SIRT1 inhibition of SREBP target gene expression plays a conserved role in regulating lipid homeostasis.
A recent study has shown that SIRT1 can function in a protein complex with the lysine-specific histone demethylase-1 (LSD1, also known as KDM1A or AOF2) to regulate the Notch signaling pathway in Drosophila (28). As the first histone demethylase discovered (29), LSD1 can catalyze the removal of mono- or di-methyl, but not tri-methyl, groups from proteins including histone H3K4 (29), H3K9 (30, 31), tumor suppressor p53 (32), and DNA methyltransferase-1 (DNMT1) (33). Biochemical studies have shown that LSD1 functions in multiprotein complexes containing CtBP1, CoREST, and HDAC1/2 (28, 34, 35). By repressing or activating gene expression, LSD1 plays important roles in cell growth and differentiation (36). Because SIRT1 regulates lipid metabolism, the newly discovered physical interaction between LSD1 and SIRT1 suggests a possible role for LSD1 in lipid metabolism. In fact, it has been shown that LSD1 is required for adipocyte differentiation (37, 38). In this study, we show that LSD1 regulates SREBP1-mediated lipogenic gene expression at multiple steps.
EXPERIMENTAL PROCEDURES
Materials
Anti-LSD1 (Invitrogen in Fig. 1A and Thermo Scientific in other figures), anti-SIRT1 (Cell Signaling and Millipore), anti-SREBP1 (Santa Cruz), anti-FAS (Cell Signaling), anti-Flag (Sigma), anti-HA (Covance), and anti-β-tubulin (Invitrogen) antibodies were used. trans-2-Phenylcyclopropylamine was purchased from Enzo Life Sciences.
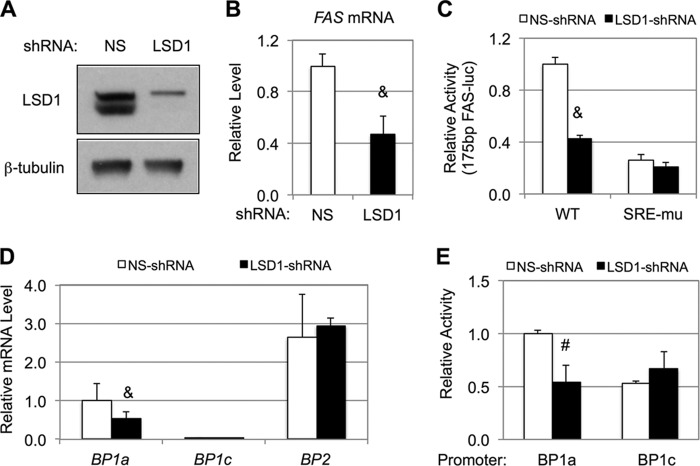
FIGURE 1.
LSD1 regulates lipogenic gene expression in HEK293 cells. A, immunoblots show LSD1 knockdown in HEK293 cells by lentivirus-based shRNA. B–E, effects of LSD1 knockdown on the mRNA levels of FAS (B) and SREBPs (D) as detected by qRT-PCR, and the activities of the wild-type or sterol response element-mutated FAS promoters (C) and the SREBP-1a or SREBP-1c promoters (E) as measured by luciferase reporter assays. The data represent means ± S.D. (n = 3). #, p < 0.01; &, p < 0.001 versus NS.
Plasmids
All pGIPZ constructs expressing shRNA were purchased from Thermo Scientific. The human FAS promoter fragments were amplified from genomic DNA of HepG2 cells by PCR and subcloned into HindIII and KpnI sites of the pGL3 vector, and the mutations in sterol response element sites were introduced by PCR. Oligonucleotides containing 3xSRE from the human FAS promoter were annealed and subcloned into HindIII and KpnI sites of the pGL3 vector to generate 3xSRE-Luc. The luciferase constructs for the SREBP-1a, SREBP-1c, and the LDL receptor promoters were generous gifts as described previously (39–41). SREBP-1a, -1c, and -2 transactivation domains (TADs) in pcDNA3-HA-Gal4DBD were generated by subcloning the TADs from pGEX-2TN (14). To generate the shRNA-resistant LSD1, six silent mutations were introduced by PCR to replace the LSD1-shRNA (clone 2) targeting sequence AGCTAGAAGAAAAACTTCA with AACTGGAGGAGAAGCTACA in pCMV-HA-LSD1 as described previously (29). Using the shRNA-resistant LSD1 construct as the template, additional mutation of A539E was introduced to generate the catalytic inactive LSD1 (42).
Tissue Culture
HEK293 and HepG2 cells were cultured at 37 °C and 5% CO2 in high glucose and low glucose DMEM (Sigma), respectively, supplemented with 100 μg/ml of penicillin/streptomycin (Invitrogen), 5% FBS (Hyclone), and 2 mm glutamine. Primary rat or mouse hepatocytes were isolated by perfusion of a rat or mouse liver by Dr. David Neufeld (Marion Bessin Liver Research Center at Albert Einstein College of Medicine). The freshly isolated primary hepatocytes were cultured as described previously (17).
Protein Extraction and Immunoblotting
Cells were lysed in a buffer containing 50 mm Tris-HCl (pH 8.0), 420 mm NaCl, 0.1 mm EDTA, 0.5% Nonidet P-40, 0.05% SDS, 10% Glycerol, 1 mm dithiothreitol, 2.5 mm PMSF, 1 mm benzamidine, 1 mg/liter aprotinin, and 0.1 mm N-acetyl-leucyl-leucyl-norleucinal. Supernatants were collected after centrifugation at 1.5 × 104 rpm for 20 min at 4 °C. Protein concentrations of the lysates were measured with a BCA kit (Pierce). Aliquots of the lysates containing the same amount of total proteins were mixed with 5× SDS loading buffer. After briefly boiling, the proteins were resolved by NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose or PVDF membranes with an iBlot gel transfer kit (Invitrogen). The membranes were first blocked with 5% nonfat milk in 1× TBST. After incubating with specific primary antibodies with appropriate dilution for 2 h at room temperature, the membranes were washed three times with 1× TBST (10 min each) and then incubated with the HRP-conjugated secondary antibodies (1:10,000 dilution) for 1 h at room temperature. After three washes with 1× TBST (10 min each), the HRP signals were visualized with the SuperSignal West Pico kit (Pierce) and exposure to x-ray films.
Transfection and Luciferase Reporter Assays
Transfection of plasmids was performed using the transfection reagent of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For luciferase reporter assays, plasmids of firefly luciferase reporter and the control Renilla luciferase were co-transfected into the cells. After overnight incubation, cells were harvested, and luciferase activities of different treatments were measured using the dual luciferase reporter assay kit (Promega) according to the manufacturer's instructions. In luciferase reporter assays, the expression level of firefly luciferase was under the control of a test promoter in pGL3 vector, and Renilla luciferase was constantly expressed driven by a basal promoter, which is generally unaffected by various treatments. The activity of firefly luciferase was normalized by the corresponding activity of Renilla luciferase.
Lentivirus Preparation and Transduction
Lentivirus was packaged using the TransLenti Viral GIPZ Packaging System (Thermo Scientific). The day before transfection of pGIPZ plasmids, HEK293T cells were plated at a density of 5.5 × 106 cells per 100-mm dish. For each dish, 9 μg of pGIPZ and 28.5 μg of viral packaging mix were co-transfected into HEK293T cells by Arrest-In transfection reagent. Six hours after transfection, culture medium was replaced with regular DMEM. Transfection efficiency was then examined 2 days post-transfection by the percentage of GFP-positive cells. Culture medium was collected at day 3 and centrifuged at 3,000 rpm for 20 min at 4 °C. The virus-containing supernatant was kept at −80 °C in aliquots. For lentiviral transduction, the cells were seeded into 6-well plates with a density of ∼70% confluent, and the next day 100 μl of virus-containing medium was added into the regular culture medium. After overnight culture, cells were replated into 100-mm dishes and selected with puromycin until all cells were GFP-positive.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cultured cells with the TRIzol reagent (Invitrogen) and quantified with an Agilent 2100 Bioanalyzer. After destroying genomic DNA with RQ1 RNase-free DNase I (Promega), cDNA was synthesized from 2 μg of RNA by a first strand cDNA synthesis kit (GE Healthcare). The resulting cDNA was then diluted for 10 times. Each real time PCR reaction mixture contained 10 μl of Fast-start Universal SYBR Green Master mix (Roche), 1 μl of primers (250 nm each), and 9 μl of diluted cDNA. Real time PCR was performed using StepOnePlusTM real time PCR System (Applied Biosystems). The cycling parameters consisted of 95 °C incubation for 10 min for enzyme activation and DNA denaturation, followed by 40 amplification cycles consisting of 95 °C for 15 s and 60 °C for 1 min. Data analysis was performed by the Software provided by StepOnePlusTM Real time PCR System (Applied Biosystems). Cyclophilin B was used as the invariant control. The sequences of PCR primers for human genes are listed in Table 1, and other primers were described previously (17).
TABLE 1.
PCR primer information for human genes
| Gene name | Primer sequences | |
|---|---|---|
| For qRT-PCR | ||
| FAS | Forward | 5′-AACTTGCAGGAGTTCTGGGAC |
| Reverse | 5′-TTGGGGTGGACTCCGAAGA | |
| SREBP-1a | Forward | 5′-GCTGCTGACCGACATCGAA |
| Reverse | 5′-ATGTGGCAGGAGGTGGAGAC | |
| SREBP-1c | Forward | 5′-GGAGCCATGGATTGCACTTT |
| Reverse | 5′-ATGTGGCAGGAGGTGGAGAC | |
| SREBP-2 | Forward | 5′-AGGCAGGCTTTGAAGACGAA |
| Reverse | 5′-GTACATCGGAACAGGCGGAT | |
| SIRT1 | Forward | 5′-GACTCCAAGGCCACGGATAG |
| Reverse | 5′-TGTTCGAGGATCTGTGCCAA | |
| Cyclophilin B | Forward | 5′-AACGCAACATGAAGGTGCTC |
| Reverse | 5′-CAAGATCACCCGGCCTACA | |
| LSD1 | Forward | 5′-GCCTGAAGAACCATCGGGGCA |
| Reverse | 5′-AGTCGGCTCTGGAAAGCTGCG | |
| For quantitative PCR of ChIP | ||
| FAS promoter | Forward | 5′-GTCCCCGGGAAGCTGCTAAG |
| Reverse | 5′-CGGGGTTACTGCCGGTCATC | |
| Luciferase for G4BE | Forward | 5′-CCACCATGGAAGACGCCAAA |
| Reverse | 5′-AGGAACCAGGGCGTATCTCT | |
Chromatin Immunoprecipitation Assay
For ChIP analysis, HA-nSREBP1a was co-transfected with the control system (HAG4DBD-VP16TAD and 2xGBE-luc) (3:1:1) to either nonspecific (NS)-shRNA or LSD1-shRNA-treated stable HEK293 cells. After culture for 24 h, cells were cross-linked in 10% formaldehyde for 10 min before being treated with glycine and resuspended in the lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris, pH 8.1) containing a protease inhibitor mixture (1 mm PMSF and 1 μg/ml each of aprotinin and leupeptin). The sample was sonicated on ice until the cross-linked chromatin DNA was sheared to an average length of ∼500 bp. The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.0% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, and 150 mm NaCl) containing a protease inhibitor mixture. The sample was precleaned with protein A/G-Sepharose and then incubated with anti-HA or control IgG antibody overnight at 4 °C. The antibody complexes were recovered with protein A/G-Sepharose at 4 °C for 1 h. The immunoprecipitated DNA within the complexes was released with reverse cross-linking buffer, extracted with phenol-chloroform, precipitated with ethanol, and analyzed with the SYBR Green-based qPCR. Cycle threshold (Ct) values were obtained for input (1% of starting chromatin), control ChIP, and anti-HA ChIP samples of the same treatment. The results were normalized by the transfection control (HA-G4DBD-VP16TAD binding to GBE). The sequences of qPCR primers are listed in Table 1.
Immunostaining
HEK293 cells were transfected with plasmids expressing Flag-tagged nSREBP-1a (amino acids 1–490) and LSD1-shRNA (or NS-shRNA as control) in a ratio of 1:2 by weight. In pGIPZ plasmids, a GFP and shRNA are under the control of the same CMV promoter, and therefore, GFP signals indicate the expression levels of shRNA. After 24 or 48 h of incubation, cells were fixed with 4% formaldehyde (Fisher) in 1× PBS for 15 min at room temperature, permeabilized with 0.25% Triton-100 (Sigma-Aldrich), and blocked with 10% BSA (Fisher) in 1× PBS for 30 min at 37 °C. The samples were incubated with monoclonal anti-Flag M2 antibody (Sigma) in 3% BSA (1:1,000) overnight at 4 °C, then washed with 1× PBS for three times, and incubated with Alexa Flour 594-labeled secondary antibody (Invitrogen) in 3% BSA (1:1,000) for 45 min at 37 °C in the dark. Nuclei were stained with Prolong Antifade reagent with DAPI (Invitrogen). Images were collected using a fluorescence microscope.
Co-immunoprecipitation
HA-tagged wild-type LSD1 was co-transfected with either Flag-tagged nuclear form of GFP, nuclear form of SREBP-1a, or SIRT1 into HEK293 cells. After ∼36 h of incubation, cell lysates were prepared in immunoprecipitation buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, 1 mm dithiothreitol, 2.5 mm PMSF, 1 mm benzamidine, 1 mg/liter aprotinin, and 0.1 mm N-acetyl-leucyl-leucyl-norleucinal. The Flag-tagged proteins were immunoprecipitated from the lysates using anti-Flag antibody. After washing five times with the immunoprecipitation buffer, Flag-tagged proteins were eluted with 3× Flag peptide in the immunoprecipitation buffer. Proteins in elutes were detected by immunoblotting using anti-Flag or anti-HA antibody.
Animal Care and Treatment
The mouse experiments conformed to the protocols approved by the Animal Care and Use Committees of Albert Einstein College of Medicine in accordance with the National Institutes of Health guidelines. Male db/db and wild-type mice at the age of 8 weeks were purchased from The Jackson Laboratory. Upon arrival, the mice were maintained under a 12-hour dark cycle with free access to water and standard mouse diet for at least 1 week before experiments.
Triglyceride Measurement
The amount of triglyceride in hepatocytes was detected by an adipogenesis assay kit (Biovision) and normalized by the total amount of cellular proteins.
Statistical Analyses
The data were presented as the means ±S.D., and the significance of the differences between the two groups was evaluated using Student's t test. The p value <0.05 was considered significant.
RESULTS
LSD1 Regulates the Expression of FAS and SREBP-1a
To examine whether LSD1 could regulate lipogenic gene expression, we knocked down LSD1 in HEK293 cells using lentivirus-based LSD1-specific shRNA (Fig. 1A) and found that the mRNA levels of the FAS gene were significantly lower than those in control cells treated with NS-shRNA (Fig. 1B). Because SREBP-1a and -1c are key transcriptional activators for the FAS promoter, we then analyzed the effects of LSD1 knockdown on the activity of the 175-bp proximal promoter of the human FAS gene, which contains the SREBP1-responsive elements. As shown in Fig. 1C, LSD1 knockdown decreased the wild-type FAS promoter activity, and the inhibitory effect was similar to that on the endogenous FAS gene expression. In contrast, when the SREBP1-responsive elements were destroyed by point mutations, the promoter activity was expectedly decreased by ∼70%, and more importantly, the effect of LSD1 knockdown vanished (Fig. 1C), suggesting that LSD1 is required for activating the FAS promoter in HEK293 cells and that such regulation is likely SREBP1-dependent.
To understand the mechanisms underlying LSD1 regulation of the FAS promoter, we examined all three SREBP transcripts in HEK293 cells by qRT-PCR. As shown in Fig. 1D, the SREBP-1a transcript was more than 30-fold more abundant than SREBP-1c, whereas the SREBP-2 transcript was about 2-fold higher than SREBP-1a. Interestingly, knockdown of LSD1 resulted in a significant decrease of SREBP-1a transcripts but had no effects on SREBP-1c or SREBP-2 (Fig. 1D). Similarly, LSD1 had little effect on SREBP-1c in primary hepatocytes (data not shown). To determine whether LSD1 regulates SREBP-1a mRNA expression at the transcription level, we examined the effect of LSD1 knockdown on the promoter activity of human SREBP-1a gene (39) by luciferase reporter assays. Consistent with the mRNA data, LSD1 knockdown similarly decreased the human SREBP-1a promoter activity, although it did not affect the human SREBP-1c promoter (Fig. 1E), indicating a specific role for LSD1 on the SREBP-1a promoter in HEK293 cells.
LSD1 Regulates the Abundance of Nuclear SREBP-1
Next, we examined the effect of LSD1 knockdown on endogenous SREBP-1 proteins. Although the commercial SREBP-1 antibody detects both SREBP-1a and SREBP-1c, the observed signals were presumably from SREBP-1a (Fig. 2A), because the SREBP-1a transcripts were much more abundant than SREBP-1c in this case (Fig. 1D). Consistent with the mRNA data in Fig. 1D, LSD1 knockdown significantly decreased the amount of endogenous SREBP-1a precursor proteins in HEK293 cells (Fig. 2, A and B). Interestingly, the nuclear form of SREBP-1a was nearly unaffected by LSD1 knockdown (Fig. 2, A and B). Because recent studies show that SIRT1 regulates the abundance of nuclear SREBP transcription factors (20–22) and LSD1 forms a complex with SIRT1 (28), the discrepancy between the precursor and nuclear SREBP-1a protein levels raised a possibility that LSD1 may also negatively regulate SREBP-1a protein stability. To test this possibility, we knocked down LSD1 or SIRT1 in HEK293 cells by specific shRNA. HA-tagged nuclear form of SREBP-1a was co-transfected with the control, a fusion protein of Gal4-DNA-binding domain (G4DBD) and Myb transactivation domain (as the transfection control). As shown in Fig. 2C, SIRT1 knockdown increased nuclear SREBP-1a levels in HEK293 cells, confirming the previous reports (20–22). Interestingly, LSD1 knockdown similarly caused a significant accumulation of overexpressed nuclear SREBP-1a proteins (Fig. 2C).
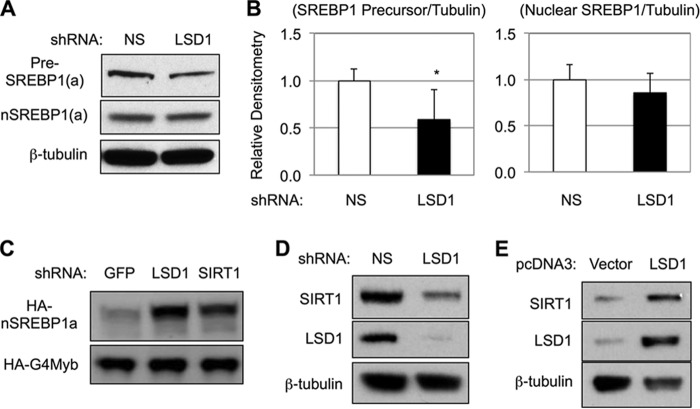
FIGURE 2.
LSD1 regulates nuclear SREBP-1 and SIRT1 protein levels in HEK293 cells. A, immunoblots show the effects of LSD1 knockdown on endogenous SREBP-1 precursor (pre-SREBP1) and nuclear form (nSREBP1). B, densitometry analysis of precursor and nuclear SREBP-1 proteins in HEK293 cells after LSD1 knockdown. The data represent means ± S.D. (n = 4). *, p < 0.05 versus NS. C, immunoblots using anti-HA antibody show the effects of the indicated shRNA on co-transfected HA-tagged nuclear SREBP-1a (HA-nSREBP1a) and G4DBD fused Myb-TAD (HA-G4Myb, the control). D and E, immunoblots show the effects of LSD1 knockdown (D) or overexpression with HA-tagged LSD1 (E) on endogenous SIRT1 protein levels.
Because LSD1 complexes with SIRT1 (28), they may stabilize each other in those cells. To examine this possibility, LSD1 was first knocked down in HEK293 cells by transfection of specific shRNA. As shown in Fig. 2D, knockdown of LSD1 significantly decreased the protein levels of SIRT1 as measured by immunoblotting. The effect was likely specific to LSD1, because similar results were obtained when a total of four independent shRNA constructs were examined (data not shown), and LSD1 knockdown also caused a decrease of SIRT1 proteins in HepG2 cells (data not shown). Conversely, when HA-tagged wild-type LSD1 proteins were overexpressed in HEK293 cells, the protein levels of SIRT1 were increased (Fig. 2E). Thus, LSD1 and SIRT1 protein levels are positively correlated with each other in HEK293 cells. Moreover, LSD1 knockdown had no effects on SIRT1 mRNA levels (data not shown), further suggesting a post-transcriptional mechanism. Considering that LSD1 regulates SIRT1, which in turn negatively regulates the abundance of nuclear SREBP-1a (20–22), these results suggest that LSD1 knockdown-caused accumulation of nuclear SREBP-1a is likely due to the decrease of SIRT1 proteins.
LSD1 Is Required for SREBP1-mediated Transcription
The discrepancy between the amount of mature/nuclear SREBP-1a (Fig. 2A) and its target gene levels (Fig. 1B) upon LSD1 knockdown suggested that LSD1 also affects the nuclear SREBP-1a-induced transcription. To test this possibility, we examined the effects of overexpressed nuclear SREBP-1a on the SREBP1-responsive promoters by luciferase reporter assays. As expected, overexpression of nuclear SREBP-1a in control cells activated the synthetic SREBP1-responsive promoter containing three tandem sterol response elements from the human FAS gene promoter (Fig. 3A). In contrast, knockdown of LSD1 completely abolished SREBP1a-induced activation (Fig. 3A), suggesting that the accumulated nuclear SREBP-1a proteins were transcriptionally inactive in the absence of LSD1. Similarly, in LSD1 knockdown cells, overexpressed SREBP-1a displayed a decreased ability to activate the 1-kb promoter of the human FAS gene (Fig. 3B), when compared with the effects in control cells. Taken together, these results demonstrate that LSD1 is required for nuclear SREBP-1a to efficiently activate transcription of its target genes in HEK293 cells.
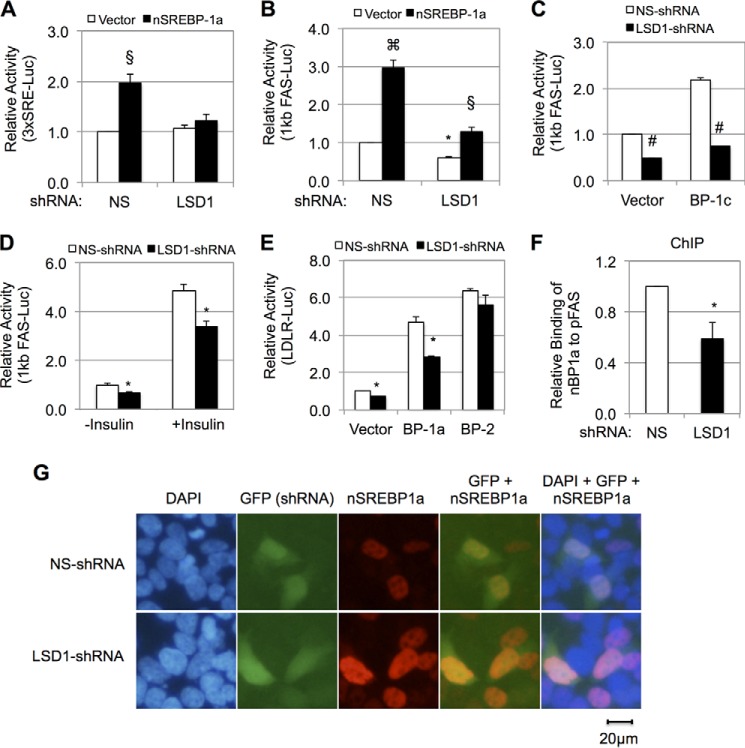
FIGURE 3.
LSD1 is required for SREBP1-mediated transcription in mammalian cells. A–C and E, luciferase reporter assays detect the effects of LSD1 knockdown in HEK293 cells using lentivirus-based shRNA on SREBP-1a-induced activation of a basal promoter that contains three tandem SRE sites (A) or the 1-kb human FAS gene promoter (B), SREBP-1c-induced activation of the 1-kb human FAS gene promoter (C), and SREBP-1a or SREBP-2 -induced activation of the human LDLR gene promoter (E). D, effects of LSD1 knockdown and insulin (200 nm, 18 h) on the FAS promoter activity in HEK293 cells. F, ChIP-qPCR shows the effects of LSD1 knockdown on HA-tagged nuclear SREBP-1a binding to the FAS promoter in HEK293 cells. HA-tagged G4DBD-VP16-TAD and 2xGBE-Luc were co-transfected as the internal control. G, immunostaining for overexpressed Flag-nSREBP-1a (amino acids 1–490) when co-transfected with LSD1-shRNA or NS-shRNA (control) in HEK293 cells. The nuclei were visualized with DAPI staining. The data represent means ± S.D. (n = 3). *, p < 0.05; #, p < 0.01 versus NS; §, p < 0.05; clover symbol, p < 0.01 versus vector.
Because SREBP-1a and -1c are isoforms produced from the same gene (SREBF-1), and they only differ at the N terminus (43), we asked whether LSD1 could also regulate SREBP1c-mediated gene expression. For this purpose, we first knocked down LSD1 in HEK293 cells by shRNA and then co-transfected a nuclear form of SREBP-1c with the firefly luciferase reporter that is under the control of the human FAS promoter. As shown in Fig. 3C, overexpression of nuclear SREBP-1c increased the FAS promoter activity as expected, whereas knockdown of LSD1 significantly decreased the basal activity of the FAS promoter. More importantly, LSD1 knockdown abolished SREBP1c-induced activation of the FAS promoter (Fig. 3C), suggesting that LSD1 is also required for SREBP1c-mediated transcription in HEK293 cells. In addition, LSD1 knockdown suppressed the FAS promoter to a similar degree in the absence or presence of insulin (Fig. 3D). Moreover, LSD1 knockdown also significantly decreased nuclear SREBP-1a-induced, but not SREBP-2-induced, activation of the LDL receptor promoter (Fig. 3E). Together, our results demonstrate that LSD1 is required for nuclear SREBP1-activated gene transcription.
LSD1 Regulates SREBP1a Binding to the FAS Promoter
Three possible mechanisms could explain the decreased ability of nuclear SREBP-1a/1c in activating target gene promoters: 1) the activity of SREBP-1a/1c TADs is decreased, 2) LSD1 is required for the DNA binding of nuclear SREBP-1, and/or 3) LSD1 regulates the localization of nuclear SREBP-1. First, we asked whether LSD1 affected the TAD activity of SREBP-1a. To test this possibility, we generated an artificial transcription factor by fusing G4DBD to human SREBP1a-TAD and tested its ability to activate a synthetic promoter containing two tandem GBEs. LSD1 knockdown did not significantly alter the ability of this fusion transcription factor to activate the GBE-containing promoter (data not shown). Similarly, LSD1 knockdown did not significantly affect the activity of SREBP1c-TAD or SREBP2-TAD (data not shown). Thus, LSD1 does not regulate the TADs of SREBP transcription factors.
Next, we asked whether LSD1 knockdown could affect the DNA binding affinity of nuclear SREBP-1a proteins. For that purpose, we performed ChIP analyses of overexpressed HA-tagged nuclear SREBP-1a. To normalize the efficiency of both transfection and ChIP, we co-transfected HA-SREBP-1a with HA-tagged G4DBD-VP16-TAD and its binding target 2xGBE luciferase reporter into HEK293 cells. As shown in Fig. 3F, despite the accumulation of this transcription factor in LSD1 knockdown cells (Fig. 2C), it displayed a significant lower DNA binding affinity to the FAS promoter. Thus, our data suggest that LSD1 is required for the efficient binding of SREBP-1a to the FAS promoter.
To determine whether the decreased DNA binding of accumulated nSREBP-1a was due to the change of nSREBP-1a localization, we performed immunostaining. As shown in Fig. 3G, the N-terminal fragment (amino acids 1–490) of SREBP-1a is exclusively localized in the nucleus, consistent with previous reports, and LSD1-shRNA did not significantly change such pattern. Therefore, LSD1 does not regulate the localization of nuclear SREBP-1a proteins.
The Demethylase Activity of LSD1 Is Involved in Regulating Lipogenic Gene Expression
To eliminate the potential off target effects of shRNA, we generated a construct expressing shRNA-resistant LSD1 by introducing silent mutations (Fig. 4A). As shown in Fig. 4B, the wild-type shRNA-resistant LSD1 could rescue LSD1 knockdown-caused inhibition of the FAS promoter activity in a dose-dependent manner, demonstrating a specific effect of LSD1 in regulating the FAS promoter. Because LSD1 is a lysine-specific histone demethylase (29), we asked whether the demethylase activity of LSD1 was required for regulating the FAS gene. For that purpose, we mutated the alanine 539 residue to glutamic acid (named as LSD1-AE) on top of the shRNA-resistant LSD1, because the mutation at alanine 539 of LSD1 completely abolishes the demethylase enzymatic activity of LSD1 (42). We then tested the effects of LSD1-AE on the FAS promoter activity by luciferase reporter assays. As shown in Fig. 4A, the wild-type and enzyme-dead LSD1 proteins were expressed at a similar level in HEK293 cells. Surprisingly, overexpression of LSD1-AE dose-dependently inhibited the FAS promoter activity in both NS-shRNA and LSD1-shRNA-treated HEK293 cells (Fig. 4C). Thus, the enzyme-dead LSD1 proteins may function in a dominant-negative manner. In agreement with these data, LSD1 inhibition by tranylcypromine (trans-2-phenylcyclopropylamine) (44) decreased the promoter activity of human FAS as examined by luciferase reporter assays (Fig. 4D). Together, these results show that the demethylase activity of LSD1 is involved in regulating lipogenic gene expression in mammalian cells.
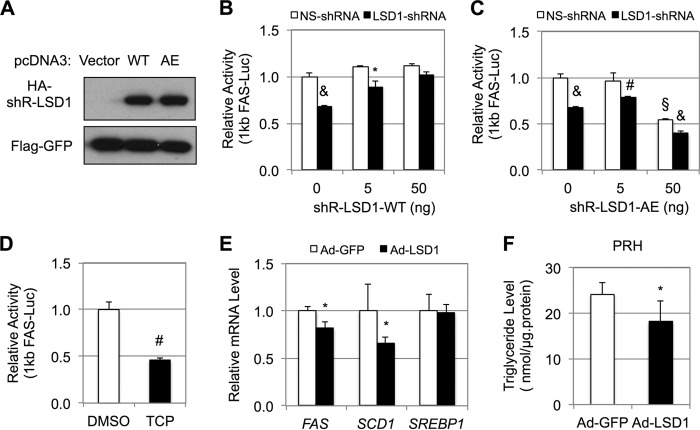
FIGURE 4.
LSD1 regulation of lipogenic gene expression requires its demethylase activity. A, immunoblots for HA-tagged WT or A539E (AE) LSD1 proteins that are resistant to shRNA#2. B and C, effects of the indicated doses of LSD1-WT (B) or -AE (C) on LSD1 knockdown-induced inhibition of the FAS promoter as examined by luciferase reporter assays. D, effects of trans-2-phenylcyclopropylamine (TCP) (0.2 mm) on the activity of the FAS promoter after 18 h of treatment. E and F, effects of LSD1 overexpression by adenovirus in primary rat hepatocytes on indicated gene expression by qRT-PCR (E) and triglyceride levels (F). The data represent means ± S.D. (n = 3). *, p < 0.05; #, p < 0.01; &, p < 0.001 versus NS or GFP; §, p < 0.05 versus vector.
LSD1 Regulates Lipid Accumulation in Hepatocytes
A possible dominant-negative effect of LSD1 overexpression was also observed in primary rat hepatocytes, where overexpression of LSD1 by adenovirus caused a significant decrease of SREBP1 target genes, such as FAS and stearoyl-CoA desaturase-1 (SCD1) (Fig. 4E). Consistent with our data in Fig. 1D, overexpression of LSD1 did not affect the mRNA levels of SREBP-1c in primary rat hepatocytes (Fig. 4E). Consistent with the decrease of lipogenic gene expression, overexpression of LSD1 significantly decreased triglyceride levels in primary rat hepatocytes (Fig. 4F).
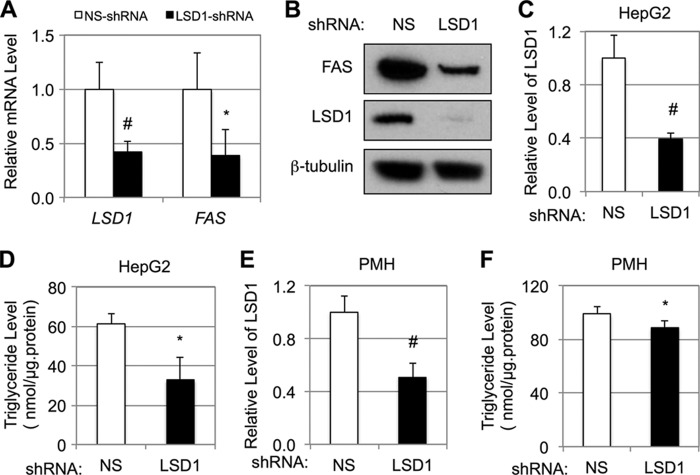
To eliminate the potential complications of long-time depletion of LSD1, we acutely knocked down LSD1 by transient transfection of the LSD1-shRNA plasmid from Fig. 2D. As shown in Fig. 5A, acute knockdown of LSD1 also resulted in a significant decrease of endogenous FAS mRNA levels in HEK293 cells as detected by qRT-PCR. Consistent with the mRNA data, acute knockdown of LSD1 also decreased FAS proteins as examined by immunoblotting (Fig. 5B). In HepG2 cells, knockdown of LSD1 (Fig. 5C) significantly decreased triglyceride accumulation (Fig. 5D). Similarly, in primary mouse hepatocytes, knockdown of LSD1 (Fig. 5E) also significantly decreased the cellular triglyceride levels (Fig. 5F). Taken together, our results suggest that LSD1 is required for triglyceride accumulation in hepatocytes.
FIGURE 5.
LSD1 regulates triglyceride levels in hepatocytes. A and B, effects of transfection with NS or LSD1-shRNA plasmids in HEK293 cells on the mRNA levels of indicated genes in HEK293 by qRT-PCR (A) and the protein levels of endogenous FAS by immunoblotting (B). C–F, effects of LSD1 knockdown in HepG2 cells (C) or primary mouse hepatocytes (E) on the triglyceride levels (D and F). The data represent means ± S.D. (n = 3). *, p < 0.05; #, p < 0.01 versus NS.
DISCUSSION
In this study, we have shown a novel role for LSD1 in regulating fatty acid biosynthesis through the SREBP-1 transcription factors in mammalian cells. We show that LSD1 is specifically involved in activating the SREBP-1a promoter in HEK293 cells, controlling the levels of SREBP-1a precursors. When the mature SREBP-1 proteins are translocated into nucleus, LSD1 regulates their stability, likely through controlling the abundance of SIRT1. Although LSD1 does not affect the activities of SREBP-TADs and localization of nuclear SREBP-1, it is required for nuclear SREBP-1 binding to the FAS promoter. Through the regulation of SREBP-1 at multiple steps, LSD1 collectively supports the expression of SREBP1 target genes in mammalian cells. In addition, the demethylase activity of LSD1 is involved in regulating lipogenic gene expression. As a result, LSD1 regulates the cellular triglyceride levels in cultured hepatocytes. These observations show that LSD1 is a novel regulator of SREBP1-mediated lipid metabolism in mammalian cells.
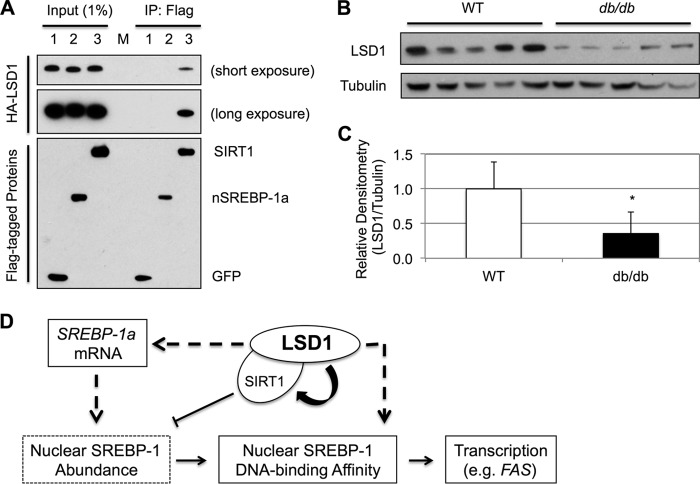
By removing the methyl groups from histone H3K4 (29), H3K9 (30), or other non-histone proteins such as p53 (32), LSD1 plays a critical role on the transcription of genes involved in cell growth and differentiation (36). The function of LSD1 in lipid metabolism has started to be appreciated recently by the results showing that LSD1 regulates adipogenesis (37, 38). Recently, we show that LSD1 physically and functionally interacts with SIRT1 (28). In this study, we show that the protein levels of LSD1 were positively correlated with those of SIRT1. Because LSD1 did not regulate SIRT1 gene expression, LSD1 and SIRT1 proteins may simply stabilize each other through their physical interaction, like in cases of other protein complexes. Consistent with recent studies showing that SIRT1 negatively controls nuclear SREBP protein levels (20–22), LSD1 knockdown caused a significant accumulation of nuclear SREBP-1a proteins, likely because of an indirect mechanism through SIRT1, because we did not detect a stable interaction between LSD1 and SREBP-1a by co-immunoprecipitation (Fig. 6A). However, this mechanism is overridden by the positive role of LSD1 on the transcription of SREBP-1a and the DNA binding affinity of nuclear SREBP-1 proteins, resulting in an overall decrease of lipogenic gene expression when LSD1 is knocked down. It will be interesting to examine whether other biological functions of SIRT1 are also regulated by LSD1 in the future.
FIGURE 6.
LSD1 regulation of SREBP-1. A, HA-tagged wild-type LSD1 is co-transfected with either Flag-tagged GFP, nuclear form of SREBP-1a, or SIRT1 in HEK293 cells. After ∼36 h of incubation, cell lysates were prepared. The Flag-tagged proteins were immunoprecipitated from the lysates after extensive wash and detected by immunoblotting using anti-Flag or anti-HA antibody. B, immunoblots for the protein levels of LSD1 in livers from 12-week-old male wild-type or db/db mice as indicated. β-Tubulin served as the loading control. C, densitometry analysis of the data in B. The data represent means ± S.D. (n = 5). *, p < 0.05 versus WT. D, the model. In mammalian cells, LSD1 is required for the expression of SREBP-1a, contributing to the pool of nuclear SREBP-1 proteins. Although LSD1 promotes nuclear SREBP-1 degradation indirectly through SIRT1, it is also required for the efficient binding of SREBP-1 to the target gene promoters. Collectively, LSD1 functions to activate lipogenic gene expression in mammalian cells.
Multiple lines of evidence in this study demonstrate that LSD1 is required for lipogenic gene expression in mammalian cells. Although we could not exclude the involvement of other transcription factors, LSD1 regulation of the FAS gene transcription is largely SREBP1-dependent, because LSD1 knockdown nearly abolished SREBP-1a- or SREBP-1c-induced activation of multiple promoters (Fig. 3). Mutagenesis analysis of the SREBP-responsive sites in the FAS promoter further supports the roles of SREBP-1a/1c in LSD1 regulation of FAS. Thus, LSD1 functions to activate SREBP-1 in mammalian cells. Moreover, the demethylase activity of LSD1 is required for this function. We have recently shown that resveratrol, curcumin, and quercetin can inhibit the LSD1 activity (45). These compounds are known to have beneficial effects on lipid homeostasis (20, 46–48). Based on this study, the effects of these compounds on lipid homeostasis may be partially through inhibiting LSD1.
Interestingly, the protein levels of hepatic LSD1 in db/db mice, one of the popular obese models, were lower than those in wild-type mice (Fig. 6, B and C). It is well known that db/db mice display higher levels of SREBP-1c target genes in the liver and thus a higher rate of de novo lipogenesis. Taking the results from this study, it is possible that the overactivation of SREBP-1c may have been partially suppressed in db/db mice because of the down-regulation of LSD1.
Consistent with the previous report (49), we detected a higher level of SREBP-1a mRNA as compared with SREBP-1c in HEK293 and HepG2 cells (data not shown). Thus, it is conceivable that the effects of LSD1 on endogenous SREBP target genes are primarily through SREBP-1a in HEK293 and HepG2 cells, whereas SREBP-1c is the predominant form of SREBP-1 in primary rat and mouse hepatocytes. Nevertheless, LSD1 is required for both SREBP-1a/1c-activated gene transcription (Fig. 3). Although LSD1 knockdown also decreases the promoter activity of LDLR in HEK293 cells, such regulation is through SREBP-1a, because LSD1 knockdown did not affect SREBP-2-mediated activation (Fig. 3D).
Our data suggest that LSD1 regulates SREBP-1a/1c at several steps. First, LSD1 is required for activating the SREBP-1a promoter, but it does not affect SREBP-1c and SREBP-2 (Fig. 1D). Previous studies have shown that the SREBP-1a promoter is primarily controlled by the transcription factor Sp1 (39). Therefore, LSD1 may regulate the SREBP-1a promoter through Sp1. Interestingly, it has been shown that Sp1 is methylated at the lysine residues, and LSD1 can functionally de-methylate Sp1 (50). Second, consistent with the mRNA data, LSD1 knockdown significantly decreased the amount of SREBP-1 precursor proteins in HEK293 cells; however, the nuclear form of SREBP-1 was unaffected (Fig. 2, A and B). This inconsistency is likely because LSD1 negatively controls the abundance of nuclear SREBP-1a (Fig. 2C), although LSD1 may also control the SREBP maturation process. Third, the difference between nuclear SREBP-1 protein levels and transcriptional functions led us to discover that LSD1 is also required for SREBP-1a binding to the FAS promoter (Fig. 3F). According to the data in Fig. 3C, it is likely that LSD1 also supports SREBP-1c binding to the FAS promoter. The underlying mechanism(s) for LSD1 regulating DNA binding affinity of SREBP1 transcription factors currently remain unclear. LSD1 may indirectly control the expression of certain genes that regulate the DNA binding affinity of nuclear SREBP-1 proteins. It has been shown that proto-oncogene FBI-1 modulates the DNA binding affinity of SREBP-1 in mammalian cells (51). It will be interesting to test such a possibility in future experiments.
In summary, our results show that LSD1 is required for lipid biosynthesis through the mechanisms of activating SREBP-1 transcription factors (Fig. 6D). This finding may have potential implications for treating metabolic diseases as well as cancer.
Acknowledgments
We thank Drs. Jeffrey E. Pessin, Nicholas Sibinga, and Rajat Singh for critical comments of this study and members of Dr. Pessin's laboratory for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK093623 and P60-DK020541 (to the Einstein Diabetes Center). This work was also supported by American Diabetes Association Grant 7-11-BS-173.
- SREBP
- sterol regulatory element-binding proteins
- TAD
- transactivation domain
- qRT-PCR
- quantitative RT-PCR
- NS
- nonspecific
- GBE
- Gal4-DBD-binding element.
REFERENCES
- 1. Osborne T. F., Espenshade P. J. (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 23, 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeon T. I., Osborne T. F. (2012) SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xiaoli, Yang F. (2013) Mediating lipid biosynthesis: Implications for cardiovascular disease. Trends Cardiovasc. Med. 23, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. (1997) Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99, 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Im S. S., Yousef L., Blaschitz C., Liu J. Z., Edwards R. A., Young S. G., Raffatellu M., Osborne T. F. (2011) Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 13, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horton J. D., Goldstein J. L., Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., Wang X. (1993) SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U.S.A. 90, 11603–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., Brown M. S. (1993) SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75, 187–197 [PubMed] [Google Scholar]
- 9. Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. (1994) SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62 [DOI] [PubMed] [Google Scholar]
- 10. Amemiya-Kudo M., Shimano H., Hasty A. H., Yahagi N., Yoshikawa T., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Sato R., Kimura S., Ishibashi S., Yamada N. (2002) Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43, 1220–1235 [PubMed] [Google Scholar]
- 11. Zhang Y., Xiaoli, Zhao X., Yang F. (2013) The mediator complex and lipid metabolism. J. Biochem. Pharmacol. Res. 1, 51–55 [PMC free article] [PubMed] [Google Scholar]
- 12. Giandomenico V., Simonsson M., Grönroos E., Ericsson J. (2003) Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 23, 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Näär A. M., Beaurang P. A., Zhou S., Abraham S., Solomon W., Tjian R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398, 828–832 [DOI] [PubMed] [Google Scholar]
- 14. Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z. Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A. C., Wagner G., Näär A. M. (2006) An ARC/mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 [DOI] [PubMed] [Google Scholar]
- 15. Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., Ericsson J. (2005) Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 [DOI] [PubMed] [Google Scholar]
- 16. Bengoechea-Alonso M. T., Ericsson J. (2009) A phosphorylation cascade controls the degradation of active SREBP1. J. Biol. Chem. 284, 5885–5895 [DOI] [PubMed] [Google Scholar]
- 17. Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., Zhou J., Sun Y., Yang E. S., Liu L. P., Vaitheesvaran B., Bridges L., Kurland I. J., Strich R., Ni J. Q., Wang C., Ericsson J., Pessin J. E., Ji J. Y., Yang F. (2012) Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122, 2417–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaw M., Cohen P., Alessi D. R. (1997) Further evidence that the inhibition of glycogen synthase kinase-3beta by IGF-1 is mediated by PDK1/PKB-induced phosphorylation of Ser-9 and not by dephosphorylation of Tyr-216. FEBS Lett. 416, 307–311 [DOI] [PubMed] [Google Scholar]
- 19. Chang H. C., Guarente L. (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walker A. K., Yang F., Jiang K., Ji J. Y., Watts J. L., Purushotham A., Boss O., Hirsch M. L., Ribich S., Smith J. J., Israelian K., Westphal C. H., Rodgers J. T., Shioda T., Elson S. L., Mulligan P., Najafi-Shoushtari H., Black J. C., Thakur J. K., Kadyk L. C., Whetstine J. R., Mostoslavsky R., Puigserver P., Li X., Dyson N. J., Hart A. C., Näär A. M. (2010) Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24, 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. You M., Liang X., Ajmo J. M., Ness G. C. (2008) Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G892–G898 [DOI] [PubMed] [Google Scholar]
- 22. Ponugoti B., Kim D. H., Xiao Z., Smith Z., Miao J., Zang M., Wu S. Y., Chiang C. M., Veenstra T. D., Kemper J. K. (2010) SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 285, 33959–33970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodgers J. T., Puigserver P. (2007) Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Erion D. M., Yonemitsu S., Nie Y., Nagai Y., Gillum M. P., Hsiao J. J., Iwasaki T., Stark R., Weismann D., Yu X. X., Murray S. F., Bhanot S., Monia B. P., Horvath T. L., Gao Q., Samuel V. T., Shulman G. I. (2009) SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc. Natl. Acad. Sci. U.S.A. 106, 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007) SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28, 91–106 [DOI] [PubMed] [Google Scholar]
- 26. Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M. H. (2008) Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X., Li X. (2009) Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulligan P., Yang F., Di Stefano L., Ji J. Y., Ouyang J., Nishikawa J. L., Toiber D., Kulkarni M., Wang Q., Najafi-Shoushtari S. H., Mostoslavsky R., Gygi S. P., Gill G., Dyson N. J., Näär A. M. (2011) A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell 42, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 30. Metzger E., Wissmann M., Yin N., Müller J. M., Schneider R., Peters A. H., Günther T., Buettner R., Schüle R. (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 [DOI] [PubMed] [Google Scholar]
- 31. Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 [DOI] [PubMed] [Google Scholar]
- 33. Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., Chen T. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 [DOI] [PubMed] [Google Scholar]
- 34. Shi Y., Sawada J., Sui G., Affar el B., Whetstine J. R., Lan F., Ogawa H., Luke M. P., Nakatani Y. (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
- 35. Lee M. G., Wynder C., Cooch N., Shiekhattar R. (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 [DOI] [PubMed] [Google Scholar]
- 36. Forneris F., Binda C., Battaglioli E., Mattevi A. (2008) LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem. Sci. 33, 181–189 [DOI] [PubMed] [Google Scholar]
- 37. Hino S., Sakamoto A., Nagaoka K., Anan K., Wang Y., Mimasu S., Umehara T., Yokoyama S., Kosai K., Nakao M. (2012) FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat. Commun. 3, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Musri M. M., Carmona M. C., Hanzu F. A., Kaliman P., Gomis R., Párrizas M. (2010) Histone demethylase LSD1 regulates adipogenesis. J. Biol. Chem. 285, 30034–30041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang C., Shin D. J., Osborne T. F. (2005) A simple promoter containing two Sp1 sites controls the expression of sterol-regulatory-element-binding protein 1a (SREBP-1a). Biochem. J. 386, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dif N., Euthine V., Gonnet E., Laville M., Vidal H., Lefai E. (2006) Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem. J. 400, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castoreno A. B., Wang Y., Stockinger W., Jarzylo L. A., Du H., Pagnon J. C., Shieh E. C., Nohturfft A. (2005) Transcriptional regulation of phagocytosis-induced membrane biogenesis by sterol regulatory element binding proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 13129–13134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stavropoulos P., Blobel G., Hoelz A. (2006) Crystal structure and mechanism of human lysine-specific demethylase-1. Nat. Struct. Mol. Biol. 13, 626–632 [DOI] [PubMed] [Google Scholar]
- 43. Hua X., Wu J., Goldstein J. L., Brown M. S., Hobbs H. H. (1995) Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics 25, 667–673 [DOI] [PubMed] [Google Scholar]
- 44. Lee M. G., Wynder C., Schmidt D. M., McCafferty D. G., Shiekhattar R. (2006) Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13, 563–567 [DOI] [PubMed] [Google Scholar]
- 45. Abdulla A., Zhao X., Yang F. (2013) Natural polyphenols inhibit lysine-specific demethylase-1. J. Biochem. Pharmacol. Res. 1, 56–63 [PMC free article] [PubMed] [Google Scholar]
- 46. Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zingg J. M., Hasan S. T., Meydani M. (2013) Molecular mechanisms of hypolipidemic effects of curcumin. Biofactors 39, 101–121 [DOI] [PubMed] [Google Scholar]
- 48. Boots A. W., Haenen G. R., Bast A. (2008) Health effects of quercetin: from antioxidant to nutraceutical. Eur. J. Pharmacol. 585, 325–337 [DOI] [PubMed] [Google Scholar]
- 49. Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001) Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 50. Chuang J. Y., Chang W. C., Hung J. J. (2011) Hydrogen peroxide induces Sp1 methylation and thereby suppresses cyclin B1 via recruitment of Suv39H1 and HDAC1 in cancer cells. Free Radic. Biol. Med. 51, 2309–2318 [DOI] [PubMed] [Google Scholar]
- 51. Choi W. I., Jeon B. N., Park H., Yoo J. Y., Kim Y. S., Koh D. I., Kim M. H., Kim Y. R., Lee C. E., Kim K. S., Osborne T. F., Hur M. W. (2008) Proto-oncogene FBI-1 (Pokemon) and SREBP-1 synergistically activate transcription of fatty-acid synthase gene (FASN). J. Biol. Chem. 283, 29341–29354 [DOI] [PMC free article] [PubMed] [Google Scholar]