Highlights
-
•
The novel peptide SL3 is a strong binder of the virulence-determining protein ESAT-6.
-
•
SL3 caused debilitating effects on mycobacterial growth and morphology.
-
•
SL3 led to accelerated clearance of M.tb and lowered immune response in a pre-clinical mouse model.
-
•
Microarray analysis of the mutant strain demonstrated a wide-scale transcriptional disruption caused by SL3 in M.tb.
Abbreviations: AMADID, agilent microarray design identifiers; CFU, colony forming units; MOI, multiplicity of infection; RD1, region of difference 1
Keywords: Antimycobacterial peptides, ESAT6, Mycobacterium tuberculosis, Protein–protein interaction
Abstract
Tuberculosis (TB) is a huge global burden, with new and resistant strains emerging at an alarming rate, necessitating an urgent need for a new class of drug candidates. Here, we report that SL3, a novel 33-amino acid peptide, causes debilitating effects on mycobacterial morphology. Treatment with SL3 drastically inhibits the growth of Mycobacterium tuberculosis in vitro as well as in a pre-clinical mouse model for M.tb infection. Microarray analysis of SL3-expressing strain demonstrates wide-scale transcriptional disruption in M.tb. We therefore believe that SL3 and similar peptides may herald a new approach towards discovering new molecules for TB therapy.
1. Introduction
Mycobacterium tuberculosis is remarkably skillful in surviving inside host macrophages and altering the macrophagial gene expression for its own benefit. This involves an intricate networking of crucial protein–protein interactions [1–3]. One interesting approach for combating tuberculosis is to target mycobacterial proteins and virulence factors [4]. One key protein in mycobacterial pathogenesis is the virulence determinant protein ESAT-6 that is encoded by region of difference 1 (RD1), the region absent from Bacillus Calmette–Guérin (BCG) and many attenuated strains of M.tb [5–8]. The ∼9.5 kb RD1 encodes for ESAT-6, its cognate binder CFP10, and a secretion system [9]. ESAT-6 is a potent human T cell antigen and a putative vaccine candidate that also contributes to virulence, mediates cellular cytolysis, and inhibits immune response by host cells [10,11]. In addition, ESAT-6 reduces the antigen presenting efficiency of macrophages by decreasing IFN-γ induced expression of MHC class II [12]. Due to its helix-turn-helix structure and hydrophobic nature, ESAT-6 possesses membrane lytic activity, an attribute that helps mycobacteria escape phagolysosome and disperse [13]. Additionally, ESAT-6 has also been postulated to play a role in macrophagial aggregation and granuloma formation [14]. Impeding ESAT-6 and its interactions may therefore provide anti-tubercular therapeutics.
ESAT-6 forms a 1:1 heterodimeric complex with CFP10 and, once secreted, dissociates under phagosomal acidic conditions [15,16]. Previously, we had shown that a 33 amino acid peptide, SL3, isolated from lung cDNA library, binds ESAT6 potently [17]. Our aim was to sequester free ESAT-6 such that it is rendered incapable in performing its functions. We report here that SL3 (amino acid sequence: AAARIRHEGVFLLIGNSCFSLPRNGPQLLLLAW∗), both exogenously and endogenously, causes debilitating effects on mycobacterial morphology and extra-as well as intracellular growth. Effects of SL3 were also studied in a mouse model of TB. We observed that SL3 led to an accelerated clearance of M.tb from lungs and spleen. We therefore believe that the present study opens up a new path for peptide-based anti-TB therapeutics and merits further exploration.
2. Materials and methods
2.1. Effect of SL3 on in vitro growth
M.tb strains H37Rv/SL3 (expressing SL3-His6X endogenously; cloning described in Supplementary information), H37Rv/pVV16 (possessing only the plasmid control), ΔRD1 (RD1-deficient H37Rv mutant), H37Rv + SL3 (SL3 peptide added exogenously to mycobacterial cultures) and H37Rv + DMSO (vehicle control) were inoculated in triplicates in vitro and growth recorded spectrophotometrically for 18 days at 630 nm as described earlier [18]. SL3-His6X peptide (GenScript, Hong Kong) was added to 7H9 medium exogenously (H37Rv + SL3) at a final concentration of 10 μg/ml. ΔRD1, H37Rv + DL1 (addition of an unrelated peptide at same concentration; sequence provided in Supplementary Fig. 1a) and H37Rv + DMSO (addition of equivalent amount of DMSO, used to dissolve SL3-His6X peptide) were used as controls. Due to precipitation of the peptide SL3-His6X at concentrations >10 μg/ml, higher concentrations could not be used. Another ESAT6 binding peptide, HCL2 – part of a separate study, was also analyzed for its effects on mycobacterial growth during this experiment (unpublished results).
2.2. Electron microscopy and colony morphology
Effects of SL3 on M.tb cellular morphology was determined by Transmission Electron Microscopy as described earlier [18]. Colony morphology of H37Rv/SL3 strain was also observed and compared with control H37Rv. Photographs of representative colonies were taken using a Panasonic 14 Megapixel Camera.
2.3. Infection of THP-1 cells by M. tuberculosis in presence of SL3 peptide
A previously described experimental protocol [19] has been briefed in Supplementary information. For intracellular survival studies, the ESAT-6 binder HCL2 – part of a separate study – was analyzed alongside (unpublished results).
2.4. In vivo immune response studies
2.4.1. Mice
BALB/c female mice at 6–8 weeks of age were used throughout this study following institutional ethical committee guidelines. All animal experiments were conducted in accordance with guidelines approved by the Institutional Animals Ethics Committee of ICGEB, New Delhi, India and Department of Biotechnology (DBT), Government of India, also specifically approved the study. Mice were housed under barrier conditions in a Biosafety Level III laboratory.
BALB/c mice were infected with ∼110 CFU of H37Rv and H37Rv/SL3 using an aerosol chamber. Mice were sacrificed at different time points and cytokine profile and T lymphocytes proliferation were assessed as described earlier [20]. For CFU counts lung and spleen were harvested at different time points and processed as described previously [20].
2.5. Total RNA isolation and microarray analysis
Total RNA was isolated using a protocol described previously [21] as detailed in Supplementary information. Custom H37Rv 8x15k array designed by iLifediscoveries Ltd. (Agilent microarray design identifiers [AMADID] 033693; Agilent) were used with two-color labeling and oligonucleotide probe length of 60mers. The total number of probes used was 15,744. Hybridization was carried out for 16 h, at 10 rpm and 65 °C. Agilent DNA Microarray Scanner was used for scanning. Microarray results were verified by using RT-PCR analysis, as described in Supplementary information.
2.6. Statistical analysis
All experiments were repeated thrice and in triplicates. Mean values were calculated with standard deviation (STDEV) unless stated otherwise. Student’s T-test was performed to compare two groups; p < 0.05 was considered significant.
3. Results
3.1. Endogenous expression of SL3 shows bacteriocidal effects
The expression of SL3 peptide was confirmed by C-terminal GFP-tag fusion (pVVGFPHis6X) inside M.tb H37Rv cells followed by fluorescence microscopy (Fig. 1a). A significant decrease was observed in mycobacterial growth in the presence of SL3. Endogenously expressed peptide reduced the growth by as much as 45% (indicated by blue dotted line) (Fig. 1b), thus indicating the antimycobacterial nature of SL3. H37Rv/pVV16, H37Rv/GFP and ΔRD1 controls displayed the normal growth pattern. As indicated by the electron micrographs in Fig. 1c, M.tb H37Rv/SL3 cells showed clear disintegration of mycobacterial cell wall and change in cell shape (indicated by arrows). Moreover, H37Rv/SL3 colony morphology appeared smooth as compared with control H37Rv (Fig. 1d). Cellular and colony morphology studies on SL3 expressing H37Rv suggested that, in addition to ESAT-6, SL3 might also be involved in binding to other cytoplasmic and cell wall proteins. In addition, intracellular survival of H37Rv/SL3 inside THP-1 cells was drastically reduced after 72 h as compared with control strains H37Rv/pVV16, H37Rv/GFP and ΔRD1 (Fig. 1e), suggesting potential SL3 activity on phagocytosed bacteria.
Fig. 1.
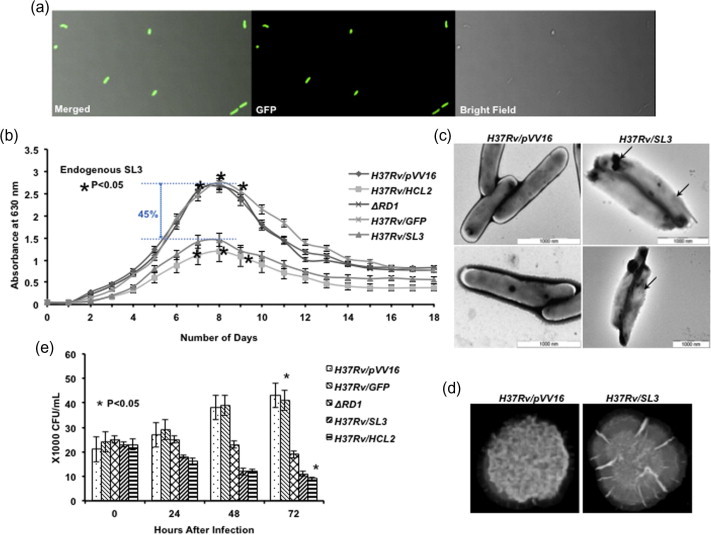
Endogenous expression of SL3 shows bacteriocidal effects. (a) Expression of peptide SL3-His6X in H37Rv cells. (b) Significant reduction was observed in mycobacterial growth in the presence of endogenous SL3. Growth reduction was found to be statistically significant (Student’s T-test, ∗p < 0.05). (c) Transmission electron micrographs of mycobacterial cells showing effects of endogenous SL3 on cell wall integrity and cell shape. (d) Representative colony images depicting change in colony surface. (e) Significant reduction was found in mycobacterial count inside THP-1 cells in the presence of endogenous SL3. All experiments were conducted in triplicates and repeated at least thrice. (t = 0) Defines the time at the beginning of phagocytosis. All readings were found to be statistically significant by applying Student’s T-test (∗p < 0.05).
3.2. SL3 induces bacterial killing
A major limitation in the development of new drugs against M.tb is the entry of the molecule through the mycolic acid cell wall so as to reach its target, making the screening for new antimycobacterial compounds an arduous and stringent exercise [22]. ESAT-6, however, is a secretory protein and SL3 does not need to penetrate the cell wall in order to reach its target. Moreover, the bacteriocidal potential of SL3 remained unchanged when administered exogenously. A 30% (in comparison to vehicle/DMSO control; blue dotted line) and 46% (in comparison to wt H37Rv; red dotted line) reduction in growth was observed when SL3 was added to M.tb culture media, while H37Rv + DL1 showed uninterrupted growth and H37Rv + DMSO demonstrated minor dip in growth due to DMSO (Fig. 2a). Similarly, CFU counts for studies where SL3 was added exogenously showed a drastic reduction in M.tb intracellular survival (Fig. 2b). Any possible cytotoxic effects of SL3 on THP-1 cells were studied by MTT assay at 0, 24, 48 and 72 h after addition of 10 μg/ml SL3 peptide. No cytotoxicity was observed (data not shown). It should also be pointed out that in presence of exogenous SL3, H37Rv + SL3 cells appeared completely devoid of cell wall (indicated by arrows) (Fig. 2c).
Fig. 2.
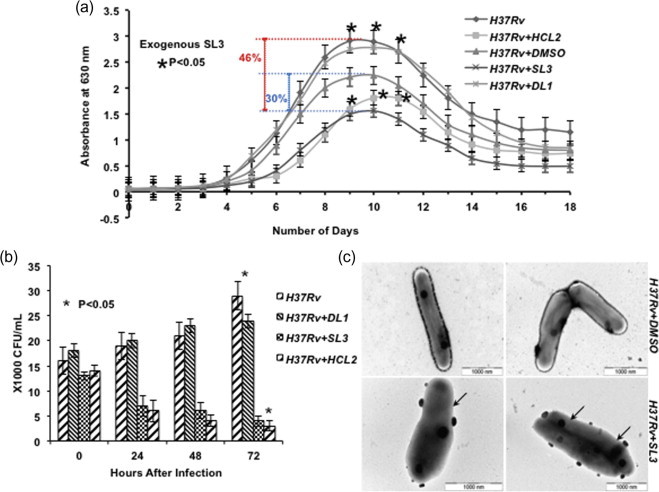
SL3 peptide addition induces bacterial killing. (a) Significant reduction was observed in mycobacterial growth in the presence of exogenous SL3. Growth reduction was found to be statistically significant (Student’s T-test, ∗p < 0.05). (b) Significant reduction was found in mycobacterial count inside THP-1 cells in the presence of exogenous SL3 added to RPMI media. All experiments were conducted in triplicates and repeated at least thrice. (t = 0) defines the time at the beginning of phagocytosis. All readings were found statistically significant by applying Student’s T-test (∗p < 0.05). (c) Transmission electron micrographs of mycobacterial cells showing effects of exogenous SL3 on cell wall integrity and cell shape (indicated by arrows).
3.3. M. tuberculosis H37Rv/SL3 establish infection
We observed a significant reduction in bacterial burden in animals that were infected with H37Rv/SL3 compared with that of control H37Rv infected animals. A complete clearance of M.tb was registered within 10–12 days post-infection (data not shown). We further analyzed immunological parameters in these animals. At 10 days post-infection, in vivo proliferation of immune cells was assessed in H37Rv/SL3 infected animals. It was observed that numbers of CD4, NK1.1 and Sca1+ were reduced compared to control animals that were infected with H37Rv (Supplementary Fig. 1b). However, there was no significant difference with regard to CD8+ T cells (Supplementary Fig. 1b). CD11B and CD11C levels were found to be higher in H37Rv/SL3 infected mice as compared to control mice (Supplementary Fig. 1b). At 10 days post-infection, reduced Alveolar cell proliferation and CD4+ T cell proliferation was observed in whole lung T cells by BrdU incorporation in H37Rv/SL3 infected mice (Supplementary Fig. 1c and d). CD69 expression in CD4+ T cell populations was assessed to determine the early activation of T helper subset and a significantly lower expression was found in case of mice infected with H37Rv/SL3 (Supplementary Fig. 1e). To check for possible SL3 cytotoxicity in host cells we performed the viability test in mice lung lymphocytes. No toxicity was observed (Supplementary Fig. 2a). To check the status of regulatory T cells in lungs of H37Rv/SL3 infected mice, we gated CD4+CD25+ T cells and studied the percentage of FoxP3 expression and found lowered regulatory T cells as compared to control mice (Supplementary Fig. 2b).
To check the cytokine milieu in H37Rv/SL3 and H37Rv infected mice, we isolated the cells from lungs and cultured them with Phorbol-12-myristate-13-acetate (PMA), Ionomycin and M.tb lysate/complete soluble antigen (CSA) in appropriate concentrations overnight. Subsequently, after 6 h of Brefeldin A (BFA) treatment, we carried out intracellular staining for IL-4, IFN-γ, IL-17, IL-9, IL-12, TNF-α, IL-6, TGF-β and IL-22. We found no difference in IL-22 and TNF-α, but obtained a significant downregulation of IL-4, IFN-γ, IL-17, IL-9, IL-12, IL-6 and TGF-β secretion in H37Rv/SL3 infected mice (Supplementary Fig. 2c). Almost all the immune parameters, irrespective of their types, were reduced in H37Rv/SL3 infected animals suggesting that the strain is poorly immunogenic, which might be because of faster depletion of antigens. Nonetheless, it is clear that introduction of such a peptide imparts anti-mycobacterial activities by accelerated antigen clearance and cell wall disintegration even though it possesses little or no immunomodulatory activity.
3.4. Effect of SL3 on global gene profile of M. tuberculosis
Microarray experiments were performed to study the effects of SL3 on mycobacterial growth and cellular morphology. To identify the genes that were differentially expressed in presence of SL3, gene expression profiles of M.tb expressing SL3 (H37Rv/SL3) and wild type H37Rv were compared. Genes having a fold change >1.5 and a p value <0.05 were considered significant. Detailed results have been deposited at NCBI’s Gene Expression Omnibus [23] and are accessible through GEO: GSE51085 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51085).
We found differential expression of 1025 genes, of which 716 were found to be significantly upregulated and 309 genes significantly downregulated (please see Supplementary Fig. 3a). The differentially expressed genes were compared with the known essential genes for in vitro growth as well as growth within macrophages, and with genes related to ESX1 secretion system [24,25]. Out of the 716 upregulated genes, 125 were found to be essential for mycobacterial growth and survival while 2 were essential as well as involved in ESX1 secretion system (Fig. 3a). On the other hand, out of the 309 downregulated genes, 83 were found to be essential for mycobacterial growth and survival, 2 were involved in ESX1 secretion system while 2 were essential as well as involved in ESX1 secretion system (Fig. 3b). Detailed lists for genes in each subset of Venn diagram have been provided in Supplementary Table 1. Out of the 127 upregulated essential genes, 64 genes code for uncharacterized and putative proteins, 32 for bacterial metabolism, 4 for ribosomal proteins, 8 are involved in mycobacterial biosynthetic pathways, 2 in mycobacterial secretion system, 2 chaperones and various other proteins involved in t-RNA synthesis, translation and transcriptional regulation, 3 in DNA repair, conserved membrane proteins and transporters. Essential genes that were downregulated contain 33 putative and uncharacterized genes, 2 genes coding for bacterial biosynthetic pathways, 2 coding for gyrase and helicases, 5 for ribosomal proteins, 17 for enzymes involved in bacterial metabolism, 7 genes are involved in mycobacterial secretion system, 4 code for proteins that are part of the proteosomal degradation system, 5 are involved in oxidative phosphorylation, 2 for chaperones and other proteins involved in t-RNA synthesis, translation, transcription, DNA replication and repair (Supplementary Table 1).
Fig. 3.
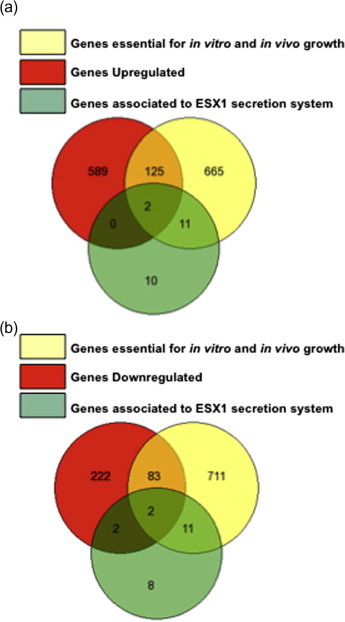
SL3 alters M. tuberculosis gene expression profile. (a) Comparison with Rubin’s lists and a list of ESX1 secretion system associated genes with genes that were found to be upregulated in microarray experiments. (b) Comparison with Rubin’s lists and ESX1 secretion system associated genes with genes that were found to be downregulated in microarray experiments.
3.5. Quantitative PCR
To validate our microarray results, we carried out real time PCR analysis of some of the identified genes. The genes chosen for such analysis were acpM (Rv2244) and cfp10 (Rv3874). The 16S rRNA gene was used as an internal control to compare the relative expression levels of these genes between treated (H37Rv/SL3) and control (H37Rv/pVV16) samples. Both the genes showed corresponding fold change values in RT-PCR experiment comparable to microarray results (Supplementary Fig. 3b). acpM and cfp10 demonstrated fold change values of −1.81 and −1.91 respectively in microarray experiment.
4. Discussion
ESAT-6 is a potent virulence determinant and an ideal target molecule to combat TB [10,13,14,26,27,11]. Previously, we reported that SL3, a short fragment from human lung cDNA library, interacts strongly with ESAT-6 possibly through hydrophobic interactions, such that even CFP10, the natural partner of ESAT-6 is not able to displace SL3 once the latter is bound to ESAT-6 [17].
In the present study, we have shown that SL3 inhibits the growth of M.tb in axenic cultures. We used two approaches to investigate the effect of SL3: endogenous expression and exogenous addition of the peptide to growing cultures. Growth curve analysis revealed a significant reduction in mycobacterial growth under both conditions. As demonstrated in our previous studies [18], ESAT-6 binders interfere with cellular morphology and cell integrity of the bacteria. In agreement with this, our electron microscopy data showed distinct alteration in cell morphology (Fig. 1c and 2c). Likewise, mycobacteria expressing SL3 formed a distinctly smoothened colony. However, expression or addition of SL3 to Escherichia coli cultures had no effect on bacterial growth and cellular integrity (data not shown). These results could be indicative of a major shift in mycobacterial secretory and cell wall biosynthetic pathways, leading to observed morphological defects brought about by the presence of SL3. Furthermore, SL3 addition to Mycobacterium smeg culture did not show any significant differences in growth pattern during the onset of log phase with some growth deprivation in late growth and stationary phase (Supplementary Fig. 3c). As previously hypothesized, proteins secreted by ESX-1 system of M. smeg might coat the bacterial cell surface thereby interfering with formation of specific cell–cell contacts [28]. It appears that M. smeg ESX-1 system ascertains a prominent role in the late log phase/early stationary phase, which is when the suppressive role of SL3 peptide on M. smeg is realized. However, this correlation requires better understanding of the working mechanism as well as role of M. smeg ESX-1 system that we are lacking currently. As M.tb is an intracellular pathogen, it is important to validate anti-mycobacterial activity of SL3 on the phagocytosed pathogen inside host macrophagial cells. We found that SL3 hinders survival of the M.tb inside human macrophagial THP-1 cells (Figs. 1e and 2b).
These results provided a basis for studying the anti-tubercular effects of SL3 in a preclinical mouse model system. We examined the mycobacterial survival and immune response in BALB/c mice infected with mycobacteria expressing SL3. Remarkably, SL3 expressing strain was cleared rapidly leading to antigenic loss as evident by the lack of immune response activation. We found reduced numbers of CD4, NK1.1 and Sca1+ and lowered CD69 expression in CD4+ populations in H37Rv/SL3 infected mice. SL3 was found to be non-toxic in mice lung lymphocytes and a lowered Treg cell population confirmed that lack of immune response is not because of regulatory T cells. A similar downregulation was observed in the protective cytokine production in H37Rv/SL3 infected mice. These data suggests that the expression of SL3 in H37Rv attenuates its virulence and reduces its ability to mount an immune response comparable to that of the virulent H37Rv strain due to early clearance of H37Rv/SL3 strain.
The results were initially intriguing, since ESAT-6 is not one of the factors that control mycobacterial growth [27]. Microarray analysis was performed to screen differentially expressed genes in the presence of SL3. We found differential expression of 1025 genes, of which 716 were significantly upregulated and 309 genes significantly downregulated (Supplementary Fig. 3a). Previous studies have identified sets of genes essential for in vitro growth of M.tb as well as for survival within macrophages [24,25]. We found a number of crucial mycobacterial genes upregulated in the presence of SL3, including genes coding for metabolic enzymes like Rv0118c, cysN/cysC, Rv1338, Rv2996c, Rv3540c, chaperones and DNA repair enzymes (clpB, dnaJ 2, Exodeoxyribonuclease III, Uracil-DNA and UvrD/Rep helicase). Importantly, we found that SL3 expression led to upregulation of mycobacterial peptidoglycan and mycolyl-arabinogalactan-peptidoglycan complex biosynthesis pathways (aftA and Glycosyl transferase genes, mycolyl-arabinogalactan–peptidoglycan complex biosynthesis genes, Rv2981c, Rv2153c and Rv2155), in addition to mycolate biosynthesis pathway (PCAA). This upregulation in the biosynthesis of cell wall components can be correlated with the electron-micrographs. M.tb affected by SL3 appears to be compensating for the loss of cell wall by making these components in excess. Similar observations have been made earlier with Staphylococcus aureus in response to cell wall-active antibiotics [29]. SL3 also upregulated integral membrane protein genes responsible for aminoglycosides/tetracycline-transport, as also EspA, that is essential for M.tb growth and associated with the ESX-1 secretion system [30]. Furthermore, we also found several genes downregulated, including genes coding for metabolic enzymes like rfbA, fadD30, hemC, ribA2, pgk, Transketolase Tkt, acn, hisC1, hisB, ACPM, pyrH, 2 genes involved in mycolic acid biosynthesis (Rv2361c and glfT1) and to our surprise, genes coding for proteins involved in DNA replication like gyrB, dnaB (Rv0058) and polA (Rv1629). This could be the reason behind the growth inhibition in the presence of SL3. SL3 also affected expression of factors involved in protein synthesis or translation, like the elongation factor Tuf, initiation factor InfC, translation initiation factor InfB, and initiation factor InfA, an observation that might explain the altered cellular and colony morphology of mutant M.tb. The major effect of SL3 seemed to be on secretion pathways of M.tb. SL3 expression repressed the genes coding for Rv1410c, TatC and Rv3921c proteins from the bacterial secretion system; EccA1 and PPE68 (essential for growth and associated with ESX1 secretion system); and EspF and ESAT-6 cognate binder CFP10 (non-essential but associated with ESX1 secretion system). Microarray data suggests that SL3 could hinder expression and secretion of crucial mycobacterial proteins that might be responsible for reduced pathogenesis of H37Rv. However, whereas SL3 showed adverse effects on the growth of M.tb, it had no visible effect on the growth of E. coli, strengthening our belief that SL3 is interacting with molecule(s) specific to the mycobacterial cell.
In conclusion, this study demonstrates adverse effects of SL3 on in vitro as well as in vivo growth and pathogenesis of M.tb, and concomitant lack of immune response. We believe that this study, that demonstrates the ability of a novel peptide to affect major mycobacterial cellular processes, will provide the basis for further exploration of SL3 functions and its implications in designing a host of peptide-based inhibitors against M.tb.
Disclosure
The authors declare no financial or commercial conflict of interest.
Acknowledgements
We thank Akash Saini for help in electron microscopy experiments and Prof. Belisle for the kind gift of pVV16 mycobacterial shuttle vector. The use of ICGEB TCAF bio-safety lab is gratefully acknowledged. For the study presented here ICGEB internals grants were utilized.
Appendix A. Supplementary data
Supplementary Figure 1.
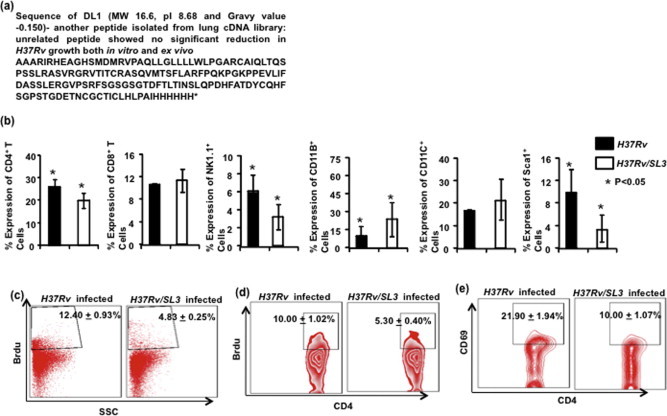
(a) Sequence of control peptide DL1 used as an unrelated peptide control’ in this study. (b) The percentage of Lung T lymphocytes in H37Rv/SL3 and H37Rv infected mice expressing CD4, CD8, Sca1 and NK1.1 is shown in the bar diagram with mean ± STDEV and Student’s T-test (∗p < 0.05). Increased number of Professional APCs, cells expressing CD11B, and CD11C in H37Rv/SL3 infected mice compared to H37Rv infected mice at day 10 after infection, with mean ± STDEV and Student’s T-test (∗p < 0.05). (c and d) T cell proliferation in vivo by Brdu incorporation. Data shown here is representative of three independent experiments with three mice in each group and represents the mean values (∗p < 0.05). (e) The percentage of cells expressing CD69 among CD4+ T cells is shown in the dot-plot with mean having Student’s T-test (∗p < 0.05).
Supplementary Figure 2.
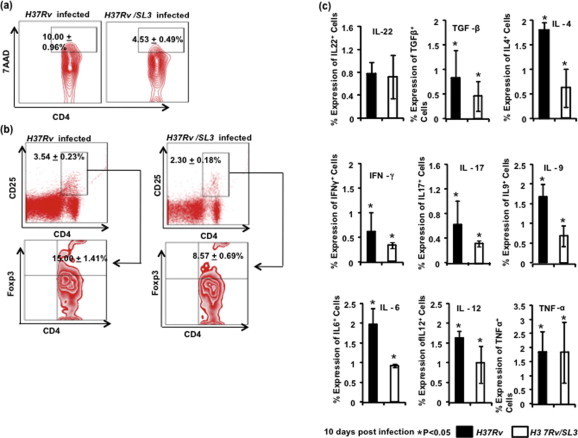
(a) Cell death in CD4+ lung lymphocytes in H37Rv/SL3 and H37Rv infected mice. Data showed in Contour plot with mean having Student’s T-test (∗p < 0.05). (b) The percentage of cells expressing CD25 among CD4+ T cells is shown in the dot-plot with mean having Student’s T-test (∗p < 0.05). CD4+CD25+ T cells gated population was taken to show Regulatory T cells. Data shown in Contour plot with mean having Student’s T-test (∗p < 0.05). (c) Total cytokine production by lung lymphocytes represented through a bar diagram. Data shown with mean ± STDEV; Student’s T-test was applied for estimating significance between two parameters (∗p < 0.05).
Supplementary Figure 3.
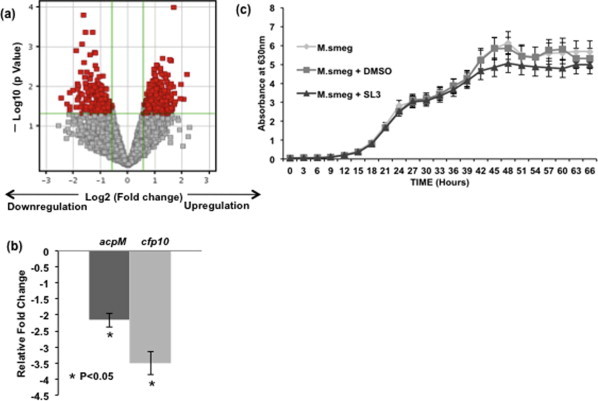
(a) Volcano plot depicting extent of up- and downregulated genes with SL3 expression along with fold change and p-value cut-offs. (b) Relative fold change in the transcript levels of ACPM (fold change: – 2.16) and CFP10 (fold change: – 3.51) genes in Real-time PCR experiment. (c) M. smeg, M. smeg +SL3 and M. smeg + DMSO were inoculated in triplicates in vitro and growth was recorded spectrophotometrically for 66 h at 630 nm. SL3 addition did not show any significant differences in growth pattern during the initial log phase.
This document file contains supplementary data.
(a) Breakdown of upregulated genes into (i) essentiality as defined by Rubin and co-workers and (ii) essential genes associated with the ESX1 secretion system (b) Breakdown of downregulated genes into (i) essentiality as defined by Rubin and co-workers, (ii) essential genes associated with the ESX1 secretion system and (iii) non-essential genes associated with the ESX1 secretion system.
References
- 1.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohde K.H., Veiga D.F.T., Caldwell S., Balázsi G., Russell D.G. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 2012;8:e1002769. doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang H., Chen A. Interaction of Mycobacterium tuberculosis ESAT6 protein with ADAM9 protein. Afr. J. Microbiol. Res. 2011;5:919–923. [Google Scholar]
- 4.Alksne L. Virulence as a target for antimicrobial chemotherapy. Expert Opin. Investig. Drugs. 2002;11:1149–1159. doi: 10.1517/13543784.11.8.1149. [DOI] [PubMed] [Google Scholar]
- 5.Mahairas G.G., Sabo P.J., Hickey M.J., Singh D.C., Stover C.K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr M.A., Wilson M.A., Gill W.P., Salamon H., Schoolnik G.K., Rane S., Small P.M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 7.Gordon S.V., Brosch R., Billault A., Garnier T., Eiglmeier K., Cole S.T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 8.Brodin P., Eiglmeier K., Marmiesse M., Billault A., Garnier T., Niemann S., Cole S.T., Brosch R. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 2002;70:5568–5578. doi: 10.1128/IAI.70.10.5568-5578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S.T., Brosch R., Parkhill J. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Barnes P.F., Dobos-Elder K.M., Townsend J.C., Chung Y.T., Shams H., Weis S.E., Samten B. ESAT-6 inhibits production of IFN-gamma by Mycobacterium tuberculosis-responsive human T cells. J. Immunol. 2009;182:3668–3677. doi: 10.4049/jimmunol.0803579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samten B., Wang X., Barnes P.F. Mycobacterium tuberculosis ESX-1 system-secreted protein ESAT-6 but not CFP10 inhibits human T-cell immune responses. Tuberculosis. 2009;89:S74–S76. doi: 10.1016/S1472-9792(09)70017-4. [DOI] [PubMed] [Google Scholar]
- 12.Kumar P., Agarwal R., Siddiqui I., Vora H., Das G., Sharma P. ESAT6 differentially inhibits IFN-gamma-inducible class II transactivator isoforms in both a TLR2- dependent and – independent manner. Immunol. Cell Biol. 2011;90:411–420. doi: 10.1038/icb.2011.54. [DOI] [PubMed] [Google Scholar]
- 13.de Jonge M.I., Pehau-Arnaudet G., Fretz M.M., Romain F., Bottai D., Brodin P., Honoré N., Marchal G., Jiskoot W., England P., Cole S.T., Brosch R. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkman H.E.V., Clay H., Beery D., Chang J.C., Sherman D.R., Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renshaw P.S., Panagiotidou P., Whelan A., Gordon S.V., Hewinson R.G., Williamson R.A., Carr M.D. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. J. Biol. Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- 16.Meher A.K., Bal N.C., Chary K.V., Arora A. Mycobacterium tuberculosis H37Rv ESAT-6–CFP-10 complex formation confers thermodynamic and biochemical stability. FEBS J. 2006;273:1445–1462. doi: 10.1111/j.1742-4658.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- 17.Tharad M., Samuchiwal S.K., Bhalla K., Ghosh A., Kumar K., Kumar S., Ranganathan A. A three-hybrid system to probe in vivo protein–protein interactions: application to the essential proteins of the RD1 complex of M. tuberculosis. PLoS One. 2011;6:e27503. doi: 10.1371/journal.pone.0027503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar K., Tharad M., Ganapathy S., Ram G., Narayan A., Khan J.A., Pratap R., Ghosh A., Samuchiwal S.K., Kumar S., Bhalla K., Gupta D., Natarajan K., Singh Y., Ranganathan A. Phenylalanine-rich peptides potently bind ESAT6, a virulence determinant of Mycobacterium tuberculosis, and concurrently affect the pathogen’s growth. PLoS One. 2009;4:e7615. doi: 10.1371/journal.pone.0007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar D., Nath L., Kamal M.A., Varshney A., Jain A., Singh S., Rao K.V. Genome wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Tousif S., Singh Y., Prasad D.V., Sharma P., Van Kaer L., Das G. T cells from programmed death-1 deficient mice respond poorly to Mycobacterium tuberculosis infection. PLoS One. 2011;6:e19864. doi: 10.1371/journal.pone.0019864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao A., Ram G., Saini A.K., Vohra R., Kumar K., Singh Y., Ranganathan A. Synthesis and selection of de novo proteins that bind and impede cellular functions of an essential mycobacterial protein. Appl. Environ. Microb. 2007;73:1320–1331. doi: 10.1128/AEM.02461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harth G., Horwitz M.A. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J. Exp. Med. 1999;189:1425–1435. doi: 10.1084/jem.189.9.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 25.Sassetti C.M., Rubin E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo S., Xue R., Li Y., Wang S.M., Ren L., Xu J.J. The CFP10/ESAT6 complex of Mycobacterium tuberculosis may function as a regulator of macrophage cell death at different stages of tuberculosis infection. Med. Hypotheses. 2012;78:389–392. doi: 10.1016/j.mehy.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Derrick S.C., Morris S.L. The ESAT6 protein of Mycobacterium tuberculosis induces apoptosis of macrophages by activating caspase expression. Cell. Microbiol. 2007;9:1547–1555. doi: 10.1111/j.1462-5822.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 28.Flint J.L., Kowalski J.C., Karnati P.K., Derbyshire K.M. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utaida S., Dunman P.M., Macapagal D., Murphy E., Projan S.J., Singh V.K., Jayaswal R.K., Wilkinson B.J. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 30.Fortune S.M., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document file contains supplementary data.
(a) Breakdown of upregulated genes into (i) essentiality as defined by Rubin and co-workers and (ii) essential genes associated with the ESX1 secretion system (b) Breakdown of downregulated genes into (i) essentiality as defined by Rubin and co-workers, (ii) essential genes associated with the ESX1 secretion system and (iii) non-essential genes associated with the ESX1 secretion system.


