Background: Intricate regulation of the growth cone cytoskeleton controls growth cone dynamics.
Results: Loss of CRMP4 disrupts growth cone cytoskeletal dynamics, growth cone expansion, and axon growth.
Conclusion: CRMP4 regulates both the actin and microtubule growth cone cytoskeleton.
Significance: CRMP4 plays a critical role in regulating cytoskeletal dynamics underlying growth cone properties.
Keywords: Axon, Cell Biology, Cytoskeleton, Microfilaments, Microtubule, Neurodevelopment, Neuroscience, CRMP, Growth Cone
Abstract
Coordinated control of the growth cone cytoskeleton underlies axon extension and guidance. Members of the collapsin response mediator protein (CRMP) family of cytosolic phosphoproteins regulate the microtubule and actin cytoskeleton, but their roles in regulating growth cone dynamics remain largely unexplored. Here, we examine how CRMP4 regulates the growth cone cytoskeleton. Hippocampal neurons from CRMP4−/− mice exhibited a selective decrease in axon extension and reduced growth cone area, whereas overexpression of CRMP4 enhanced the formation and length of growth cone filopodia. Biochemically, CRMP4 can impact both microtubule assembly and F-actin bundling in vitro. Through a structure function analysis of CRMP4, we found that the effects of CRMP4 on axon growth and growth cone morphology were dependent on microtubule assembly, whereas filopodial extension relied on actin bundling. Intriguingly, anterograde movement of EB3 comets, which track microtubule protrusion, slowed significantly in neurons derived from CRMP4−/− mice, and rescue of microtubule dynamics required CRMP4 activity toward both the actin and microtubule cytoskeleton. Together, this study identified a dual role for CRMP4 in regulating the actin and microtubule growth cone cytoskeleton.
Introduction
Dynamic growth cones at the tip of neuronal processes control axonal growth and guidance during both development and regeneration. Rearrangements of the actin cytoskeletal network at the growth cone leading edge, exploratory extension of microtubules into the peripheral domain, and protrusion of stabilizing microtubules behind extending lamellipodia and filopodia are all critical for directed outgrowth (1). Overlap between the actin and microtubule networks in the transition domain of the growth cone plays a key role in generating force required for neurite extension (2). Thus, fully understanding how the actin and microtubule cytoskeletons are coordinated within neuronal growth cones is an important aspect of understanding growth cone behavior and axon guidance. To date, a limited number of proteins have been described to regulate both the actin and microtubule cytoskeleton in neurons. Spectraplakin family members interact with both F-actin and microtubules and are robustly expressed in the nervous system (3). The Drosophila spectraplakin family member Short Stop is important in many types of neurons for growth of axons, dendrites, and synapses (4–6). The mammalian spectraplakin family member ACF7/MACF1 both organizes microtubules and regulates filopodial formation in part through binding to the microtubule plus-end-binding protein 1 (EB1)2 (7, 8). Similarly, the F-actin-associated protein Drebrin binds to EB3 at the tips of microtubules that invade filopodia, and this interaction is required for appropriate neuritogenesis in cultured embryonic cortical neurons (9).
Members of the dihydropyrimidinase-like or collapsin response mediator protein (CRMP) family of cytosolic phosphoproteins are excellent candidates to dually regulate actin and microtubule cytoskeletal rearrangements underlying growth cone dynamics and neurite extension. CRMP family members (CRMP1–5) play important roles in neuronal differentiation, axonal growth, guidance, axon/dendrite specification, and microtubule organization including establishing the microtubule asymmetry that underlies protein sorting axons and dendrites (10–20). CRMP alleles each produce two transcripts with the long (CRMPL) isoforms being long amino-terminal variants of the originally identified short (CRMPS) isoforms. All CRMP family members can bind to tubulin in vitro and in the brain (21–23). CRMP2 regulates endocytosis and promotes microtubule stabilization and assembly (21, 24–26). In addition, the CRMP4 isoform binds directly to F-actin and organizes F-actin into bundles in vitro. CRMP4 is also the sole CRMP isoform that can bind to RhoA, a key regulator of the actin cytoskeleton (27, 28). In zebrafish, CRMP4 function is important for proper positioning of neural crest cells and caudal primary motor neurons, whereas in the mouse, CRMP4 supports proper bifurcation of apical dendrites (29–31).
The subcellular roles of CRMP family proteins in regulating the axonal growth cone cytoskeleton have not been explored. Here, we focus on elucidating the role of CRMP4 in regulating the actin and microtubule growth cone cytoskeleton in hippocampal neurons and the impact of its activity on growth cone dynamics and extension. We found that CRMP4 is responsible for organizing both the actin and microtubule structure within hippocampal growth cones and that this activity underlies growth cone morphology and axon extension.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used for immunohistochemistry and Western blotting: rabbit CRMP4 (1:500 for immunohistochemistry and 1:7500 for Western blotting; Ref. 28), mouse polyclonal DPYSL3 (1:1000; Abnova, Taiwan), mouse βIII-tubulin (1:1000 dilution; Covance, Princeton, NJ), mouse mCherry (1:200; Abcam, La Jolla, CA), mouse actin (1:2000; Millipore, Bedford, MA), mouse GAPDH (1:1000; Abcam), Alexa Fluor secondary antibodies (1:1000 dilution; Invitrogen), and HRP-coupled IgG antibodies (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA). F-actin was labeled with rhodamine- or fluorescein-labeled phalloidin (1:250; Invitrogen).
Constructs
The HSV-CRMP4S-V5 construct was described previously (28). CRMP4S, CRMP4ΔN300 (amino acids 300–570), CRMP4ΔC50 (amino acids 1–520), CRMP4ΔC100 (amino acids 1–470), and CRMP4Δ470–520 (deletion of amino acids 470–520) DNAs were amplified by PCR from the pcDNA3.1/V5-His-CRMP4S construct and subcloned into the pHSV-PUC-IRES-GFP vector using SalI and XbaI restriction sites. To generate glutathione S-transferase (GST) fusion proteins, CRMP4S, CRMP4ΔN300, CRMP4ΔC50, CRMP4ΔC100, and CRMP4Δ470–520 DNA were amplified by PCR and subcloned into the pGEX-2T vector using EcoRI and BamHI restriction sites. GST-Fascin was a generous gift from Dr. Yun Huang (32). To generate HSV-EB3-mCherry, EB3 was amplified by PCR from the CMV-EB3-GFP vector, a generous gift from Dr. N. Galjart, and subcloned into the pHSV-PUC vector using HindIII and XbaI restriction sites. mCherry was inserted into the XbaI/BamHI site of HSV-EB3 to generate HSV-EB3-mCherry (33).
Preparation of Herpes Simplex Viruses
pHSVPrPUC plasmids were transfected into 2-2 Vero cells that were superinfected with 5dl 1.2 herpes simplex virus (HSV) helper virus 1 day later. Recombinant virus was amplified though three passages and stored at −80 °C as described previously (34).
Hippocampal Cell Culture
Primary cultures of dissociated hippocampal neurons were prepared as described previously with some modifications (35). Briefly, the hippocampus was dissected from E18 mouse embryos and dissociated, and neurons were plated in Neurobasal medium (supplemented with B-27, 0.5 mm glutamine, and 100 units/ml penicillin/streptomycin (Invitrogen)) onto glass coverslips coated with 0.1% poly-l-lysine (Sigma-Aldrich) at a density of 105 cells/coverslip in 12-well plates.
Immunofluorescence
Neurons were fixed for 20 min with 4% paraformaldehyde in phosphate-buffered saline (PBS) and 40% sucrose at room temperature. Following fixation, neurons were permeabilized in 0.1% Triton X-100 and 0.1% sodium citrate in PBS for 5 min and then placed in blocking buffer (5% BSA) for 1 h. Primary antibodies were diluted in a blocking buffer solution and incubated with the cells overnight at 4 °C. Following extensive washing, samples were incubated with fluorophore-conjugated secondary antibodies for 1 h.
Analysis of Neurite Outgrowth and Growth Cone Morphology
Neurite outgrowth was assessed from neurons stained with βIII-tubulin using the NIH ImageJ program and with the tracing algorithm contained in the NeuronJ plug-in as described previously (36, 37). Axon growth was defined as the length of the longest neurite from each cell. For growth cone measurements, the proximal boundary of the growth cone was defined as the point where it first splayed from the axon, and area measurements included all portions of the growth cone including the filopodia and lamellipodia. The growth cone surface area and the length of filopodia were measured with ImageJ. Filopodial length was measured from the base to their tip and following all branches. Filopodia were counted if they were greater than 2 μm in length and positively stained by phalloidin.
Purification of Recombinant Proteins from Escherichia coli
GST fusion proteins were expressed in E. coli strain BL21 by induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 25 °C. Bacterial pellets were resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 1 mm DTT, protease inhibitor mixture (Roche Applied Science)). Bacteria were then lysed by sonication (three times at 50% amplitude for 30 s). The clarified lysate obtained after centrifugation was incubated with glutathione-agarose beads (GE Healthcare) for 2 h at 4 °C. Beads were then washed five times with 10 bead volumes of washing buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT) and eluted with 20 mm glutathione in elution buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT) followed by concentration and buffer exchange to tubulin polymerization buffer (10 mm Tris-HCl, pH 7.5, 2 mm MgCl2, 1 mm DTT) with an Amicon Ultra-4 30,000 Centricon column concentrator (Millipore). All samples were aliquoted, snap frozen in liquid nitrogen, and stored at −80 °C.
Tubulin Polymerization Assay
Polymerization of tubulin was performed as described previously (38). Briefly, 20 μm GST fusion proteins were dialyzed against 0.1 m PEM buffer (80 mm PIPES, pH 6.9, 1 mm EGTA, 1 mm MgCl2) and mixed with 1 mm GTP and 1 mg/ml tubulin, resulting in a stoichiometry of ∼2 CRMP:1 tubulin. Polymerization was initiated by placing the mixture immediately into 0.2-ml cuvettes at 35 °C, and turbidity changes were assessed at 350 nm using a CARY 300 UV-visible spectrophotometer (Varian Inc., Palo Alto, CA).
Actin Bundling Assay
GST fusion proteins were dialyzed against F buffer (25 mm Hepes, pH 7.5, 100 mm KCl, 0.2 mm CaCl2, 2 mm MgCl2, 2 mm EGTA, 0.2 mm ATP, 1 mm dithiothreitol), clarified by centrifugation, and mixed with 5 μm F-actin in F buffer for 1 h at room temperature. The mixture was then subjected to centrifugation at 10,000 × g for 30 min. Equal aliquots of the supernatant and the pellet solubilized in SDS sample buffer were analyzed on a Coomassie gel. For visualization of actin bundles, F-actin (final concentration, 5 μm) was mixed with 2 μm GST fusion proteins, incubated for 30 min on ice in F buffer, and stained with rhodamine phalloidin. Samples were applied to poly-l-lysine-coated glass coverslips and fixed with paraformaldehyde.
Spatial-Temporal Image Correlation Spectroscopy
E18 hippocampal neurons were cultured to 3 DIV and transduced for 3 h with HSV-EB3-mCherry alone or together with HSV-GFP-CRMP4 constructs for 6–12 h. EB3 movements in growth cones were captured by time lapse microscopy for 215 s at an imaging rate of 2.4 frames/s, and videos were analyzed by spatial-temporal image correlation spectroscopy to generate vector maps of the labeled molecules (39). For each 16 × 16-pixel region of interest selected every third frame from the image series, vector maps were superimposed on the micrographs and revealed the direction and speed of anterograde EB3 movements over a 45-s integration time period.
Generation and Characterization of CRMP4-deficient Mice
The C57BL/6 ES cells with the CRMP4 gene trapped by a retroviral gene trap vector at intron 3 were obtained from the Texas A&M Institute for Genomic Medicine (College Station, TX). The insertion results in incorrect splicing such that all exons downstream of the insertion site are not expressed. The ES cells were injected into eight-cell embryos isolated from timed pregnant BALB/c females. Injected eight-cell embryos were cultured to blastocysts and then transferred into the uterus of pseudopregnant females for development into individual pups. Heterozygous mutants were obtained by breeding chimeric mice to C57BL/6 females, and the mutation was backcrossed to C57BL/6 two more times before undergoing homozygosis. Brain sections from wild-type and CRMP4-deficient mice were Nissl-stained as described previously (40). Genomic DNA was amplified with two pairs of primers to genotype the mice to detect wild-type and mutant alleles (wild-type/mutant forward primer, 5′-GACGGTTCACTCTAGCACAC-3′; wild-type reverse primer, 5′-TCCACCTGATCTGGGGCG-3′; mutant reverse primer, 5′-CTTGCAAAATGGCGTTACTTAAGC-3′).
Statistical Analysis
Statistical significance values were obtained by performing Student's t test for pairwise comparisons and by one-way ANOVA with Tukey's post hoc test for multiple comparisons. Data are representative of at least three independent experiments.
RESULTS
CRMP4 Promotes Growth Cone Spreading and Axon Outgrowth
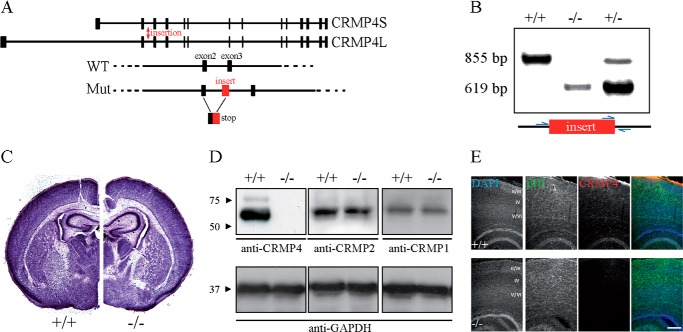
Previous reports have demonstrated that overexpression of CRMP2 or CRMP4 promotes axon elongation in rat hippocampal neurons (20, 42). To investigate the physiologic contribution of CRMP4 to outgrowth of hippocampal neurons, we generated CRMP4−/− mice from a CRMP4-targeted embryonic stem cell clone (Fig. 1, A and B). CRMP4−/− mice had no anatomical or macroscopic changes in gross brain anatomy, consistent with a recently published report describing an independently generated CRMP4-null line (Fig. 1C and Ref. 29). CRMP4 proteins levels were undetectable in CRMP4−/− mice by Western blotting or immunofluorescent staining with an anti-CRMP4 antibody (Fig. 1, D and E). Other CRMP family members did not exhibit compensatory regulation in the brain of CRMP4-null mice (Fig. 1D).
FIGURE 1.
Generation and characterization of CRMP4−/− mice. A, structure of mouse CRMP4 (Dpysl3) gene on chromosome 18 illustrating exons 1–14. The Omnibank 76 trapping vector with a splice acceptor sequence was inserted after exon 2 of the CRMP4 gene (red arrow). This insertion results in incorrect splicing of CRMP4 mRNA such that all downstream exons are not expressed. B, genotyping of CRMP4 mice using PCR amplification of DNA isolated from tails with specific primers (blue arrows). The wild-type allele yields an 855-bp fragment, whereas the knock-out allele yields a 619-bp fragment. C, Nissl staining of 50-μm-thick coronal sections from P5 mice at the level of the ventromedial hypothalamus. Brain cytoarchitecture is similar in the CRMP4−/− and CRMP4+/+ littermate controls. D, Western blot analysis of brain lysates from P5 CRMP4+/+ and CRMP4−/− mice with anti-CRMP4, anti-CRMP1, anti-CRMP2, or anti-GAPDH antibodies. E, sagittal sections of P5 mouse brain from wild-type CRMP4+/+ and CRMP4−/− mice stained with DAPI and anti-βIII-tubulin and anti-CRMP4 antibodies. Scale bar, 250 μm.
When dissociated hippocampal neurons from CRMP4-null mice were grown in culture, they exhibited diminished outgrowth and small growth cones (Fig. 2, A and E). Quantification of outgrowth from all neuronal processes or specifically from axons revealed that CRMP4 loss of function specifically impaired axonal growth, whereas total neurite outgrowth remained unaffected (Fig. 2, B and C). Hippocampal growth cones from CRMP4−/− mice were characterized by a condensed central domain, numerous filopodia, and small or absent lamellipodia (Fig. 2E). To fully characterize the defect, we categorized the growth cones as blunt ended (collapsed with no visible filopodia or lamellipodia), filopodial (growth cones with numerous filopodia and a small or absent lamellipodial veil), or lamellipodial (well spread growth cones with elaborate lamellipodia; Fig. 2D). In cultures from CRMP4+/+ mice, the majority of growth cones were lamellipodial or filopodial in nature, whereas blunt end growth cones were rare (Fig. 2F). In the CRMP4−/− mice, there was a significant increase in the number of filopodial growth cones and loss of lamellipodial growth cones (Fig. 2, E and F). Furthermore, the growth cones from CRMP4−/− mice were significantly smaller in area (Fig. 2G). Growth cones overexpressing CRMP4 were similar in size to control GFP-transduced neurons (Fig. 2, H and J); however, the number of growth cones with a filopodial phenotype was significantly increased (Fig. 2, H and I). Together, these data support a physiological role for CRMP4 in promoting axon extension and indicate that CRMP4 plays a role in maintaining an expanded lamellipodial growth cone.
FIGURE 2.
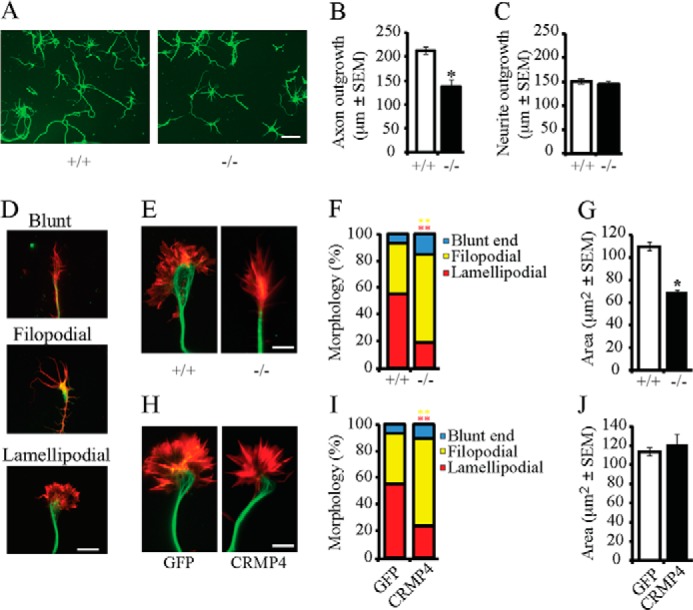
CRMP4 regulates axon outgrowth and growth cone size. A, hippocampal neurons from CRMP4+/+ and CRMP4−/− mice were cultured for 3 DIV and stained with anti-βIII-tubulin antibody. Scale bar, 25 μm. B and C, the average axon length (B) and average neurite length (C) were quantified from cultures derived from CRMP4+/+ or CRMP4−/− mice. D, hippocampal neurons stained with anti-βIII-tubulin antibody (green) and rhodamine phalloidin (red) were classified as blunt ended, filopodial, or lamellipodial. Scale bar, 5 μm. E–J, hippocampal neurons from CRMP4+/+ and CRMP4−/− mice (E–G) or wild-type mice infected with GFP or CRMP4 (H–J) were fixed at 3 DIV and co-stained with an anti-βIII-tubulin antibody and rhodamine phalloidin (E and H). Growth cone morphology (F and I) and size (G and J) were quantified. Determinations are mean ± S.E. from three experiments performed on 30 cells per experiment. *, p < 0.05; **, p < 0.01 by Student's t test compared with control. Error bars represent S.E. Scale bar, 5 μm.
Distribution of CRMP4 in Hippocampal Neurons
To investigate the potential mechanism through which CRMP4 impacts growth cone morphology and axon growth, we stained 3 DIV hippocampal neurons with an anti-CRMP4 antibody that recognizes both short and long CRMP4 isoforms (28). CRMP4L was weakly expressed in lysates from E18 hippocampal neurons; thus our staining likely reflects the expression of CRMP4S (Fig. 3A). CRMP4 was expressed in the cell body, neurites, and growth cone tips of hippocampal neurons at 3 DIV (Fig. 3B). Within the growth cone, CRMP4 was distributed in a punctate pattern in the microtubule-rich central domain and extended into the growth cone transition domain. CRMP4 staining was also present in more peripheral regions of the growth cone in close proximity to thick actin filopodial bundles (Fig. 3B). CRMP4 staining was negligible in growth cones derived from CRMP4 −/− mice (Fig. 3C). The distribution of CRMP4 in close proximity to elements of the actin and microtubule cytoskeleton raised the possibility that CRMP4 affects both cytoskeletal elements within the growth cone.
FIGURE 3.
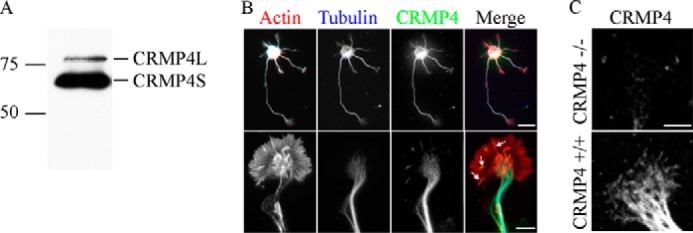
CRMP4 expression in hippocampal neurons. A, Western blot of an E18 wild-type hippocampal lysate probed with anti-CRMP4 antibody. B, 3 DIV wild-type hippocampal neurons stained with rhodamine phalloidin and anti-βIII-tubulin and anti-CRMP4 antibodies. CRMP4 is expressed in the microtubule-rich neurite and central domain and is also present in the actin-rich periphery (arrows). Scale bar, 20 (top row) and 5 μm (bottom row). C, 3 DIV hippocampal neurons from CRMP4+/+ or CRMP4−/− mice stained with anti-CRMP4 antibody. Scale bar, 5 μm.
CRMP4 Promotes Tubulin Assembly in Vitro
Other members of the CRMP family have been shown to directly affect microtubules. CRMP2 has been shown to enhance microtubule polymerization, whereas CRMP3 and CRMP5 inhibit the process (21, 22, 43–45). CRMP4 binds directly to tubulin, but its effects on microtubule assembly have not been tested (18, 21, 46). We generated GST fusion proteins for the short isoform of CRMP4 and for a series of CRMP4 deletion mutants to test the effect of CRMP4 in microtubule assembly in vitro and to map the regions of CRMP4 responsible for this activity (Fig. 4, A and B). Using a turbidimetric in vitro microtubule polymerization assay, we found that GST-CRMP4 promoted microtubule assembly at a stoichiometric ratio of 2 CRMP:1 tubulin, whereas the GST control protein had no effect (Fig. 4C). Deletion of the first 300 amino acids from the amino terminus of CRMP4 (CRMP4ΔN300) enhanced the effect on tubulin polymerization likely because this domain autoinhibited the tubulin binding site as has been reported for CRMP2 (Fig. 4C and Ref. 43). To identify the region of CRMP4 responsible for microtubule assembly, we tested a series of carboxyl-terminal deletion mutants of CRMP4 (Fig. 4A). Mutants lacking the last 100 residues (CRMP4ΔC100) failed to increase microtubule assembly, whereas deletion of the last 50 amino acids of CRMP4 (CRMP4ΔC50) had little effect. Further mapping revealed that deletion of residues 470–520 resulted in a minimal CRMP4 deletion mutant that failed to assemble microtubules (Fig. 4C), thus locating the microtubule assembly domain to those residues.
FIGURE 4.
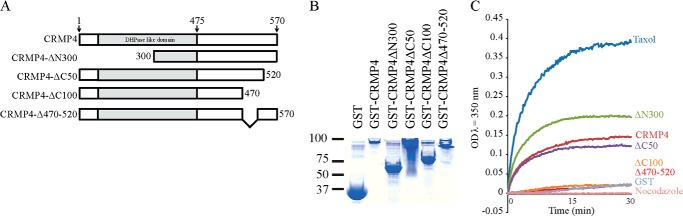
CRMP4 promotes microtubule assembly. A, schematic of the rat sequence for the short isoform of CRMP4 and mutants generated in this study. B, Coomassie-stained SDS-polyacrylamide gel loaded with individual GST-CRMP4 proteins. C, effects of GST-CRMP4 fusion proteins on microtubule assembly measured by light scattering. Purified GST fusion proteins (20 μm) as indicated were incubated with tubulin at 35 °C, and turbidity changes were assessed at 350 nm. Taxol (2.5 μm) and nocodazole (5 μm) were used as positive and negative controls, respectively. Turbidity measurements are representative of three replicates. DHPase, dihydropyrimidinase. Scale bar, 5 μm.
CRMP4 Regulates Microtubule Organization in Hippocampal Growth Cones
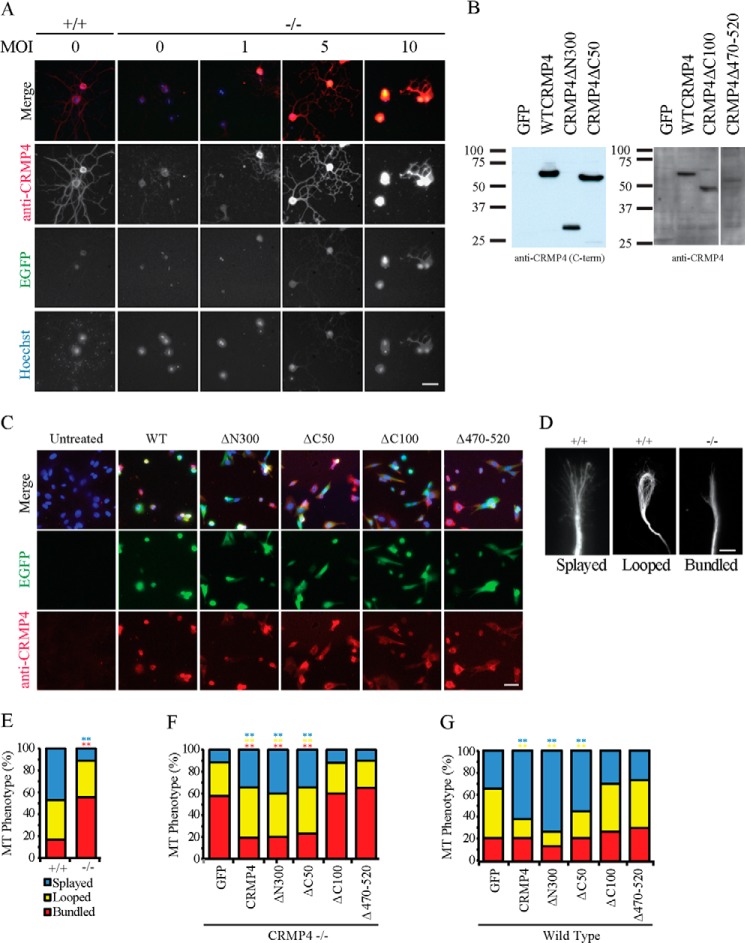
We next assessed the impact of CRMP4 on microtubule organization in growth cones. CRMP4 constructs were packaged into replication-deficient HSV. CRMP4 null neurons were transduced at increasing multiplicity of infection (MOI) to define an MOI that resulted in endogenous levels of CRMP4 expression. Neurons from CRMP4−/− mice infected with an MOI of 5 exhibited endogenous levels of wild-type CRMP4 expression in the neurites of dissociated hippocampal neuron (Fig. 5A). Lysates from transduced 3T3 cells, which do not express CRMP4 endogenously, were analyzed to verify that CRMP4 deletion mutants express at levels similar to wild-type CRMP4 (Fig. 5B) and this was further verified by immunofluorescent staining (Fig. 5C).
FIGURE 5.
CRMP4 promotes microtubule splaying through its tubulin-binding domain. A, hippocampal neurons from littermate control mice (+/+) or from CRMP4−/− mice transduced with an increasing multiplicity of infection (MOI) of wild-type CRMP4 virus were stained with anti-CRMP4 antibody. Neurons transduced with a multiplicity of infection of 5 generated wild-type levels of CRMP4 in neurites. Scale bar, 40 μm. B, Western blot of NIH 3T3 cells transduced with individual CRMP4 viruses at a multiplicity of infection of 5 and stained with an antibody raised against the carboxyl terminus (C-term) of CRMP4 (left panel) or a polyclonal CRMP4 antibody raised against full-length CRMP4 (right panel). Individual constructs were expressed at comparable levels. C, immunofluorescent staining of 3T3 cells transduced with individual CRMP4 constructs with a polyclonal CRMP4 antibody raised against full-length CRMP4. Scale bar, 50 μm. D, hippocampal neurons from CRMP4+/+ or CRMP4−/− mice were fixed, stained with anti-βIII-tubulin, and classified as having a splayed, looped, or bundled microtubule phenotype. Scale bar, 5 μm. E–G, quantitation of microtubule (MT) phenotype in hippocampal neurons from CRMP4+/+ and CRMP4−/− mice (E) or in CRMP4−/− growth cones transduced with HSV-GFP as a control or with HSV-CRMP4 rescue constructs (F) or for wild-type growth cones transduced with HSV-GFP as a control or with HSV-CRMP4 constructs (G). Determinations are mean ± S.E. from three experiments performed on 30 cells per experiment. **, p < 0.01 compared with GFP control condition by one-way ANOVA with Tukey's post hoc test. EGFP, enhanced GFP.
During axon elongation, growth cones have been reported to experience recurrent cycles of advancing, pausing, and retracting. In advancing growth cones, microtubules form a fan-shaped splayed array in the central domain that occasionally extends into the periphery. During pausing, microtubule loops are often present within the growth cone. During retraction, tight parallel bundled microtubules are present along the axon shaft and in the central domain (47, 48). In wild-type neurons, a large fraction of growth cones displayed microtubules that were either splayed or looped (Fig. 5, D and E). In contrast, bundled microtubules were much more prevalent in growth cones from CRMP4−/− mice whereas splayed microtubules were rare (Fig. 5, D and E). A normal distribution of microtubule phenotypes was restored by transducing neurons with CRMP4, CRMP4ΔN300, or CRMP4ΔC50 but not with CRMP4ΔC100 or CRMP4Δ470–520, indicating that the microtubule assembly property of CRMP4 is necessary for microtubule organization in growth cones (Fig. 5F). Likewise, CRMP4 overexpression in wild-type growth cones significantly increased the percentage of growth cones with splayed microtubules whereas reducing the number of growth cones with looped microtubules. This effect was also dependent on the microtubule assembly domain of CRMP4 (Fig. 5G).
In addition to analyzing microtubule phenotypes in fixed growth cones, we examined microtubule dynamics in dynamic growth cones by live imaging mCherry-tagged EB3 over time. EB3 decorates the growing tip of the microtubule and EB3 comets can be tracked by live imaging to assess microtubule growth (33). EB3 movements were quantified using spatial-temporal image correlation spectroscopy, which we have adapted for growth cones (Fig. 6 and Ref. 39). In wild-type neurons, the average anterograde movement of EB3 comets was 4.78 ± 0.2 μm/min. In contrast, EB3 comets in growth cones derived from CRMP4−/− mice moved significantly slower at 3.33 ± 0.6 μm/min (Fig. 6, A and B). The EB3 phenotype was rescued by transducing neurons with CRMP4 or CRMP4ΔN300, which had the strongest effect on microtubule assembly in vitro (Fig. 4C). CRMP4 mutants lacking the microtubule-binding domain (CRMP4ΔC100 and CRMP4Δ470–520) failed to rescue the KO phenotype. In an overexpression paradigm, CRMP4ΔN300, but not wild-type CRMP4, enhanced EB3 anterograde movement, suggesting that heightened microtubule assembly is required to boost the endogenous regulatory mechanisms underlying EB3 movements (Fig. 6D). Intriguingly, the CRMP4 carboxyl terminal deletion mutant (CRMP4 Δ C50), which readily assembled microtubules, also failed to rescue the rate of EB3 anterograde movement in CRMP4−/− neurons (Fig. 6C). Previous findings have ascribed actin bundling activity to the carboxyl terminus of CRMP4 in vitro, thus we further investigated the possibility that EB3 dynamics may be regulated by coordinated activity of actin and microtubules by CRMP4 (27).
FIGURE 6.
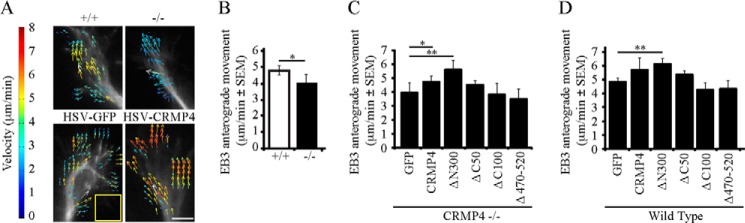
CRMP4 enhances EB3 anterograde movements. A, spatial-temporal image correlation spectroscopy analysis of E18 hippocampal neurons transduced with HSV-EB3-mCherry. Neurons were derived from CRMP4+/+ or CRMP4−/− mice or from wild-type mice co-transduced with HSV-GFP (GFP) or HSV-CRMP4 constructs. Comets were filtered to quantify anterograde movements only. Vector overlays represent the speed and direction of growth over 15 s. Scale bar, 5 μm. Yellow square, 16 × 16-pixel (6 × 6-μm) region of interest. B–D, quantitation of EB3 anterograde movements in CRMP4+/+ versus CRMP4−/− mice (B), in CRMP4−/− mice transduced with CRMP4 rescue constructs (C), or in wild-type mice transduced with GFP or CRMP4 constructs (D). Determinations are mean ± S.E. from three experiments performed on 10 growth cones per experiment. *, p < 0.05; **, p < 0.01 by t test (B) or one-way ANOVA with Tukey's post hoc test compared with GFP control (C and D). Error bars represent S.E.
CRMP4 Regulates the Actin Cytoskeleton in Hippocampal Growth Cones
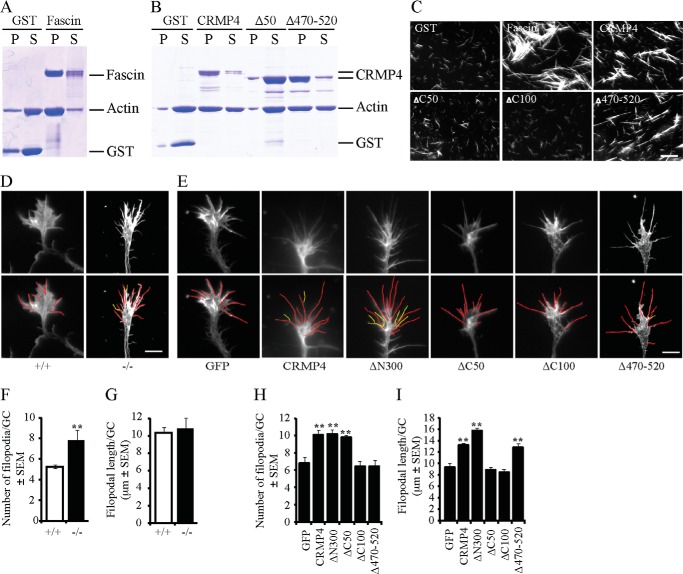
We performed actin co-sedimentation assays to fully map the region of CRMP4 responsible for the F-actin bundling activity. When centrifuged at low speed, F-actin failed to pellet in the presence of GST control protein; however, full-length CRMP4, as well as the control actin-bundling protein Fascin, markedly enhanced the amount of polymerized actin in the pellet fraction (Fig. 7, A and B, and Ref. 49). We found that a CRMP4 mutant with 50 residues deleted from the carboxyl terminus (CRMP4ΔC50) lost its capacity to pellet F-actin, whereas the CRMP4Δ470–520 mutant, which failed to assemble microtubules, retained its capacity to bundle F-actin (Fig. 7B). We confirmed the F-actin bundling activity of CRMP4 by microscopic analysis of the in vitro bundled F-actin (Fig. 7C). In the presence of the control GST protein, F-actin failed to bundle. The addition of full-length CRMP4, CRMP4Δ470–520, or Fascin enhanced visible F-actin bundles, whereas CRMP4ΔC50 or CRMP4ΔC100, each lacking residues 520–570, failed to bundle F-actin. Together, these findings identify residues 520–570 as critical for F-actin bundling and residues 470–520 as critical for microtubule assembly and demonstrate that these two activities are separable. Moreover, CRMP4 requires both F-actin bundling activity and microtubule assembly activity to rescue rates of EB3 transport, suggesting that CRMP4 may coordinate the two cytoskeletal elements in the growth cone (Fig. 6C).
FIGURE 7.
CRMP4 bundles the actin cytoskeleton. A and B, Coomassie-stained SDS-polyacrylamide gels loaded with supernatant (S) and pellet (P) fractions of F-actin that was incubated with purified GST fusion proteins and subjected to low speed centrifugation. GST-Fascin, an F-actin-bundling protein, was used as a positive control (A). Full-length CRMP4 and CRMP4Δ470–520 promotes actin sedimentation, whereas CRMP4Δ50 lacks this activity (B). C, micrograph of rhodamine phalloidin-stained F-actin following incubation with GST fusion proteins. Scale bar, 10 μm. D and E, hippocampal neurons from CRMP4+/+ or CRMP4−/− mice (D) or from wild-type mice transduced with GFP control virus or individual CRMP4 viruses (E) were fixed and stained with rhodamine phalloidin. Traces outline filopodial extensions (red) and filopodial branches (yellow) on individual growth cones used for quantitation. Scale bar, 5 μm. F–I, quantitation of the number and length of filopodia from CRMP4+/+ and CRMP4−/− mice (F and G) or wild-type mice transduced with GFP (as a control) or with CRMP4 constructs (H and I). Values are mean ± S.E. from three experiments performed on 30 cells per experiment. **, p < 0.01 by t test (F and G) or one-way ANOVA with a Tukey's post hoc test compared with GFP control (H and I). Error bars represent S.E.
To evaluate the role of CRMP4 in regulating the actin cytoskeleton in hippocampal growth cones, we examined cultured hippocampal growth cones from CRMP4−/− mice stained with rhodamine phalloidin. The actin cytoskeleton was collapsed in the CRMP4−/− growth cones, although it was not possible to distinguish whether this was a direct actin phenotype or a consequence of the smaller growth cones in the CRMP4-null mice (Fig. 7D). We also noted that the number of actin-rich filopodia was significantly increased in CRMP4−/− growth cones, but there was no parallel effect on filopodial length (Fig. 7, D, F, and G). This apparent increase in the number of filopodia appears to be a result of lamellipodial shrinkage without a corresponding retraction of the filopodia. CRMP4 overexpression increased filopodial numbers in wild-type neurons, dependent on its microtubule assembly properties (Fig. 7, E and H). In the case of wild-type CRMP4 overexpression, the increase in filopodial number was accompanied by an increase in filopodial length, distinguishing this phenotype from the loss of function phenotype (Fig. 7, G and I). Intriguingly, CRMP4ΔC50 and CRMP4ΔC100 constructs failed to increase filopodial length, implicating the F-actin bundling activity of CRMP4 in promoting filopodial length (Fig. 7I). Together, these results suggest that CRMP4 loss of function results in an apparent increase in filopodia number without affecting their length, likely reflecting lamellipodial shrinkage without a parallel retraction of filopodia. CRMP4 overexpression enhances filopodial formation through its microtubule assembly properties and promotes filopodial extension through its F-actin bundling activity.
CRMP4 Activity toward Microtubules Regulates Growth Cone Size and Axon Outgrowth
To explore the relative contribution of CRMP4 microtubule and F-actin bundling activities to axonal growth and morphology, we examined axon outgrowth and growth cone morphology in hippocampal neurons from CRMP4−/− mice transduced with CRMP4 rescue constructs (Fig. 8). Full-length CRMP4 and CRMP4 constructs that retained their ability to assemble microtubules rescued the diminished axon outgrowth (Fig. 8, A and B) and growth cone size (Fig. 8, C and D) phenotypes that we observed in the CRMP4−/− cultures. The CRMP4ΔC50 construct, which failed to bundle actin but retained activity toward microtubules, rescued both neurite outgrowth and growth cone size to the same extent as full-length CRMP4, indicating that the activity of CRMP4 toward microtubules is critical for maintaining axon outgrowth and growth cone size.
FIGURE 8.
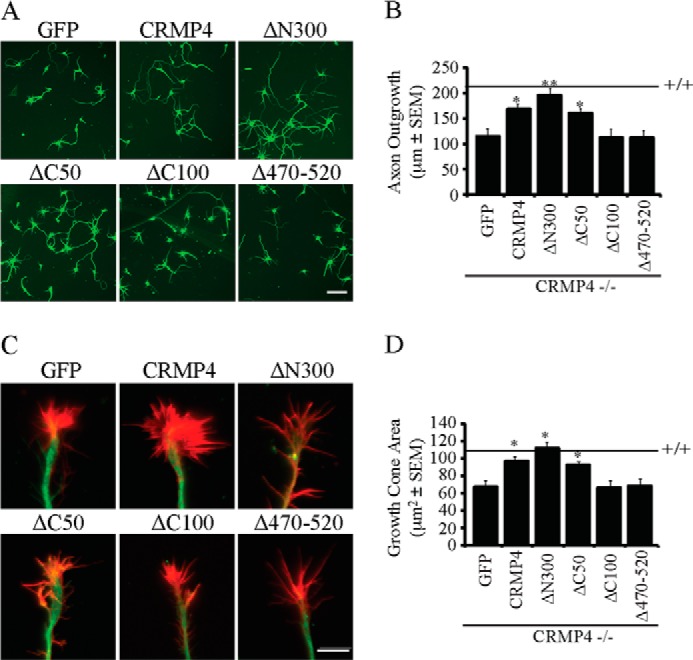
CRMP4 activity toward microtubules promotes growth cone expansion and axon outgrowth. A and C, hippocampal neurons from CRMP4−/− mice transduced with GFP or CRMP4 rescue constructs and stained with anti-βIII-tubulin (A) or anti-βIII-tubulin and rhodamine phalloidin (C). Scale bar, 25 (A) and 5 μm (C). B and D, quantitation of axon outgrowth per cell (B) and growth cone area per cell (D) for cultures derived from CRMP4−/− mice and transduced with GFP or CRMP4 rescue constructs. Values are mean ± S.E. and are from three experiments performed on 30 cells per experiment. Statistical analysis was performed by one-way ANOVA with a Tukey's post hoc test compared with GFP control. *, p < 0.05; **, p < 0.01. Error bars represent S.E.
DISCUSSION
Growth cone motility and morphology are controlled through regulatory and structural interactions between the actin and microtubule cytoskeleton. Here, we demonstrate that CRMP4 promotes both microtubule assembly and actin bundling both in vitro and in neuronal growth cones. CRMP4 activity toward microtubules regulates growth cone size and neurite outgrowth, whereas CRMP4 activity toward actin bundling is important for filopodial extension and may be more important for acute growth cone rearrangements in response to guidance cues. These findings identify CRMP4 as one of the few identified proteins that are important for physiologically regulating both microtubule and actin elements.
CRMP4 Promotes Growth Cone Spreading and Axon Outgrowth
We found that CRMP4 loss of function limited axon outgrowth and that this can be attributed to its effect on microtubules. This finding is consistent with a report that CRMP4 overexpression promotes axon extension from hippocampal neurons (20). The selective effect of CRMP4 loss of function on axon growth is reminiscent of the ability of CRMP2 overexpression in hippocampal neurons to promote the formation of multiple tau-1-positive/MAP-2-negative axonal processes (42). However, the uniform distribution of CRMP4 throughout axon and dendritic processes in hippocampal neurons differs from that of CRMP2, which is enriched in the distal parts of growing axons and growth cones (42). The distribution of CRMP4 suggests additional roles in dendritic formation, and indeed, hippocampal neurons from CRMP4−/− mice exhibit enhanced dendritic branching in culture with no change in total dendritic length (29). In vivo, CA1 pyramidal neurons from CRMP4−/− mice exhibit a proximal bifurcation of their apical dendrites perhaps in part as a result of desensitization to Semaphorin3A signaling (29). The mild phenotype in the hippocampus of the CRMP4 −/− mice suggests that there is functional compensation that promotes hippocampal neurite outgrowth perhaps by other CRMP family members (29). A more extensive analysis of hippocampal development in the CRMP4 −/− mice will be necessary to reveal any early developmental defects. Intriguingly, adult sensory neurons from CRMP4−/− mice also become desensitized to myelin repellents, raising the possibility that CRMP4-dependent cytoskeletal rearrangements may also be critical for axonal responses to the environment following nerve injury (17, 28, 37, 50). Understanding how CRMP4 affects axon growth may suggest additional strategies for targeting CRMP4 for neuronal repair (28).
Previous studies have demonstrated that CRMP2 and CRMP4 interact with tubulin heterodimers, and here, we extend these observations to show that CRMP4 can promote microtubule assembly and that this activity is critical for its effect on axon extension (21). CRMP4 activity toward microtubules also rescued the growth cone collapse phenotype exhibited in CRMP4−/− neurons, suggesting that disrupted cytoskeletal architecture in the central domain of the growth cone is responsible for this collapse.
CRMP4 Regulates Actin and Microtubule Dynamics
Microtubule and actin dynamics must be intricately coordinated in growth cones to mediate axon extension and turning (51, 52). CRMP4 distribution is strongest in the microtubule-rich central region of the growth cone, consistent with its ability to regulate microtubule dynamics. The evidence suggests that its role in microtubule dynamics is fundamental to regulating growth cone morphology and neurite outgrowth. It is striking that within the central domain of the growth cone CRMP4 was distributed in a rather punctate pattern. A punctate pattern of CRMP4L expression had been initially reported in dorsal root ganglion growth cones with partial colocalization with the synaptic vesicle marker SV2, leading to speculation that CRMP4 regulates exocytosis in the growth cone (15). Here, we posit that a population of CRMP4 may be decorating the tips of extending microtubules to facilitate interactions between the actin and microtubule cytoskeleton. The only phenotype that we found to be dependent on both the actin and microtubule activities of CRMP4 is EB3 comet extension. Although this does not seem to play a major role in growth cone morphology or neurite extension, it raises the possibility that this interaction may be important for local and acute rearrangements of the growth cone in response to guidance cues that signal through CRMP family members including Semaphorin 3A (29). This would be consistent with a critical role for microtubules and more specifically dynamic microtubules in mediating growth cone turning responses (51–54).
CRMP4 Regulates the Actin Cytoskeleton in Hippocampal Growth Cones
Although a population of CRMP1 and CRMP2 may also be distributed to actin-rich regions of the growth cone, the residues responsible for the F-actin bundling activity of CRMP4 are 35–59% identical to other CRMP family members and between 42 and 76% similar (23, 55). Thus, actin bundling activity may be unique to CRMP4. Furthermore, the CRMP4 isoform may be uniquely situated for F-actin bundling activity because it is the only isoform that can bind to RhoA, a critical regulator of cytoskeletal dynamics (28). The importance of CRMP4 activity toward actin in mediating filopodial extension provides the first demonstration that CRMP4 activity toward actin is physiologically relevant in neurons. The phenotype is consistent with the filopodial extension phenotype reported in E13 chick dorsal root ganglion neurons overexpressing the long isoform of CRMP4 (28). It is intriguing that the effects of CRMP4 overexpression on filopodia number were dependent on CRMP4 activity toward microtubules, whereas the effects on filopodial length were fully dependent on CRMP4 activity toward actin. This indicates that CRMP4 effects on microtubules regulate the genesis of filopodia, whereas its effects on actin promote filopodial extension. The phenotype differs from other actin-bundling proteins such as Fascin, which increases both the number and length of filopodia in growth cones (32, 49, 56). Recent studies of the F-actin protein Drebrin, which couples actin to microtubules through binding to EB3, have demonstrated that Drebrin phosphorylation regulates its interaction with actin to dynamically regulate coupling between the actin and microtubule networks. Bacterially expressed recombinant CRMP4 bundles actin, demonstrating that phosphorylation is not critical to its activity; nonetheless, CRMP4 is extensively phosphorylated, and its phosphorylation status regulates its interactions with both microtubules and RhoA, raising open questions about how physiological CRMP4 phosphorylation may impact growth cone dynamics (37, 41).
Acknowledgment
We thank Peter McPherson for comments on the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research and the McGill Program in Neuroengineering and by a Canada Research Chair (to A. E. F.).
- EB
- end-binding protein
- CRMP
- collapsin response mediator protein
- HSV
- herpes simplex virus
- DIV
- day(s) in vitro
- ANOVA
- analysis of variance
- CRMPL
- CRMP long isoform
- CRMPS
- CRMP short isoform.
REFERENCES
- 1. Vitriol E. A., Zheng J. Q. (2012) Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron 73, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hur E. M., Saijilafu, Zhou F. Q. (2012) Growing the growth cone: remodeling the cytoskeleton to promote axon regeneration. Trends Neurosci. 35, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee S., Kolodziej P. A. (2002) Short Stop provides an essential link between F-actin and microtubules during axon extension. Development 129, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 4. Prokop A., Uhler J., Roote J., Bate M. (1998) The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. J. Cell Biol. 143, 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S., Nahm M., Lee M., Kwon M., Kim E., Zadeh A. D., Cao H., Kim H. J., Lee Z. H., Oh S. B., Yim J., Kolodziej P. A., Lee S. (2007) The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development 134, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 6. Lee S., Harris K. L., Whitington P. M., Kolodziej P. A. (2000) short stop is allelic to kakapo, and encodes rod-like cytoskeletal-associated proteins required for axon extension. J. Neurosci. 20, 1096–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez-Soriano N., Travis M., Dajas-Bailador F., Gonçalves-Pimentel C., Whitmarsh A. J., Prokop A. (2009) Mouse ACF7 and Drosophila short stop modulate filopodia formation and microtubule organisation during neuronal growth. J. Cell Sci. 122, 2534–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alves-Silva J., Sánchez-Soriano N., Beaven R., Klein M., Parkin J., Millard T. H., Bellen H. J., Venken K. J., Ballestrem C., Kammerer R. A., Prokop A. (2012) Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). J. Neurosci. 32, 9143–9158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geraldo S., Khanzada U. K., Parsons M., Chilton J. K., Gordon-Weeks P. R. (2008) Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 10, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 10. Maniar T. A., Kaplan M., Wang G. J., Shen K., Wei L., Shaw J. E., Koushika S. P., Bargmann C. I. (2012) UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat. Neurosci. 15, 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hedgecock E. M., Culotti J. G., Thomson J. N., Perkins L. A. (1985) Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158–170 [DOI] [PubMed] [Google Scholar]
- 12. Goshima Y., Nakamura F., Strittmatter P., Strittmatter S. M. (1995) Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature 376, 509–514 [DOI] [PubMed] [Google Scholar]
- 13. Quach T. T., Duchemin A. M., Rogemond V., Aguera M., Honnorat J., Belin M. F., Kolattukudy P. E. (2004) Involvement of collapsin response mediator proteins in the neurite extension induced by neurotrophins in dorsal root ganglion neurons. Mol. Cell. Neurosci. 25, 433–443 [DOI] [PubMed] [Google Scholar]
- 14. Quach T. T., Mosinger B., Jr., Ricard D., Copeland N. G., Gilbert D. J., Jenkins N. A., Stankoff B., Honnorat J., Belin M. F., Kolattukudy P. (2000) Collapsin response mediator protein-3/unc-33-like protein-4 gene: organization, chromosomal mapping and expression in the developing mouse brain. Gene 242, 175–182 [DOI] [PubMed] [Google Scholar]
- 15. Quinn C. C., Chen E., Kinjo T. G., Kelly G., Bell A. W., Elliott R. C., McPherson P. S., Hockfield S. (2003) TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J. Neurosci. 23, 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L. H., Strittmatter S. M. (1996) A family of rat CRMP genes is differentially expressed in the nervous system. J. Neurosci. 16, 6197–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagai J., Goshima Y., Ohshima T. (2012) CRMP4 mediates MAG-induced inhibition of axonal outgrowth and protection against Vincristine-induced axonal degeneration. Neurosci. Lett. 519, 56–61 [DOI] [PubMed] [Google Scholar]
- 18. Lin P. C., Chan P. M., Hall C., Manser E. (2011) Collapsin response mediator proteins (CRMPs) are a new class of microtubule-associated protein (MAP) that selectively interacts with assembled microtubules via a Taxol-sensitive binding interaction. J. Biol. Chem. 286, 41466–41478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ong Tone S., Dayanandan B., Fournier A. E., Mandato C. A. (2010) GSK3 regulates mitotic chromosomal alignment through CRMP4. PLoS One 5, e14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cole A. R., Causeret F., Yadirgi G., Hastie C. J., McLauchlan H., McManus E. J., Hernández F., Eickholt B. J., Nikolic M., Sutherland C. (2006) Distinct priming kinases contribute to differential regulation of collapsin response mediator proteins by glycogen synthase kinase-3 in vivo. J. Biol. Chem. 281, 16591–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 22. Gu Y., Ihara Y. (2000) Evidence that collapsin response mediator protein-2 is involved in the dynamics of microtubules. J. Biol. Chem. 275, 17917–17920 [DOI] [PubMed] [Google Scholar]
- 23. Yuasa-Kawada J., Suzuki R., Kano F., Ohkawara T., Murata M., Noda M. (2003) Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur. J. Neurosci. 17, 2329–2343 [DOI] [PubMed] [Google Scholar]
- 24. Arimura N., Menager C., Fukata Y., Kaibuchi K. (2004) Role of CRMP-2 in neuronal polarity. J. Neurobiol. 58, 34–47 [DOI] [PubMed] [Google Scholar]
- 25. Wang L. H., Strittmatter S. M. (1997) Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J. Neurochem. 69, 2261–2269 [DOI] [PubMed] [Google Scholar]
- 26. Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 27. Rosslenbroich V., Dai L., Baader S. L., Noegel A. A., Gieselmann V., Kappler J. (2005) Collapsin response mediator protein-4 regulates F-actin bundling. Exp. Cell Res. 310, 434–444 [DOI] [PubMed] [Google Scholar]
- 28. Alabed Y. Z., Pool M., Ong Tone S., Fournier A. E. (2007) Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J. Neurosci. 27, 1702–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niisato E., Nagai J., Yamashita N., Abe T., Kiyonari H., Goshima Y., Ohshima T. (2012) CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus. Dev. Neurobiol. 72, 1447–1457 [DOI] [PubMed] [Google Scholar]
- 30. Tanaka H., Morimura R., Ohshima T. (2012) Dpysl2 (CRMP2) and Dpysl3 (CRMP4) phosphorylation by Cdk5 and DYRK2 is required for proper positioning of Rohon-Beard neurons and neural crest cells during neurulation in zebrafish. Dev. Biol. 370, 223–236 [DOI] [PubMed] [Google Scholar]
- 31. Morimura R., Nozawa K., Tanaka H., Ohshima T. (2013) Phosphorylation of Dpsyl2 (CRMP2) and Dpsyl3 (CRMP4) is required for positioning of caudal primary motor neurons in the zebrafish spinal cord. Dev. Neurobiol. 73, 911–920 [DOI] [PubMed] [Google Scholar]
- 32. Yang S., Huang F. K., Huang J., Chen S., Jakoncic J., Leo-Macias A., Diaz-Avalos R., Chen L., Zhang J. J., Huang X. Y. (2013) Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 288, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stepanova T., Slemmer J., Hoogenraad C. C., Lansbergen G., Dortland B., De Zeeuw C. I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. (2003) Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fournier A. E., Kalb R. G., Strittmatter S. M. (2000) Rho GTPases and axonal growth cone collapse. Methods Enzymol. 325, 473–482 [DOI] [PubMed] [Google Scholar]
- 35. Brewer G. J., Torricelli J. R., Evege E. K., Price P. J. (1993) Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567–576 [DOI] [PubMed] [Google Scholar]
- 36. Meijering E., Jacob M., Sarria J. C., Steiner P., Hirling H., Unser M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 [DOI] [PubMed] [Google Scholar]
- 37. Alabed Y. Z., Pool M., Ong Tone S., Sutherland C., Fournier A. E. (2010) GSK3β regulates myelin-dependent axon outgrowth inhibition through CRMP4. J. Neurosci. 30, 5635–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heo K., Ha S. H., Chae Y. C., Lee S., Oh Y. S., Kim Y. H., Kim S. H., Kim J. H., Mizoguchi A., Itoh T. J., Kwon H. M., Ryu S. H., Suh P. G. (2006) RGS2 promotes formation of neurites by stimulating microtubule polymerization. Cell. Signal. 18, 2182–2192 [DOI] [PubMed] [Google Scholar]
- 39. Hebert B., Costantino S., Wiseman P. W. (2005) Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 88, 3601–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paul C. A., Beltz B., Berger-Sweeney J. (2008) The nissl stain: a stain for cell bodies in brain sections. CSH Protoc. 2008, pdb.prot4805 [DOI] [PubMed] [Google Scholar]
- 41. Arimura N., Ménager C., Kawano Y., Yoshimura T., Kawabata S., Hattori A., Fukata Y., Amano M., Goshima Y., Inagaki M., Morone N., Usukura J., Kaibuchi K. (2005) Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol. Cell Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 43. Chae Y. C., Lee S., Heo K., Ha S. H., Jung Y., Kim J. H., Ihara Y., Suh P. G., Ryu S. H. (2009) Collapsin response mediator protein-2 regulates neurite formation by modulating tubulin GTPase activity. Cell. Signal. 21, 1818–1826 [DOI] [PubMed] [Google Scholar]
- 44. Brot S., Rogemond V., Perrot V., Chounlamountri N., Auger C., Honnorat J., Moradi-Améli M. (2010) CRMP5 interacts with tubulin to inhibit neurite outgrowth, thereby modulating the function of CRMP2. J. Neurosci. 30, 10639–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aylsworth A., Jiang S. X., Desbois A., Hou S. T. (2009) Characterization of the role of full-length CRMP3 and its calpain-cleaved product in inhibiting microtubule polymerization and neurite outgrowth. Exp. Cell Res. 315, 2856–2868 [DOI] [PubMed] [Google Scholar]
- 46. Rembutsu M., Soutar M. P., Van Aalten L., Gourlay R., Hastie C. J., McLauchlan H., Morrice N. A., Cole A. R., Sutherland C. (2008) Novel procedure to investigate the effect of phosphorylation on protein complex formation in vitro and in cells. Biochemistry 47, 2153–2161 [DOI] [PubMed] [Google Scholar]
- 47. Fu X., Brown K. J., Yap C. C., Winckler B., Jaiswal J. K., Liu J. S. (2013) Doublecortin (Dcx) family proteins regulate filamentous actin structure in developing neurons. J. Neurosci. 33, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Conde C., Cáceres A. (2009) Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 10, 319–332 [DOI] [PubMed] [Google Scholar]
- 49. Edwards R. A., Bryan J. (1995) Fascins, a family of actin bundling proteins. Cell Motil. Cytoskeleton 32, 1–9 [DOI] [PubMed] [Google Scholar]
- 50. Gordon-Weeks P. R., Fournier A. E. (2014) Neuronal cytoskeleton in synaptic plasticity and regeneration. J. Neurochem. 129, 206–212 [DOI] [PubMed] [Google Scholar]
- 51. Buck K. B., Zheng J. Q. (2002) Growth cone turning induced by direct local modification of microtubule dynamics. J. Neurosci. 22, 9358–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williamson T., Gordon-Weeks P. R., Schachner M., Taylor J. (1996) Microtubule reorganization is obligatory for growth cone turning. Proc. Natl. Acad. Sci. U.S.A. 93, 15221–15226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Challacombe J. F., Snow D. M., Letourneau P. C. (1996) Actin filament bundles are required for microtubule reorientation during growth cone turning to avoid an inhibitory guidance cue. J. Cell Sci. 109, 2031–2040 [DOI] [PubMed] [Google Scholar]
- 54. Challacombe J. F., Snow D. M., Letourneau P. C. (1997) Dynamic microtubule ends are required for growth cone turning to avoid an inhibitory guidance cue. J. Neurosci. 17, 3085–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Higurashi M., Iketani M., Takei K., Yamashita N., Aoki R., Kawahara N., Goshima Y. (2012) Localized role of CRMP1 and CRMP2 in neurite outgrowth and growth cone steering. Dev. Neurobiol. 72, 1528–1540 [DOI] [PubMed] [Google Scholar]
- 56. Cohan C. S., Welnhofer E. A., Zhao L., Matsumura F., Yamashiro S. (2001) Role of the actin bundling protein fascin in growth cone morphogenesis: localization in filopodia and lamellipodia. Cell Motil. Cytoskeleton 48, 109–120 [DOI] [PubMed] [Google Scholar]