Background: Acid β-glucosidase is trafficked to the lysosome by LIMP-2.
Results: A unique 11-amino acid sequence on acid β-glucosidase was critical for its LIMP-2-dependent targeting to the lysosome.
Conclusion: This sequence is essential for oligosaccharide-independent targeting of synthesized acid β-glucosidase to the lysosome.
Significance: Modification of this sequence has basic/therapeutic implications for Gaucher disease and its comorbidities (e.g. Parkinson disease).
Keywords: Enzyme Kinetics, Inborn Error of Metabolism, Intracellular Trafficking, Lysosomal Storage Disease, Protein Sorting, Enterovirus 71, Glucocerebrosidase, Lysosomal Targeting
Abstract
The acid β-glucosidase (glucocerbrosidase (GCase)) binding sequence to LIMP-2 (lysosomal integral membrane protein 2), the receptor for intracellular GCase trafficking to the lysosome, has been identified. Heterologous expression of deletion constructs, the available GCase crystal structures, and binding and co-localization of identified peptides or mutant GCases were used to identify and characterize a highly conserved 11-amino acid sequence, DSPIIVDITKD, within human GCase. The binding to LIMP-2 is not dependent upon a single amino acid, but the interactions of GCase with LIMP-2 are heavily influenced by Asp399 and the di-isoleucines, Ile402 and Ile403. A single alanine substitution at any of these decreases GCase binding to LIMP-2 and alters its pH-dependent binding as well as diminishing the trafficking of GCase to the lysosome and significantly increasing GCase secretion. Enterovirus 71 also binds to LIMP-2 (also known as SCARB2) on the external surface of the plasma membrane. However, the LIMP-2/SCARB2 binding sequences for enterovirus 71 and GCase are not similar, indicating that LIMP-2/SCARB2 may have multiple or overlapping binding sites with differing specificities. These findings have therapeutic implications for the production of GCase and the distribution of this enzyme that is delivered to various organs.
Introduction
Acid β-glucosidase (d-glucosyl-N-acyl-sphingosine glucohydrolase, glucocerebrosidase (GCase)2; EC 3.2.1.45) is a lysosomal exoglycosidase for β-glucose-terminated sphingolipids (1, 2). Insufficient activity of GCase is causal to the variants of Gaucher disease, a common lysosomal storage disease (3). The human and mouse genes, GBA1 and Gba1, respectively, are about 7.5 kb and contain 11 exons, which encode highly (∼86% identical/92% similar) conserved amino acid sequences (data not shown). Over 300 mutations of various types have been found in association with the variants of Gaucher disease, and some have clear prognostic value for affected patients (4). The resultant different single amino acid substitutions lead to GCases with defects in catalytic or stability properties or both (e.g. see Ref. 5). GCase is translated from mRNAs into a protein that contains two functional, in tandem, leader sequences that differ in length, either 39 or 19 amino acids (6). The preferred initiation codon is not known.
Mature human GCase is a glycoprotein of 497 amino acids that is produced by co-translational glycosylation of four of the five N-glycosylation sequences (Asn463 is not occupied), of which only Asn19 is essential for the formation of a catalytically active conformer (7). The enzyme also has properties of a membrane-associated, but not trans-membrane, protein. Unlike most soluble lysosomal proteins that are trafficked to the lysosome by the cation-independent mannose 6-phosphate receptor (CI-M6PR) system (8, 9), GCase contains little if any mannose 6-phosphate (10). Newly synthesized, unglycosylated GCase is retained in and not secreted out of mammalian cells (11). Also, GCase is not secreted from I-cell fibroblasts, which are deficient in the enzyme needed for mannose 6-phosphorylation, the targeting signal for most soluble lysosomal enzymes (8). Thus, the targeting of newly synthesized, intracellular, GCase to the lysosome is not oligosaccharide-dependent (11).
LIMP-2 (lysosomal integral membrane protein 2) has been identified as a trafficking receptor for GCase (10, 12, 13). LIMP-2 is a 478-amino acid, 85-kDa glycoprotein, which is also known as SCARB-2/CD36L2. This protein is present in the ER, Golgi, endosomal, lysosomal, and plasma membrane compartments of cells (13, 14). As the name indicates, LIMP-2 is an integral membrane protein with NH2- and COOH-terminal transmembrane domains and a luminal domain (ldLIMP-2) that binds GCase (10) and potentially other proteins. LIMP-2 binds to GCase in the ER (pH ∼6.8), and the enzyme remains bound to LIMP-2 during its transport through the Golgi, trans-Golgi network, and endosomal compartments. LIMP-2 delivers GCase to the lysosomes after an acidic pH-modulated dissociation of the receptor and ligand in the late endosomal compartment with liberation of GCase. This dissociation was thought to be mediated by LIMP-2 histidine 171 (15). During revision of this manuscript, a detailed crystallographic study of LIMP-2 proposed a model of a heterotrimeric GCase·LIMP-2·CI-M6PR complex. His150 was shown to act as a pH sensor for GCase binding rather than the previously proposed His171. Additionally, the glycosylated sugar on Asn325 of ldLIMP-2 purified from conditioned medium of HEK293S cells contained P-Man9GlcNAc2 (15, 44), suggesting a potential role for CI-M6PR in LIMP-2/GCase trafficking. However, GCase is not secreted from I-cells that have defective mannose 6-phosphorylation systems.
LIMP-2/SCARB-2 is also a scavenger receptor on the plasma membrane that binds peptide sequences of viruses, in particular enteroviruses (e.g. enterovirus 71), for internalization, lysosomal delivery, and degradation (16–19). The ligand amino acid sequence of enterovirus 71 (FY) for human LIMP-2 has been identified within VP1 between residues 152 and 178 (17) and has no homology to GCase sequences (data not shown). The corresponding receptor sequence on LIMP-2 is between amino acids 144 and 151 (15). Other LIMP-2/SCARB-2 protein ligands that bind at the plasma membrane include KCNQ1, KCNE2, and megalin (20).
Humans and mice with mutations in the LIMP-2-encoding genes (SCARB2 and Scarb2, respectively) develop characteristic neurologic and renal diseases but do not exhibit gross findings of Gaucher disease (i.e. GC storage or Gaucher cells) (20, 21). The human diseases associated with SCARB2 mutations are termed the action myoclonus-renal failure syndromes (AMRF) (21). LIMP-2-deficient cells in humans and mice exhibit excess secretion of GCase out of the cells and into plasma or culture medium but little GC accumulation in tissues (10, 21). LIMP-2 variants have also been implicated as potential modifiers in the development of Parkinson/Alzheimer diseases (20, 22, 23), as have GBA1 mutations (23–26). Disruption of appropriate trafficking of GCase to lysosomes may provide a mechanistic basis for the impact of GBA1/Gba1 mutations in the modification of α-synuclein metabolism and its role in Parkinson disease (24, 25, 27). The impacts of LIMP-2 trafficking of GCase on the expression of Gaucher disease and the impacts of GCase and LIMP-2 variants as modifiers of synucleinopathies highlight the importance of understanding the interactions of GCase and LIMP-2 and the localization of synthesized GCase to the lysosome. Here, the peptide sequence on mature human GCase that is a motif for binding to LIMP-2 has been identified, and mutations at specific amino acids are shown to alter the localization within and secretion of GCase from cells.
EXPERIMENTAL PROCEDURES
Materials
The following were from commercial sources: 4-methylumbelliferyl-β-d-glucopyranoside (Biosynth AG, Staad, Switzerland); sodium taurocholate (Calbiochem); rabbit anti-LIMP-2 polyclonal antibody, rabbit anti-LAMP1 antibody, and goat anti-actin antibody (Santa Cruz Biotechnology, Inc., Dallas, TX); goat or rabbit anti-calreticulin and -calnexin antibodies (Abcam, Cambridge, UK); NuPAGE 4–12% BisTris gel, NuPAGE MES SDS running buffer, DMEM, pBluescript vector, Dynabeads protein G immunoprecipitation kits, and BS3 chemical cross-linker (Invitrogen); BCA protein assay reagent (Pierce); pCMV-AC-GFP/YFP/cMyc expression vectors (Origene, Rockville, MD); PVDF membranes and ECL detection reagent (Amersham Biosciences); ABC Vectastain and Alkaline Phosphatase Kit II (black) (Vector Laboratory, Burlingame, CA); restriction enzymes (New England Biolabs Inc.); site-directed mutagenesis kits (Clontech or QuikChange (Stratagene)). Purified ldLIMP-2 was custom-made (Sino Biological Inc.) ImigluceraseTM was a gift from Genzyme Corp., a Sanofi company (Cambridge, MA). Rabbit anti-GCase polyclonal antibody was produced in this laboratory (28).
Methods
Deletion Constructs of GCases
The full-length human GCase cDNA in pBluescript was used as a backbone for deletion constructs. Four single cut restriction enzymes (ScaI, BstAPI, BalI, or BamHI) were used to digest the full-length cDNA to create the deletion constructs (GCase-225, GCase-150, GCase-75, and GCase-23) (Fig. 1). These were individually cloned into the pCMV6-AC-GFP vector for mammalian cell expression to provide GCase-XX in frame with GFP. All constructs were resequenced for verification.
FIGURE 1.
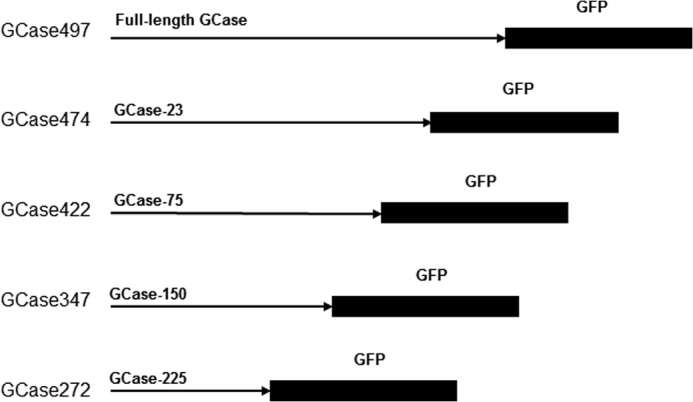
GCase-GFP transfection constructs for Gba1 null/null cells. On the left are the resultant GCases following the various deletions, indicated as GCaseZZZ to show the encoded mature amino acid contents of the expressed enzymes. The constructs are labeled GCase-XX or -XXX to designate the number of amino acids that were deleted from the -COOH end of the mature 497-amino acid sequence of the WT GCase. GFP was cloned in frame with the WT or GCase-XX or -XXX. These constructs were subsequently transfected into Gba1−/− fibroblasts.
GCase Expression Constructs for Point Mutations
Using site-directed mutagenesis kits and designed primers, all single amino acid mutation constructs were created using the full-length pCMV-GCase-GFP or YFP vectors. All constructs were sequenced for verification, and no additional mutations were found.
Immunoprecipitation of GCases with Mutations in the TKD Sequence
For immunoblots, Gba1 knock-out (Gba1−/− (i.e. null/null)) mouse fibroblasts were transfected with the GBA1 cDNA constructs (WT or mutants) and expressed for 5 days. The GCases from harvested cell lysates were purified using a GCase antibody affinity column (Dynabeads protein G cross-linked (BS3) with rabbit anti-human GCase antibody). The purified GCases were then mixed with ldLIMP-2 (1:1, mol/mol), incubated (30 min), and applied to another affinity column (Dynabeads protein G cross-linked (BS3) with rabbit anti-LIMP-2 antibody). Aliquots of each step (loading, wash, and eluent) were collected and analyzed on 12.5% SDS gels. Immunoblots were developed using anti-human GCase antibody and alkaline phosphatase-conjugated color development kits. Mock experiments with either non-LIMP-2 proteins added or no GCases added were used as controls. The immunoprecipitation data were quantified by densitometry measurements of the gels. The no-ldLIMP-2 bars represent the controls for comparison because GCase was processed through all steps without ldLIMP-2 to account for any losses of GCase; about 50% of the GCase was lost during processing.
Fluorescence Polarization Binding Studies
The DSPIIV and DDRQLLL peptides and their mutants (FITC-labeled or non-labeled) were synthesized (American Peptide Co., Vista, CA). Lyophilized peptides were solubilized in acetonitrile and then diluted at least 10,000-fold into the reactions for fluorescence polarization assessments. The reactions were done at least in duplicate at various peptide and LIMP-2 concentrations (SpectraMax M5, Molecular Devices). Fluorescence polarization data were collected and analyzed with SoftMax Pro version 5.0 software. Reaction mixtures contained 100 mm phosphate, pH 6.8, and 1 mm DTT.
Immunofluorescence Studies
Mouse Gba1−/− cultured fibroblasts were used for the host cells for all transfection experiments. The pCMV-AC-GFP mammalian cell expression vector was used for expression of various GCases that included the TKD (amino acids 407, 408, and 409, respectively) or the DSPIIV (amino acids 399–404) sequence. Directly labeled first antibodies were applied for immunofluorescence detection, including anti-human GCase, -calreticulin, -calnexin, -LIMP-2, and -Lamp1. LysoTracker and ER-Tracker (Invitrogen) were also used as indicated. Analyses were done with a Zeiss AxioVert 200 M microscope with ApoTome. Co-localization analyses to obtain Pearson indices (PIs) were conducted using a module in the Axiovision version 4.8 software.
Enzyme Assays
Cell pellets were homogenized in 0.25% sodium taurocholate and 0.25% Triton X-100. GCase activities were determined fluorometrically with 4 mm 4-methylumbelliferyl-β-d-glucopyranoside in 0.2 m/0.1 m citrate/phosphate, pH 5.6 (5, 29).
Although presented as “data not shown,” protein sequence homologies were derived using MUSCLE (30, 31). For GCase, this included 60 species representing mammals, rodents, reptiles, fish, worms, flies, and mosquitos.
RESULTS
Initial studies to localize the targeting peptide were conducted with GCase-GFP fusion constructs that had COOH-terminal deletions of GCase (Fig. 1). The constructs are designated as GCase-XX or -XXX where the Xs represent the number of GCase amino acids deleted from the COOH terminus. These constructs were transfected into and expressed in Gba1−/− mouse fibroblasts. Immunofluorescence analyses using anti-human GCase IgG (green) and LysoTracker (red) were used for co-localization of GCase to the lysosome (red) or GCase (green) and calreticulin or calnexin (red) for co-localization to the ER or Golgi, respectively (Fig. 2). The Pearson indices for these immunofluorescence studies indicated that GCase-23 and GCase-75 had similar co-localization to the lysosome as the full-length WT GCase. In comparison, GCase-150 and GCase-225 were more localized to the ER and Golgi and much less to the lysosome and had Pearson indices similar to that of the GFP control without GCase sequences (N-GFP). These studies showed that the critical GCase amino acids for targeting to the lysosome were between amino acids 347 and 422 (i.e. in the fragment between GCase-75 and GCase-150). Because LIMP-2 is the receptor for targeting of GCase to the lysosome, our working hypothesis was that the peptide sequence for binding of GCase to LIMP-2 was located within amino acids 347–422.
FIGURE 2.
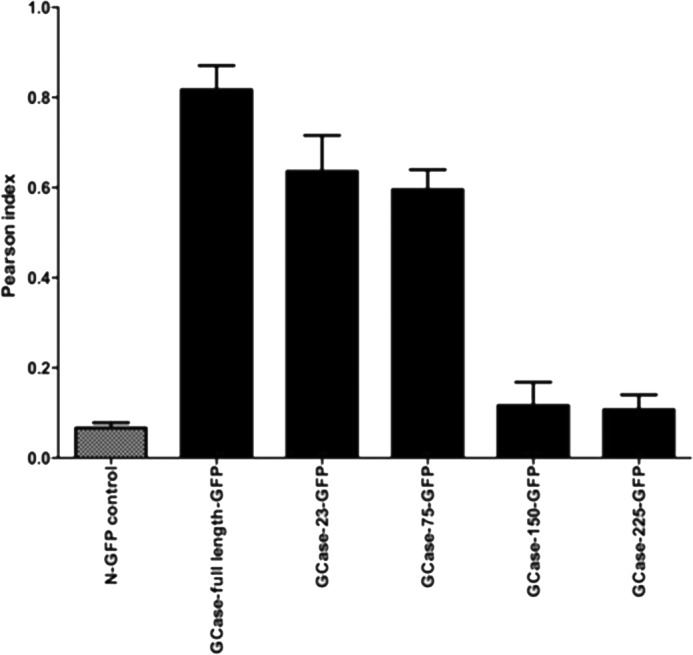
Co-localization of GCase-XX- or GCase-XXX-GFP with LysoTracker. Pearson indices were obtained (see “Methods”) for co-localization of the WT or truncated GCases using LysoTracker to label the lysosomes. The construct containing only GFP (N-GFP) showed no co-localization. Compared with the WT sequence, GCase-23 and -75 showed only small decreases in lysosomal localization. GCase-150 and -225 showed decreases in co-localization with LysoTracker to nearly background (N-GFP) levels. GCase-150 and -225 showed much more extensive retention in the ER and Golgi compared with WT, GCase-23, or GCase-75 (data not shown). Error bars, S.E.
The location of this GCase amino acid sequence is shown in a linear form in Fig. 3 (bracketed and in orange highlight). This amino acid sequence is located in Domain I of the GCase crystal structure. Analysis of this region shows that the amino acids between Asn399 and Asp409 are surface-exposed and form a loop structure that would be accessible for ligand binding to LIMP-2 (Fig. 4). However, amino acids Phe347–Asp398 and Phe411–His422 are more internal to the structure and less likely to be able to interact with LIMP-2. The likely exposed residues include the sequence DSPIIVDITKD (Fig. 4, yellow). Searches of the databases for lysosomal glucocerebrosidases from multiple species show that this sequence is highly conserved and nearly invariant in mammals and other vertebrates (data not shown); the consensus sequence is DSPIIVDIAKD. Based on these analyses, efforts were focused on the amino acid sequence between Asp399 and Asp409 (Fig. 4). Although GCase is a membrane-associated and not a transmembrane lysosomal protein, the mature GCase sequence does contain a dileucine structure that has been previously identified as important in the indirect targeting of transmembrane proteins to the lysosome (32–34). This sequence spans amino acids Asp282–Leu288 (Fig. 3). DDQRLLL (Fig. 4) is located in Domain III of GCase and was also analyzed for interactions/binding to LIMP-2 as a potential ligand sequence.
FIGURE 3.

Mature WT GCase amino acid sequence highlighting the regions targeted for mutagenesis, expression, and binding analyses. The amino acid sequence of GCase is marked with various dark red arrows indicating the number of amino acids deleted from the COOH-terminal end of the enzyme for the GFP reporter. Shown in orange is a typical dileucine sequence for indirect targeting of membrane-bound lysosomal proteins. Underlined in red are the amino acids for targeted mutagenesis and binding studies to ldLIMP-2. These sequences have very high conservation across species (data not shown).
FIGURE 4.
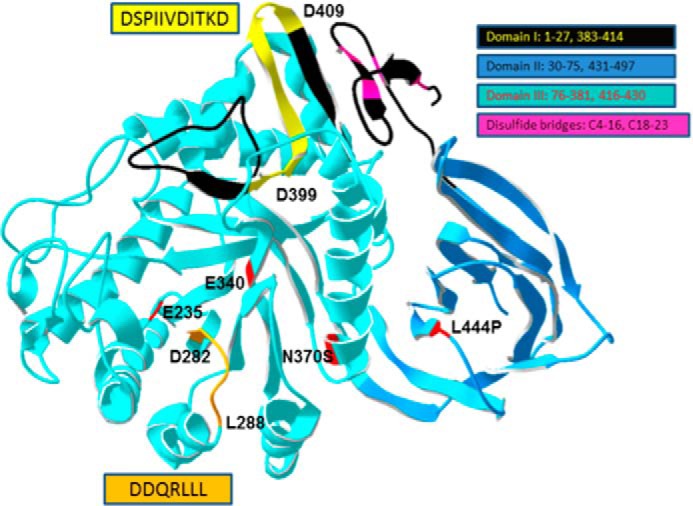
Structure of WT GCase highlighting the potential ldLIMP2 binding sequence localization. The two amino acid sequences from Fig. 3 were mapped to the crystal structure of WT human GCase. The orientation of the sequences is indicated by the amino acid numbers in the highlighted (yellow or orange) regions. The domains of GCase are shown in various indicated colors as are the disulfides. The potential ldLIMP-2 binding (yellow, Asp399–Asp409) sequence forms a surface-accessible loop in domain I (black, with the motif in yellow). The DSPIIVDITKD sequence and the DDQRLLL (orange, Asp282–Leu288 in Domain III) sequence are highly conserved in GCases from insects to humans (data not shown). The acid-base (Glu235) and nucleophile (Glu340) in catalysis and the N370S and L444P common mutations causal to Gaucher disease are shown in red (5).
LIMP-2 is a transmembrane protein whose ldLIMP-2 binds to and traffics GCase to the lysosome (10). A series of alanine-swapped mutant GCase proteins and peptides were made for cellular localization, immunoprecipitation, and secretion analyses. For the expression studies, Gba1−/− fibroblasts were used, and the GCase variants were localized to cellular compartments. A 3-amino acid (TKD) and a 6- (DSPIIV) or 11-amino acid sequence of interest (Fig. 3) were targeted for alanine mutagenesis. The various mutant GCases for the TKD sequence showed immunofluorescence lysosomal localization with wild type (TKD (PI ∼0.988)), AKA (PI < 0.2), and TAD (PI ∼0.5) to varying degrees. AAA (PI >0.1) lacked this property (i.e. the AAA mutant showed no lysosomal localization but abundant co-localization to the ER by using anti-calreticulin) (Fig. 5). Thus, AAA was expressed in the cell but was not trafficked to the lysosome. The WT (TKD) and TAD mutant had similar ER (Fig. 5). The AKA mutant showed less lysosomal localization compared with WT or the TAD mutant (Fig. 5).
FIGURE 5.
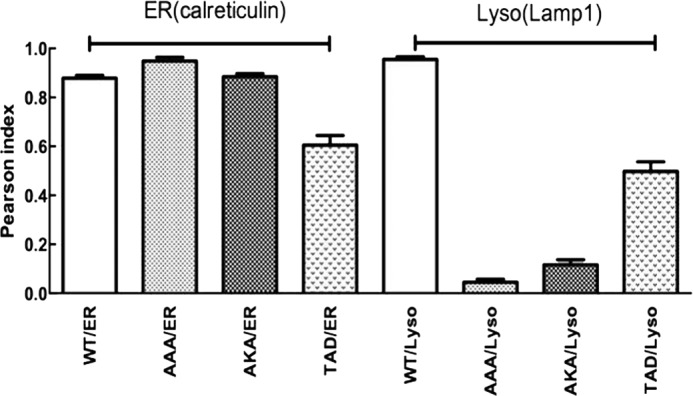
Co-localization (Pearson) indices of GCases with specific alanine substitutions in the WT TKD sequence. GCase/mutant constructs were transfected into Gba1−/− fibroblasts, and co-localization was evaluated with directly labeled anti-mouse calreticulin (rhodamine; red) antibody for ER/Golgi, directly labeled Lamp1 (rhodamine; red) for lysosome (Lyso), and directly labeled anti-human GCase (FITC; green) for GCase. The alanine (A) substitutions are as altered from the WT TKD sequence. For the AAA and AKA GCases, the majority of the intracellular enzymes co-localized to the ER/Golgi (ER), whereas the TAD GCase showed about 50% of the total intracellular enzyme localized to the lysosome (Lyso). Co-localization indices (Pearson) were calculated (n = 5) and presented as the mean ± S.E. (error bars)
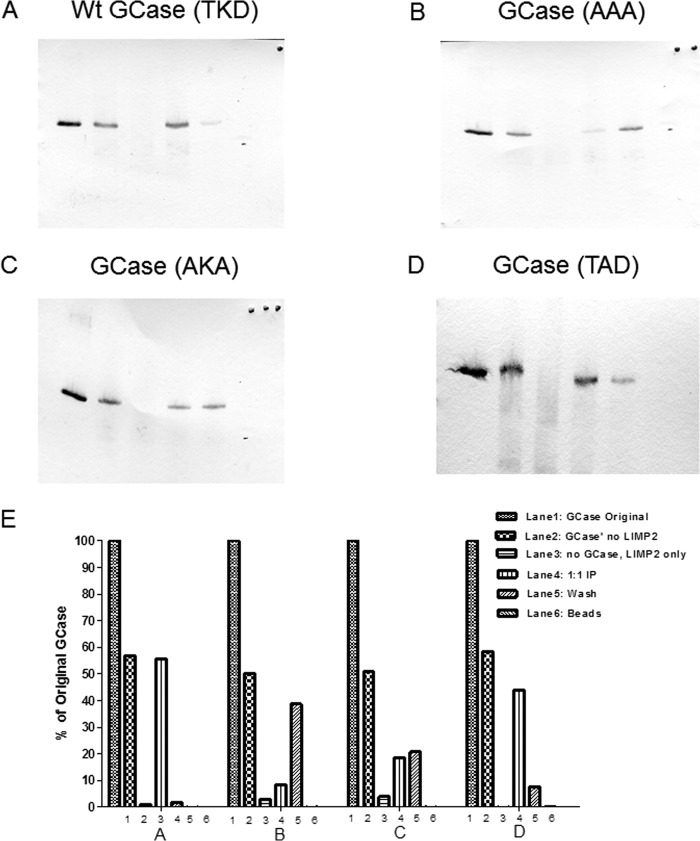
To demonstrate a correlation between the co-localization results and LIMP-2 binding, the WT and TAD variant GCases were expressed and purified by anti-GCase affinity chromatography and used to conduct in vitro immunoprecipitation studies with ldLIMP-2 (Fig. 6). These results show that WT and the TAD GCases (Fig. 6, A and D) were bound similarly by ldLIMP-2, whereas the AKA mutant (Fig. 6C) was much less bound, and the AAA GCase did not bind to ldLIMP-2 (Fig. 6B). Quantitative densitometry results are shown in Fig. 6E. These results for ldLIMP-2 binding correlated well with the co-localization analyses, which followed a similar pattern for the WT, intermediate, or absent lysosomal localization.
FIGURE 6.
Immunoprecipitation of the alanine-substituted GCases in the TKD sequence with ldLIMP-2. In A–D, lanes 1–6 correspond to the band densities indicated by the bars in E. The panels show that the WT sequence (A) had nearly complete binding with ldLIMP-2 (i.e. retention on the beads to which ldLIMP-2 was bound (lane 4) and no GCase in the wash (lane 5)). In comparison, the triple alanine mutant (B) shows essentially no binding to ldLIMP-2 (i.e. all of the GCase is in the wash (lane 5)). The AKA GCase mutant (C) showed about equal amounts of GCase in lanes 4 and 5, or about 50% binding to ldLIMP-2. The AKD GCase mutant (D) had a binding pattern that was similar to the WT sequence but with more GCase in the wash (lane 5) (i.e. somewhat less binding). The quantitative results are shown in E, in which 1, 2, 3, and 4 correspond to WT, AAA, AKA, and AKD, respectively. The results are typical of multiple experiments.
Several additional peptides were used to explore the DSPIIVDITKD region by alanine scanning mutagenesis, and a peptide from another region of GCase was used as a random control (Table 1). These peptides were synthesized with and without a fluorescent probe (FITC) covalently linked to the COOH-terminal end to assess direct binding to ldLIMP-2 by changes in fluorescence polarization (δmP).
TABLE 1.
Synthetic peptide characteristics
Peptides were custom-synthesized by American Peptide Co. The boldface letters indicate the differences from the WT sequences of DSPIIV or DDQRLLL. The final peptide in each group is an unrelated peptide in GCase.
| Sequence | GCase amino acids | Mr (FITC)a | Mr (non-labeled) | Puritya |
|---|---|---|---|---|
| % | ||||
| DSPIIV | 399–404 | 1032.2 | 642.8 | 95.3/98.0 |
| ASPIIV | 399–404 | 998.1 | 578.8 | 95.0/98.1 |
| DSPAIV | 399–404 | 990.1 | 600.7 | 96.5/98.2 |
| DSPIAV | 399–404 | 990.1 | 600.7 | 95.3/97.7 |
| DSPAAV | 399–404 | 948.1 | 514.6 | 95.4/95.1 |
| SKDVPL | 465–470 | 1047.2 | 657.8 | 96.6/98.0 |
| DDQRLLL | 282–288 | 1261.4 | 872 | 95.3/97.1 |
| ADQRLLL | 282–288 | 1226.4 | 828 | 96.1/97.6 |
| DAQRLLL | 282–288 | 1226.4 | 828 | 95.7/97.1 |
| DDQRAAA | 282–288 | 1144.2 | 745.8 | 95.9/98.0 |
| DFIARDL | 258–264 | 1247.4 | 849 | 95.1/97.5 |
a Quality control: HPLC grade, purity: 95.1–98.8%, MS analysis/desalted/lyophilized into powder.
The interactions/binding of the GCase peptides, DSPIIV variants, to ldLIMP-2 were analyzed by δmP with increasing WT or mutant peptide concentration in the presence of a fixed amount of ldLIMP-2 (Fig. 7A). The substitution of an alanine for either of the isoleucines decreased binding to ldLIMP-2 by about 50%, whereas alanine substitution for the NH2-terminal aspartate nearly eliminated the binding to ldLIMP-2. Substitution of alanines for Asp399, Ile402, and Ile403 obliterated the binding.
FIGURE 7.
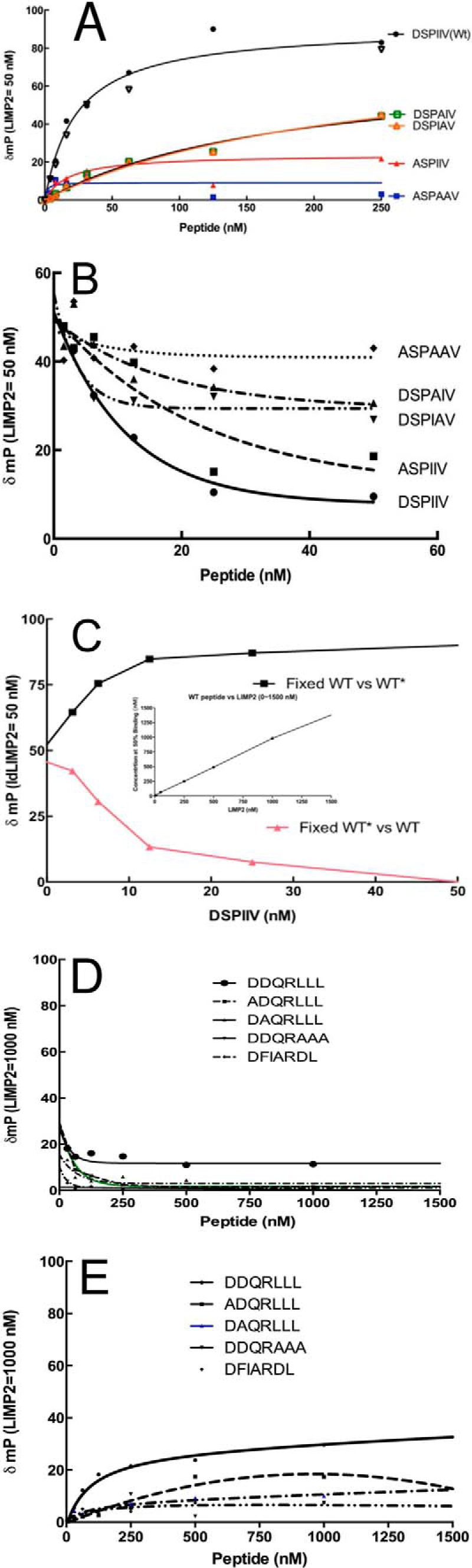
Binding and competition of fluorescence-labeled or unlabeled DSPIIV GCase peptides to ldLIMP-2. The change (δmP) in fluorescence polarization is plotted on the ordinate, and the increasing concentrations of the various peptides are shown on the abscissa. A, the concentration of ldLIMP-2 (50 nm) was fixed. With the WT (DSPIIV) peptide (● and ▿, duplicate experiments), saturation kinetics with about half-maximal binding to ldLIMP-2 was observed at 50 nm. The DSPAIV and DSPIAV mutants did not show saturation up to 250 nm, indicating their poor interaction with ldLIMP-2. The ASPAAP peptide showed background changes. The ASPIIV peptide showed δmP values slightly above background, indicating little binding to ldLIMP-2. B, to ensure that the label did not interfere/promote binding, similar studies were conducted using fluorescently labeled peptides as binders and their respective unlabeled peptides as competitors. Each of the unlabeled peptides “competed” with the labeled peptides in the expected ratios. C, labeled (WT*) and unlabeled (WT) peptides were used in complementary competition studies. Either the labeled or unlabeled WT peptides equally competed for binding to ldLIMP-2, showing that both were equally effective in binding and that the label did not change the properties of the peptide-ldLIMP-2 interaction. The inset indicates 1:1 stoichiometry and tight binding properties of DSPIIV and ldLIMP-2. D, the δmP of the WT (DDQRLLL) or variously alanine-substituted labeled peptides were plotted against the corresponding unlabeled peptide competitors. Note that the fixed ldLIMP-2 concentration was 1000 nm, or 20 times that used in Fig. 7, A and B. For all peptides, the δmP values were near or at background levels. E, binding δmP for the various peptides as in D. The signals were near background levels for all peptides using concentrations 5–10 times that for DSPIIV and with a 20 times greater LIMP-2 fixed concentration.
To ensure that the fluorescent label was not interfering with or promoting binding to ldLIMP-2, a similar experiment was conducted using unlabeled peptide to compete with the corresponding labeled peptide. The data show direct competition of the corresponding labeled and unlabeled peptides (Fig. 7B). Similarly, a direct comparison was conducted to assess the competition of either the labeled or unlabeled WT peptide for binding to ldLIMP-2 (Fig. 7C). The data show symmetry of the changes in δmP that are nearly mirror images with either the labeled or unlabeled WT peptide in competition for ldLIMP-2. These data show that the fluorescent label did not alter the binding properties to ldLIMP-2. Using the WT labeled peptide, the peptide binding to ldLIMP-2 was shown to have 1:1 stoichiometry and was tight (i.e. the molar concentrations of the peptide and ldLIMP-2 needed for 50% binding were in a 1:1 ratio) (Fig. 7C, inset).
In other binding experiments, the DDQRLLL labeled and unlabeled peptides were synthesized and tested. These peptides had little or no binding to ldLIMP-2 above background. Thus, the DDQRLLL had essentially no interaction with ldLIMP-2 (Fig. 7, D and E).
Immunoprecipitation and Competition Studies
To further evaluate the interaction of ldLIMP-2, WT GCase, and the DSPIIV peptide, competitive immunoprecipitation experiments were conducted using increasing concentrations of the WT peptide in competition with GCase for ldLIMP-2 binding (Fig. 8A). Increasing WT peptide, DSPIIV, concentrations from 0 to 5× molar excess over GCase showed that this peptide competed off GCase binding to ldLIMP-2 when GCase was first incubated with ldLIMP-2 and then the WT peptide was added. A similar experiment was conducted in which the WT peptide was preincubated with ldLIMP-2 prior to adding purified GCase (Fig. 8B); almost all GCase binding to ldLIMP 2 was prevented at 1–5× molar peptide ratios. The combined results (Fig. 8, A and B) show that the WT peptide can compete or prevent GCase binding to ldLIMP-2.
FIGURE 8.
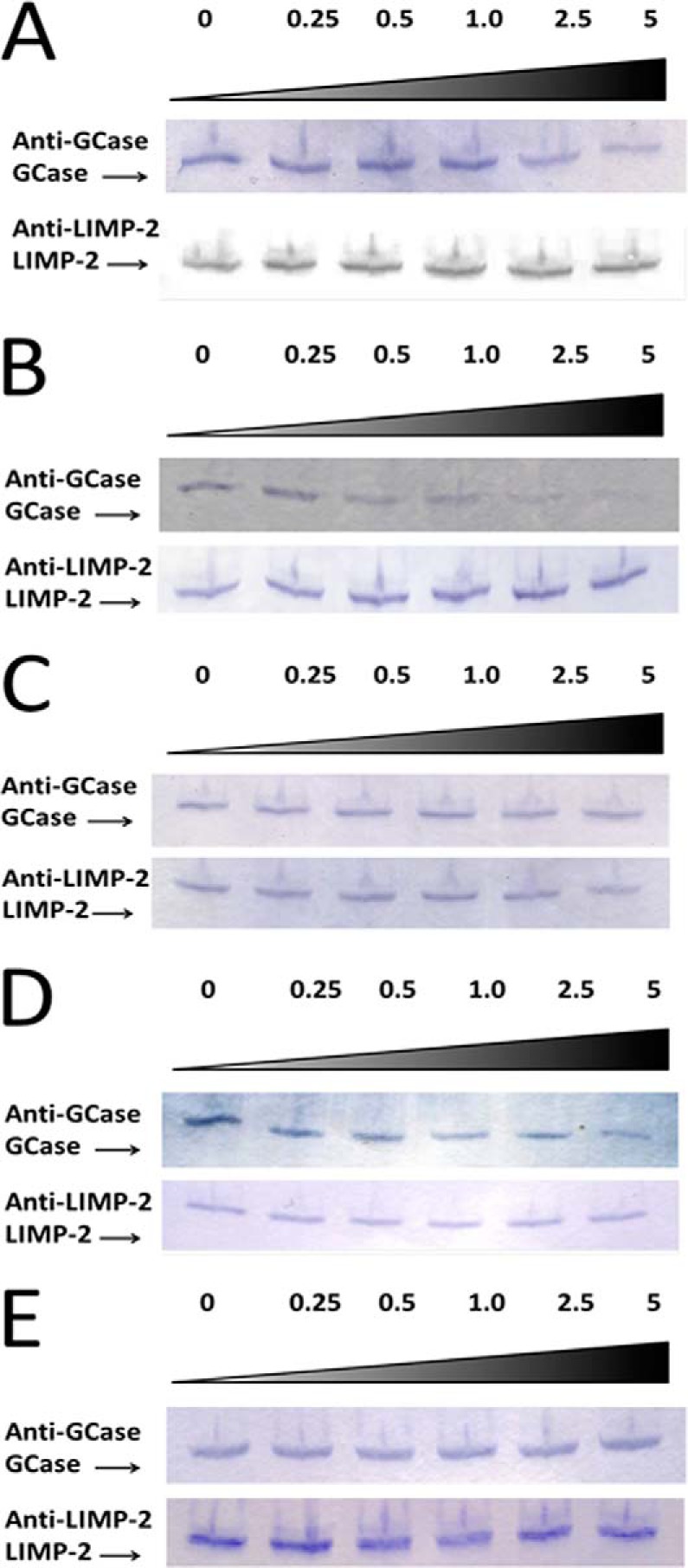
Immunoprecipitation of GCase in the presence of ldLIMP-2 and increasing molar ratios of the WT (DSPIIV) peptide. A, purified WT GCase was preincubated with purified ldLIMP-2 in a molar ratio of 1:1. Then unlabeled DSPIIV was added in varying molar excesses (0–5×; top) over GCase, incubated, and then immunoprecipitated with protein G-coupled anti-LIMP-2 antibody beads. The beads were then eluted, and the eluants were analyzed for GCase and ldLIMP-2 on Western blots using the specified antibodies. The results show decreasing amounts of bound GCase with increasing peptide molar ratios (i.e. peptide DSPIIV competed bound GCase off of ldLIMP-2. In the bottom panel, ldLIMP-2 recovery was the same at all peptide ratios. B, purified ldLIMP-2 was preincubated with unlabeled WT peptide at various molar ratios. Then, purified WT GCase was added in a 1:1 molar ratio with ldLIMP-2, incubated, and then immunoprecipitated and processed as in A. The results show that the peptide prevents the binding of ldLIMP-2 and WT GCase. C, purified WT GCase was preincubated with purified ldLIMP-2 in a molar ratio of 1:1. Then peptide DDQRLLL, a potential candidate for the GCase ligand for ldLIMP-2, was added in varying molar excesses over WT GCase, incubated, and then immunoprecipitated and processed as in A. The results show that peptide DDQRLLL did not compete GCase off of ldLIMP-2, thereby showing that this peptide did not have specificity for the GCase binding site on ldLIMP-2. DDQRLLL is a dileucine peptide outside of the localized targeting region of DSPIIVDITKD (see Fig. 3). D and E show the immunoprecipitation of GCase in the presence of ldLIMP-2 and increasing molar ratios of the WT (DDQRLLL) peptide. In D and E, ldLIMP-2 and DDQRLLL were incubated before (D) or together with (E) GCase and analyzed as in A–C. Under either condition, the peptide did not complete with GCase for ldLIMP-2 binding.
The DDQRLLL peptide from GCase was tested as a control. Although this sequence was contained in the GCase fragment (GCase-225) that did not localize to the lysosome, it does have a known motif for lysosomal targeting of transmembrane lysosomal proteins, which GCase is not. In addition, this sequence is highly conserved throughout phylogeny (data not shown). Up to a 5× molar excess of this peptide did not compete with GCase for ldLIMP-2 binding when the peptide was added after GCase and ldLIMP-2 were preincubated (Fig. 8C). In addition, when the peptide was preincubated with LIMP-2 before GCase was added or when LIMP-2, the peptide, and GCase were incubated together, there was no effect on GCase·LIMP-2 binding (Fig. 8, D and E).
pH Dependence of ldLIMP-2·GCase Binding
LIMP-2 binding to GCase is dependent upon pH in that greater binding is obtained at more neutral pH, and dissociation obtains at pH 5.6 (10, 15, 44). The WT and various mutants of the DSPIIV peptide have differential pH dependence. Substitution of either isoleucine with alanine decreases the binding at pH 6.8 by 40–50%. The ASPIIV peptide binding was decreased by about 30–40%, whereas the ASPAAV peptide was decreased by ∼90%. There is little effect on the binding at pH 5.8 of any of the substituted peptides compared with the WT peptide (Fig. 9).
FIGURE 9.
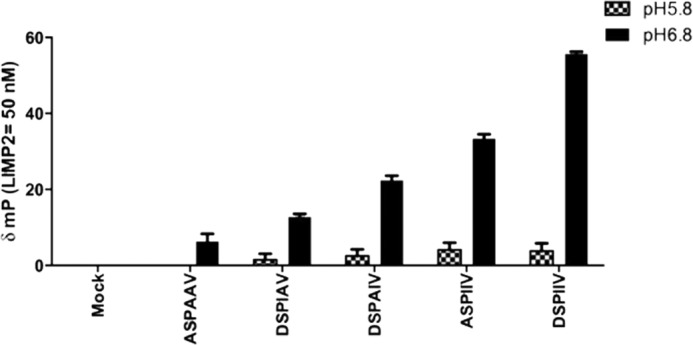
Effect of pH on binding WT and alanine-substituted DSPIIV peptides to ldLIMP-2. The binding of the different peptides to ldLIMP-2 showed little effect of alanine substitutions at pH 5.8. In comparison, incremental decreases were evident in ldLIMP-2 binding at pH 6.8, the approximate pH of the ER/cis-Golgi, as follows: the triple mutant (ASPAAV) having the lowest binding (<10% of WT), the DSPAIV (I402A) and DSPIAV (I403A) mutants being intermediate (∼40–50% of WT), and ASPIIV (D399A) having the least change (∼50–60% of WT), relative to WT. Error bars, S.E.
Effects of Mutations of DSPIIVDITKD on Cellular Co-localization and/or Secretion of GCase
The effects on cellular localization of GCase from Gba1−/− cells transfected with WT or various GCases with mutations in the target region are shown in Fig. 10A. Representative examples show that the WT or D399E GCases are directed to the lysosomes, indicating that the charge on amino acid 399 is important for this targeting. In comparison, neither the DSPAIV (I402A) nor DSPIAV (I403A) significantly localizes to the lysosome. Essentially identical results were obtained with the ASPIIV (D399A) or I402A/I403A (DSPAAV, double mutant) GCases. Confirmatory quantification by Pearson indices of co-localization to the ER/Golgi or lysosome of all of these expressed WT and mutant GCases is shown in Fig. 10B.
FIGURE 10.
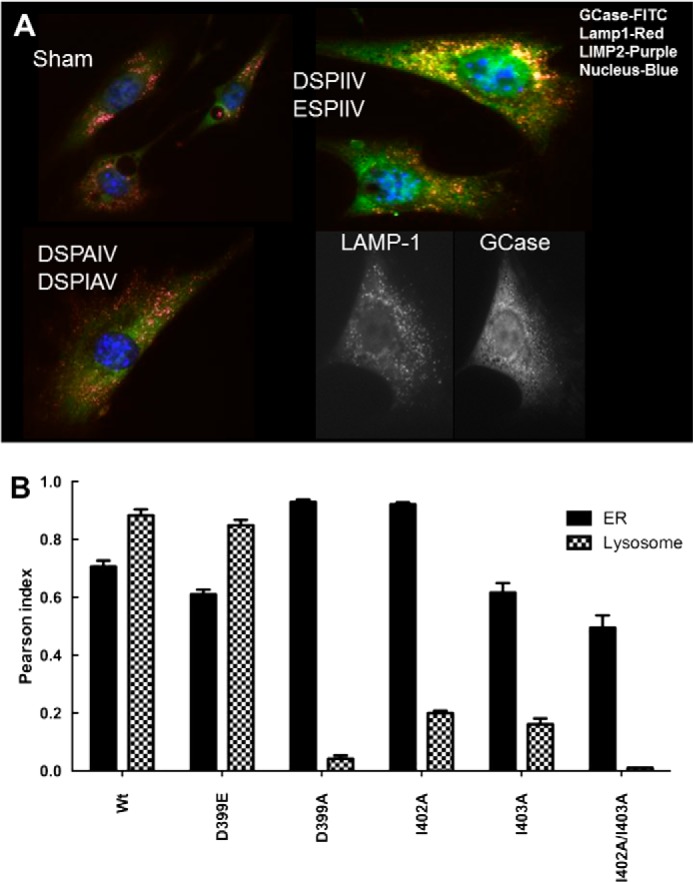
Intracellular localization of GCase DSPIIV variants in Gba1−/− cells. A, typical examples of immunofluorescence localization of DSPIIV-substituted GCase variants following transient transfections. The DSPIIV (WT) and ESPIIV show co-localization of GCase (FITC) with either Lamp1 (red) or LIMP-2 (purple) (top right), indicating that the retention of charge by Glu399 does not impact localization. The cells are typical for either Asp399 (DSPIIV) or Glu399 (ESPIIV). The DSPAIV (I402A) or DSPIAV (I403A) mutant shows no co-localization with ldLIMP-2 or Lamp1 (bottom left) (i.e. no binding to ldLIMP-2 in the cell and no localization to the lysosome (Lamp1/ldLIMP2)). The black and white images in the bottom right indicate the relative localization of LAMP-1 (lysosomes) and WT GCase (ER/Golgi and lysosomes) in Gba1−/− fibroblasts for comparison. Sham control is shown in the top left. B, Pearson indices for co-localization of the various mutants in the ER/Golgi (black) or lysosome (hatched). The substitution of Glu for Asp (WT) at 399 (ESPIIV) did not alter the co-localization compared with WT. In comparison, ASPIIV (D399A) and the double mutant, DSPAAV (I402A/I403A) showed little co-localization to the lysosome but significant retention in the ER/Golgi. The single mutants, DSPAIV (I402A) and DSPIAV (I403A), showed partial co-localization to the lysosome but greater retention in the ER/Golgi. Error bars, S.E.
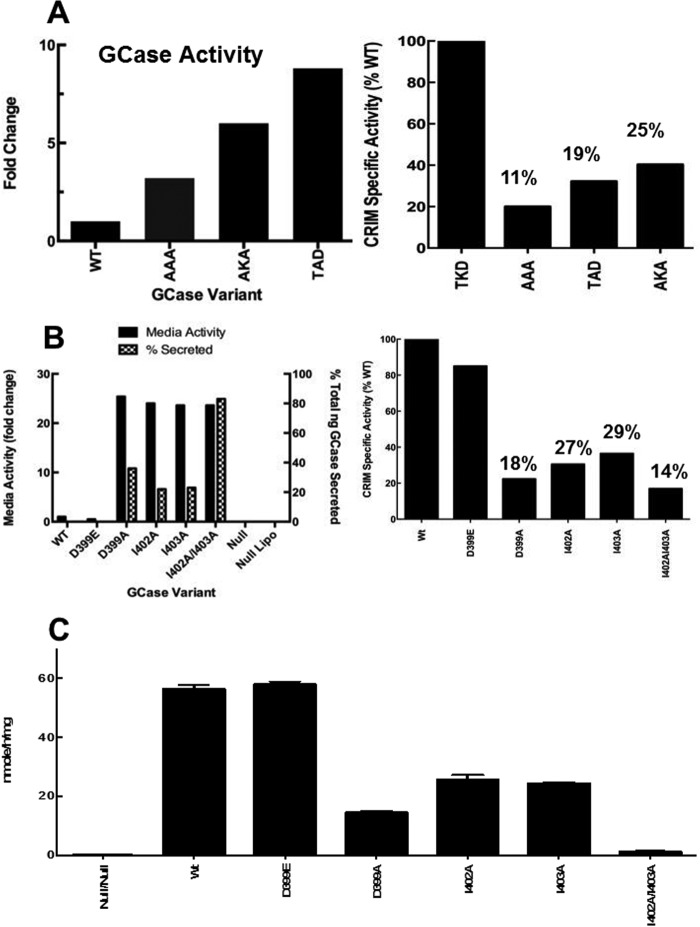
Corresponding effects on the secretion from the cells of GCases containing various mutations in DSPIIVDITDK were found by assessing the GCases in the medium and cell lysate (Fig. 11, A–C). Compared with the WT sequence (TDK), the amount of GCase activity in medium progressively increased from the AAA to AKA to TAD mutants to an ∼8-fold increase (Fig. 11A, left). The lower GCase activity in medium of the AAA mutant seemed at odds with the lack of binding of this mutant to ldLIMP-2. Immunoblots of the various mutations in the TKD sequence provided assessments of the GCase activity/amount of CRIM (i.e. CRIM-specific activity, an estimate of the catalytic rate constant relative to WT enzyme). Using this assessment, the ratios of CRIM-specific activities relative to WT (assigned 100%) for AAA, TAD, and AKA were 11, 19, and 25%, respectively (Fig. 11A, right) (i.e. 9, 5.2, and 4 times more GCase proteins were required from the respective mutants to achieve the same enzyme activity as the WT). These results imply that the levels of GCase protein secreted from the Gba1−/− fibroblasts were 4–9-fold greater than suggested by the enzyme activity measures and correspond well with the binding data (Fig. 6). Similarly for the DSPIIV sequence, all of the alanine mutant GCases showed large increases (∼25-fold) in medium activity compared with WT (i.e. increased secretion of active GCase) (Fig. 11B, left). When assessed for CRIM-specific activity, the alanine mutants had 14–29% of WT activity, implying that 7.1 (for the double mutant) to 3.4 (I403A) more GCase enzyme protein was secreted than suggested by the raw activity measures. The Gba1−/− cells transfected with the charge-conserved substituted GCase, D399E, showed no medium activity above background (i.e. zero) and a minor change in CRIM-specific activity. Using the CRIM-specific activity assessments and the total activities in lysates and medium, the percentage of secreted GCase (ng) showed that ∼83% of the I402A/I403A mutant was secreted from transfected cells (Table 2). For the other mutants, this ranged from 22 to 36%. This compares to ∼0% for the WT or D399E GCases. Immunoaffinity anti-GCase column purification of the various GCases from medium confirmed the excesses of these mutant GCases and the inability to detect WT or D399E GCases in medium (data not shown). Complementary data for the retention of GCases in cell lysates showed ∼15–20% of WT or ESPIIV (D399E) activities with the single alanine mutants (ASPIIV (D399A), DSPAIV (I402A), and DSPIAV (I403A)) and <5% with the double DSPAAV (I402A/I403A) (Fig. 11C). Thus, all of the alanine-mutated GCases that were shown to diminish ldLIMP-2 binding and poor lysosomal localization showed large increases in GCase in the medium (i.e. excess secretion).
FIGURE 11.
Secretion and retention of GCase with alanine variants in the TDK (A) or DSPIIV (B and C) sequences. A, left, activities in the medium of Gba1−/− fibroblasts following transient transfections of each of the specified WT or alanine-substituted GCases in the TDK region. Increases of 4–8-fold in GCase activities were found in the medium, with AKD showing the greatest increases and AAA the least. Right, CRIM-specific activity of WT and these same mutant GCases, demonstrating that the mutations decreased this activity relative to WT as indicated above the bars. B, left, activities in the medium following transient transfections of various DSPIIV alanine-substituted GCases and the percentage of GCase secreted. This was calculated as (ng of GCase (in medium)/(ng of GCase (in medium) + ng of GCase (in lysates))) × 100. The ESPIIV GCase did not alter secretion (i.e. little enzyme in the medium). All of the alanine substitutions in the DSPIIV sequence resulted in an ∼20–25-fold increase in GCase activity in the medium (solid bars). The transfections alone (Null Lipo) had no effect. In comparison, the amount of secreted GCase protein varied between 22 and 83% (hatched bars). Right, CRIM-specific activity of WT and these same mutant GCases, demonstrating that the mutations decreased this activity relative to WT as indicated above the bars. C, cellular lysate GCase activities from the various alanine-substituted GCases in the DSPIIV sequence as indicated. The WT and ESPIIV had equal intracellular activities of GCase, whereas the singly alanine-substituted GCases had ∼30% of WT levels. The doubly alanine-substituted GCase had little intracellular activity (i.e. nearly all of the GCase was secreted into the medium). All results are from n = 3 determinations. Error bars, S.E.
TABLE 2.
Proportion of GCase protein in cells and media
Human WT and variant GCases were transfected into Gba1−/− cells with resultant transfection efficiencies of 20.5 ± 3%. The calculations were based on the specific activities of GCase relative to a standard curve of cross-reacting immunological material (CRIM) densities on immunoblots of purified human GCase, which has a specific activity of 1.1 nmol/h/ng of GCase. Cellular and medium GCase represent total ng from each source. Results are expressed as the mean ± S.D., n = 3.
| GCase variant | Cellular GCase | Medium GCase | Total GCase | Percentage secreted |
|---|---|---|---|---|
| ng | ng | ng | % | |
| Human WT | 23 ± 4 | 0 | 23 ± 4 | 0 |
| D399A | 32 ± 4 | 18 ± 2 | 50 ± 4 | 36 |
| I402A | 38 ± 6 | 11 ± 4 | 49 ± 5 | 22 |
| I403A | 34 ± 2 | 10 ± 2 | 44 ± 3 | 23 |
| I402A/I403A | 4 ± 1 | 20 ± 3 | 24 ± 4 | 83 |
| Mouse WT | 64 ± 7 | 0 | 64 ± 7 | 0 |
DISCUSSION
The GCase binding sequence to ldLIMP-2 has been identified using heterologous expression of deletion constructs, the available GCase crystal structures, and binding and co-localization of identified peptides or mutant GCases. These studies show a complex interaction of ldLIMP-2 with the highly conserved 11-amino acid sequence, DSPIIVDITKD, within human GCase. The binding is not dependent upon a single amino acid, but its interactions with ldLIMP-2 are heavily influenced by Asp399 and the di-isoleucines, Ile402 and Ile403. Single alanine substitutions at any of these decreases binding to ldLIMP-2 and alters their pH-dependent ldLIMP-2 binding as well as diminishing the trafficking of GCase to the lysosome while significantly increasing GCase secretion. Replacement of the di-isoleucines and Asp399 with alanines obliterated binding to ldLIMP-2 and lysosomal localization. However, the retention of the charge at Asp399 (i.e. Glu399) did not alter the cellular localization or secretion of GCase. In comparison, alanine replacements in the TKD sequence showed less profound effects on ldLIMP-2 binding as single substitutions (i.e. TAD) compared with double (AKA) or triple (AAA) substituted GCases. Finally, the lack of trafficking to the lysosome of the deletion construct GCase-150, which contains the DDQRLLL sequence, and the lack of binding of DDQRLLL peptides to ldLIMP-2 eliminate this known indirect lysosomal trafficking signal from participation in GCase targeting.
The GCase 11-amino acid ligand for LIMP-2 is highly conserved throughout phylogeny with near identity at 10 of the amino acids. Thr407 of the human GCase has an alanine substituted in several mammalian GCases so that the consensus sequence across 60 species, including some insects and worms, is DSPIIVDIAKD. Also, only in mosquitoes does the Glu399 equivalent occur. The IIV motif is identical across most of the species, but in the few variants, the branched chain amino acid valine is substituted for either of the isoleucines, usually the Ile402 equivalent.
Enterovirus 71 causes foot and mouth disease by gaining access to cells after binding to LIMP-2/SCARB2 (17–19). The binding sequence on enterovirus 71 for LIMP-2/SCARB2 has been localized to VP1 amino acids 152–178 (17), but BLAST searches with the entire virus revealed no significant similarity to GCase, specifically in the 11-amino acid region (data not shown). Several other proteins (e.g. KCNQ1, KCNE2, and megalin) interact with LIMP-2/SCARB2 at the plasma membrane (20), but these do not share any homology with GCase. Other intracellular proteins that bind to LIMP-2 have not been identified. Because the LIMP-2/SCARB2 binding sequences for enterovirus 71 and GCase are not similar, LIMP-2/SCARB2 may have a structure that contains multiple binding sites with differing specificities. Such structures are present in some receptors for carbohydrates (i.e. the macrophage mannose receptor that contains multiple carbohydrate binding domains with differing affinities/specificities for mannosyloligosaccharides) (35, 36). Also, the cation-independent mannose 6-phosphate receptor/insulin-like growth factor receptor 2 has differing specificities for mannose 6-phosphate-containing glycoproteins, ILGF-2, and prorenin (8, 37). Indeed, the residues on LIMP-2/SCARB2 that are critical to GCase binding are within the same large segment of the receptor (residues 142–204) that may interact with a “canyon” for the virus and potentially the sequence of GCase binding. LIMP-2/SCARB2 residue His150 in this region appears critical to GCase binding, whereas amino acids 144–151 are essential for enterovirus 71 binding and infectivity (17, 44). Consequently, the domains of binding for these two disparate proteins with different ligand sequences occur in a region with overlapping but apparently distinct critical residues.
For understanding the biology of LIMP-2, such specificities are important as are the other intracellular proteins that may bind to LIMP-2/SCARB2. For example, mutations in LIMP-2 are causal to a human and mouse disease, termed AMRF (21). To understand the biochemical bases of this disease, it will be critical to understand which proteins interact with LIMP-2/SCARB2 and cause AMRF rather than Gaucher disease-like manifestations. However, being a heterozygote for a pathogenic LIMP-2/SCARB2 mutation potentiates the phenotype in Gaucher disease homozygotes (38). Additionally, disruptive mutations of GBA1 in the sequence critical to LIMP-2/SCARB2 may have more profound effects on the Gaucher disease phenotype than might be anticipated by the in vitro levels of residual activity because the mutant protein may never reach the lysosome. The biology of LIMP-2/SCARB2 also may facilitate the understanding of the clear relationships between GCase mutations and Parkinson disease and Lewy body dementia. Indeed, the role of GCase binding to LIMP-2/SCARB2 in the ER may be important to the proposed feed-forward mechanism of GCase/α-synuclein interaction (27) in the development and treatment of Parkinson disease and other α-synucleinopathies.
The precise role of LIMP-2 in trafficking GCase remains unexplored, but the studies here and in LIMP-2-deficient patients and mice offer some insights. GCase activity does not depend on whether it binds to LIMP-2. The poor or deficient binding of GCase variants to ldLIMP-2 in these studies does not appear to alter the enzyme activity or stability of GCase. Also, the LIMP-2 knock-out mouse cells produce active GCase, albeit at lower intracellular levels than WT (10) because of secretion from the cells. These findings suggest that, like the mannose 6-phosphate receptor, LIMP-2 does not have a chaperone/protective function. LIMP-2/GCase binding also is not needed for GCase stability vis-à-vis the stabilization by saposin C of GCase against proteolysis (39). The exact structure and role of the LIMP-2·GCase complex, potentially with the CI-M6PR as a heterotrimeric complex in GCase trafficking (44), remains to be elucidated, particularly in view of the lack of secretion of GCase in I-cell disease. Irrespective of the structure, the function of LIMP-2/SCARB2 appears to be as a delivery vehicle for intracellularly synthesized GCases to the lysosomes.
The identification of a critical sequence for GCase binding to its receptor, LIMP-2/SCARB2, may have significant therapeutic implications. These studies indicate that GCases with specific mutations that disrupt binding to its receptor, but not its activity, could provide for enhanced secretion of GCase from cells for bulk production. Selected conservative mutations could greatly enhance the secretion of normally active enzyme, suggesting the potential for enhanced production from mammalian or other systems that contain LIMP-2/SCARB2 analogues/homologues. Similarly, transplantation of specific cell types specifically expressing mutated GCases that interact poorly with LIMP-2 could supply a large amount of secreted enzyme for therapeutic effects both in local and distant parts of the body. For example, for an enzyme that is normally not secreted (i.e. GCase), such secretion, attained by cellular replacement and/or gene therapy approaches with controllable expression elements, could provide a reservoir for the supply of secreted, active enzyme for cross-correction in other cells. Such an approach would be particularly helpful in the CNS variants of Gaucher or Parkinson diseases, which are caused by or potentiated by GCase defects (26, 40) or the consequences of α-synucleinopathies (25, 41–43) because generalized distribution of the enzyme throughout the brain is currently not possible.
Acknowledgment
We thank Joyce Life-Ishmael for clerical expertise.
Note Added in Proof
In the original version of the manuscript that was published as a JBC Paper in Press, the data shown in Fig. 8C were not correct. The correct Fig. 8C is now provided. This correction does not change the interpretation of the results or the conclusions.
This work was supported, in whole or in part, by National Institutes of Health Grant DK 36729 (to G. A. G.).
- GCase
- glucocerbrosidase
- CI-M6PR
- cation-independent mannose 6-phosphate receptor
- AMRF
- action myoclonus-renal failure syndromes
- ldLIMP-2
- luminal domain LIMP-2
- ER
- endoplasmic reticulum
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PI
- Pearson index
- CRIM
- cross-reacting immunological material
- δmP
- change in fluorescence polarization.
REFERENCES
- 1. Brady R. O., Kanfer J. N., Bradley R. M., Shapiro D. (1966) Demonstration of a deficiency of glucocerebroside-cleaving enzyme in Gaucher's disease. J. Clin. Invest. 45, 1112–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brady R. O., Kanfer J. N., Shapiro D. (1965) Metabolism of glucocerebrosides. II. Evidence of an enzymatic deficiency in Gaucher's disease. Biochem. Biophys. Res. Commun. 18, 221–225 [DOI] [PubMed] [Google Scholar]
- 3. Grabowski G. A., Petsko G. A., Kolodny E. (2010) Gaucher disease. in The Metabolic and Molecular Bases of Inherited Disease (Valle D., Beaudet A. L., Vogelstein B., Kinzler K. W., Antonarakis S. E., Ballabio A., Sly W. S., eds) McGraw-Hill, New York [Google Scholar]
- 4. Grabowski G. A., Kolodny E. H., Weinreb N. J., Rosenbloom B. E., Prakash-Cheng A., Kaplan P., Charrow J., Pastores G. M., Mistry P. K. (2006) Gaucher disease: phenotypic and genetic variation. in The Metabolic and Molecular Bases of Inherited Diseases (Scriver C., Beaudet A., Sly W., Valle D., eds) McGraw-Hill, New York [Google Scholar]
- 5. Liou B., Kazimierczuk A., Zhang M., Scott C. R., Hegde R. S., Grabowski G. A. (2006) Analyses of variant acid β-glucosidases: effects of Gaucher disease mutations. J. Biol. Chem. 281, 4242–4253 [DOI] [PubMed] [Google Scholar]
- 6. Sorge J., West C., Westwood B., Beutler E. (1985) Molecular cloning and nucleotide sequence of the human glucocerebrosidase gene. Proc. Natl. Acad. Sci. U.S.A. 82, 7289–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berg-Fussman A., Grace M. E., Ioannou Y., Grabowski G. A. (1993) Human acid β-glucosidase: N-glycosylation site occupancy and the effect of glycosylation on enzymatic activity. J. Biol. Chem. 268, 14861–14866 [PubMed] [Google Scholar]
- 8. Kornfeld S., Sly W. S. (2001) I-cell disease and pseudo-Hurler polydystrophy: disorders of lysosomal enzyme phosphorylation and localization. in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A., Sly W., Valle D., eds) McGraw-Hill, New York [Google Scholar]
- 9. Reitman M. L., Kornfeld S. (1981) Lysosomal enzyme targeting: N-acetylglucosaminylphosphotransferase selectively phosphorylated native lysosomal enzymes. J. Biol. Chem. 256, 11977–11980 [PubMed] [Google Scholar]
- 10. Reczek D., Schwake M., Schröder J., Hughes H., Blanz J., Jin X., Brondyk W., Van Patten S., Edmunds T., Saftig P. (2007) LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell 131, 770–783 [DOI] [PubMed] [Google Scholar]
- 11. Leonova T., Grabowski G. A. (2000) Fate and sorting of acid β-glucosidase in transgenic mammalian cells. Mol. Genet. Metab. 70, 281–294 [DOI] [PubMed] [Google Scholar]
- 12. Saftig P., Schröder B., Blanz J. (2010) Lysosomal membrane proteins: life between acid and neutral conditions. Biochem. Soc. Trans. 38, 1420–1423 [DOI] [PubMed] [Google Scholar]
- 13. Saftig P., Klumperman J. (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 14. Ogata S., Fukuda M. (1994) Lysosomal targeting of Limp II membrane glycoprotein requires a novel Leu-Ile motif at a particular position in its cytoplasmic tail. J. Biol. Chem. 269, 5210–5217 [PubMed] [Google Scholar]
- 15. Zachos C., Blanz J., Saftig P., Schwake M. (2012) A critical histidine residue within LIMP-2 mediates pH sensitive binding to its ligand β-glucocerebrosidase. Traffic 13, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 16. Wang X., Peng W., Ren J., Hu Z., Xu J., Lou Z., Li X., Yin W., Shen X., Porta C., Walter T. S., Evans G., Axford D., Owen R., Rowlands D. J., Wang J., Stuart D. I., Fry E. E., Rao Z. (2012) A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 19, 424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen P., Song Z., Qi Y., Feng X., Xu N., Sun Y., Wu X., Yao X., Mao Q., Li X., Dong W., Wan X., Huang N., Shen X., Liang Z., Li W. (2012) Molecular determinants of enterovirus 71 viral entry: cleft around Gln-172 on VP1 protein interacts with variable region on scavenge receptor B2. J. Biol. Chem. 287, 6406–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamayoshi S., Koike S. (2011) Identification of a human SCARB2 region that is important for enterovirus 71 binding and infection. J. Virol. 85, 4937–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamayoshi S., Yamashita Y., Li J., Hanagata N., Minowa T., Takemura T., Koike S. (2009) Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801 [DOI] [PubMed] [Google Scholar]
- 20. Knipper M., Claussen C., Rüttiger L., Zimmermann U., Lüllmann-Rauch R., Eskelinen E. L., Schröder J., Schwake M., Saftig P. (2006) Deafness in LIMP2-deficient mice due to early loss of the potassium channel KCNQ1/KCNE1 in marginal cells of the stria vascularis. J. Physiol. 576, 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blanz J., Groth J., Zachos C., Wehling C., Saftig P., Schwake M. (2010) Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand β-glucocerebrosidase. Hum. Mol. Genet. 19, 563–572 [DOI] [PubMed] [Google Scholar]
- 22. Maniwang E., Tayebi N., Sidransky E. (2013) Is Parkinson disease associated with lysosomal integral membrane protein type-2?: challenges in interpreting association data. Mol. Genet. Metab. 108, 269–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lopez G., Sidransky E. (2013) Predicting parkinsonism: new opportunities from Gaucher disease. Mol. Genet. Metab. 109, 235–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westbroek W., Gustafson A. M., Sidransky E. (2011) Exploring the link between glucocerebrosidase mutations and parkinsonism. Trends Mol. Med. 17, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cullen V., Sardi S. P., Ng J., Xu Y. H., Sun Y., Tomlinson J. J., Kolodziej P., Kahn I., Saftig P., Woulfe J., Rochet J. C., Glicksman M. A., Cheng S. H., Grabowski G. A., Shihabuddin L. S., Schlossmacher M. G. (2011) Acid β-glucosidase mutants linked to Gaucher disease, Parkinson disease, and Lewy body dementia alter α-synuclein processing. Ann. Neurol. 69, 940–953 [DOI] [PubMed] [Google Scholar]
- 26. Sidransky E., Nalls M. A., Aasly J. O., Aharon-Peretz J., Annesi G., Barbosa E. R., Bar-Shira A., Berg D., Bras J., Brice A., Chen C. M., Clark L. N., Condroyer C., De Marco E. V., Dürr A., Eblan M. J., Fahn S., Farrer M. J., Fung H. C., Gan-Or Z., Gasser T., Gershoni-Baruch R., Giladi N., Griffith A., Gurevich T., Januario C., Kropp P., Lang A. E., Lee-Chen G. J., Lesage S., Marder K., Mata I. F., Mirelman A., Mitsui J., Mizuta I., Nicoletti G., Oliveira C., Ottman R., Orr-Urtreger A., Pereira L. V., Quattrone A., Rogaeva E., Rolfs A., Rosenbaum H., Rozenberg R., Samii A., Samaddar T., Schulte C., Sharma M., Singleton A., Spitz M., Tan E. K., Tayebi N., Toda T., Troiano A. R., Tsuji S., Wittstock M., Wolfsberg T. G., Wu Y. R., Zabetian C. P., Zhao Y., Ziegler S. G. (2009) Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 361, 1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazzulli J. R., Xu Y. H., Sun Y., Knight A. L., McLean P. J., Caldwell G. A., Sidransky E., Grabowski G. A., Krainc D. (2011) Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fabbro D., Grabowski G. A. (1991) Human acid β-glucosidase: use of inhibitory and activating monoclonal antibodies to investigate the enzyme's catalytic mechanism and saposin A and C binding sites. J. Biol. Chem. 266, 15021–15027 [PubMed] [Google Scholar]
- 29. Liou B., Grabowski G. A. (2012) Is E326K glucocerebrosidase a polymorphic or pathological variant? Mol. Genet. Metab. 105, 528–529 [DOI] [PubMed] [Google Scholar]
- 30. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Misra S., Puertollano R., Kato Y., Bonifacino J. S., Hurley J. H. (2002) Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933–937 [DOI] [PubMed] [Google Scholar]
- 33. Gough N. R., Zweifel M. E., Martinez-Augustin O., Aguilar R. C., Bonifacino J. S., Fambrough D. M. (1999) Utilization of the indirect lysosome targeting pathway by lysosome-associated membrane proteins (LAMPs) is influenced largely by the C-terminal residue of their GYXXφ targeting signals. J. Cell Sci. 112, 4257–4269 [DOI] [PubMed] [Google Scholar]
- 34. Barriocanal J. G., Bonifacino J. S., Yuan L., Sandoval I. V. (1986) Biosynthesis, glycosylation, movement through the Golgi system and transport to lysosomes by N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J. Biol. Chem. 261, 16755–16763 [PubMed] [Google Scholar]
- 35. Mullin N. P., Hitchen P. G., Taylor M. E. (1997) Mechanism of Ca2+ and monosaccharide binding to a C-type carbohydrate-recognition domain of the macrophage mannose receptor. J. Biol. Chem. 272, 5668–5681 [DOI] [PubMed] [Google Scholar]
- 36. Ezekowitz R. A., Sastry K., Bailly P., Warner A. (1990) Molecular characterization of the human macrophage mannose receptor: demonstration of multiple carbohydrate recognition-like domains and phagocytosis of yeasts in Cos-1 cells. J. Exp. Med. 172, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kornfeld S. (1992) Structure and function of the mannose 6-phosphate/insulin like growth factor II receptors. Annu. Rev. Biochem. 61, 307–330 [DOI] [PubMed] [Google Scholar]
- 38. Velayati A., DePaolo J., Gupta N., Choi J. H., Moaven N., Westbroek W., Goker-Alpan O., Goldin E., Stubblefield B. K., Kolodny E., Tayebi N., Sidransky E. (2011) A mutation in SCARB2 is a modifier in Gaucher disease. Hum. Mutat. 32, 1232–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun Y., Qi X., Grabowski G. A. (2003) Saposin C is required for normal resistance of acid β-glucosidase to proteolytic degradation. J. Biol. Chem. 278, 31918–31923 [DOI] [PubMed] [Google Scholar]
- 40. Bultron G., Kacena K., Pearson D., Boxer M., Yang R., Sathe S., Pastores G., Mistry P. K. (2010) The risk of Parkinson's disease in type 1 Gaucher disease. J. Inherit. Metab. Dis. 33, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sardi S. P., Clarke J., Viel C., Chan M., Tamsett T. J., Treleaven C. M., Bu J., Sweet L., Passini M. A., Dodge J. C., Yu W. H., Sidman R. L., Cheng S. H., Shihabuddin L. S. (2013) Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proc. Natl. Acad. Sci. U.S.A. 110, 3537–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sardi S. P., Singh P., Cheng S. H., Shihabuddin L. S., Schlossmacher M. G. (2012) Mutant GBA1 expression and synucleinopathy risk: first insights from cellular and mouse models. Neurodegener. Dis. 10, 195–202 [DOI] [PubMed] [Google Scholar]
- 43. Sardi S. P., Clarke J., Kinnecom C., Tamsett T. J., Li L., Stanek L. M., Passini M. A., Grabowski G. A., Schlossmacher M. G., Sidman R. L., Cheng S. H., Shihabuddin L. S. (2011) CNS expression of glucocerebrosidase corrects α-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc. Natl. Acad. Sci. U.S.A. 108, 12101–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y., Ren J., Padilla-Parra S., Fry E. E., Stuart D. I. (2014) Lysosomal sorting of β-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor. Nat. Commun. 5, 4321. [DOI] [PMC free article] [PubMed] [Google Scholar]