Abstract
A 45-year-old man with dilated cardiomyopathy presented with acute leg pain and erythema suggestive of necrotising fasciitis. Initial surgical exploration revealed no necrosis and treatment for a soft tissue infection was started. Blood and tissue cultures unexpectedly grew a Gram-negative bacillus, subsequently identified by an automated broth microdilution phenotyping system as an extended-spectrum β-lactamase producing Escherichia coli. The patient was treated with a 3-week course of antibiotics (ertapenem followed by ciprofloxacin) and debridement for small areas of necrosis, followed by skin grafting. The presence of E. coli triggered investigation of both host and pathogen. The patient was found to have previously undiagnosed liver disease, a risk factor for E. coli soft tissue infection. Whole genome sequencing of isolates from all specimens confirmed they were clonal, of sequence type ST131 and associated with a likely plasmid-associated AmpC (CMY-2), several other resistance genes and a number of virulence factors.
Background
The majority of primary soft tissue infections are due to Gram-positive organisms—empiric therapy is tailored towards these pathogens. However, immunocompromised patients may be infected with atypical pathogens. This case illustrates that the presence of such a pathogen can lead to the identification of an underlying immunosuppressing condition.
Whole genome sequencing (WGS) is a technology which is relatively new to clinical microbiology and will form a much greater part of routine practice within the next decade. In this case, if available in ‘real time’, it would have provided sufficient information to guide appropriate antimicrobial treatment, as well as additional genetic information to facilitate epidemiological comparisons (strain typing, resistance gene and virulence gene identification). In this instance however, it was not possible to link phenotype (ie, soft tissue infection) with genotype, which is likely to reflect a combination of host and pathogen factors that are still to be explored in larger genetic association studies.
Case presentation
A 45-year-old self-employed businessman presented to the emergency department with a 12 h history of right leg pain, erythema and swelling. This was associated with fevers, rigors and feeling increasingly non-specifically unwell. The patient had not had any recent infections or injuries to the leg. He had an extensive cardiac history, with known dilated cardiomyopathy, atrial fibrillation and episodes of bradycardia, for which he had been fitted with a cardiac pacemaker. He suffered from chronic peripheral oedema despite therapy with spironolactone 25 mg, bumetanide 3 mg twice daily and amiloride 5 mg. He was also on ramipiril 5 mg, omeprazole 20 mg and warfarin with a target international normalised ratio of 2.5. He admitted to excess alcohol consumption of approximately 50 units per week. Despite his underlying medical conditions he was generally well prior to admission.
On examination there was marked erythema around superficial scratches on the medial aspect of the right thigh, with erythema extending below the knee. No surgical emphysema was observed. He was noted to have marked pitting oedema to the knees in both legs and changes in keeping with chronic venous stasis. He was febrile, markedly hypotensive with a blood pressure of 50/20 mm Hg and had a paced heart rhythm of 80 bpm. His chest was clear and his heart sounds were normal.
Owing to concerns about necrotising fasciitis, the patient was urgently reviewed by the plastic surgery team. Arrangements were made for immediate debridement in theatre and postoperative management on the intensive care unit (ICU) due to his resistant hypotension.
Investigations
Initial routine haematology and biochemistry results were as follows: C reactive protein >156 (mg/dL), Na+ 137 (mmol/L), K+ 3.6 (mmol/L), urea 10.3 (mmol/L), creatinine 155 (μmol/L), bilirubin 19 (μmol/L), alanine transaminase 18 (IU/L), alkaline phosphatase 149 (μmol/L), albumin 21 (g/L); haemoglobin 12.3 (mg/dL), platelets 191×109/L, leukocytes 14.3×109/L, neutrophils 13.16×109/L, lymphocytes 0.43×109/L.
Blood and tissue culture—ESBL-producing E. coli (7/7 samples)—see figure 1 for comparative antimicrobial susceptibilities across three different methods (disc diffusion direct from blood culture, BD Phoenix automated broth microdilution, and prediction from genomic data).
Figure 1.
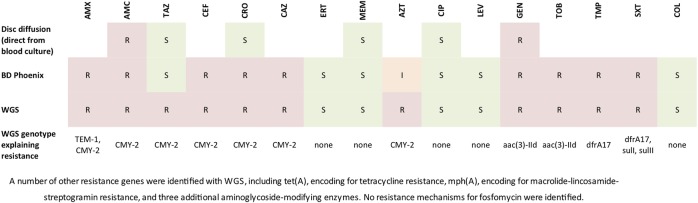
Antibiotic susceptibilities (phenotypic and predicted from genotype) of the identified pathogen, Escherichia coli.
Serological investigations: HIV antigen/antibody negative; Hepatitis B surface antigen negative; Hepatitis C antibody negative.
Liver ultrasound—18 cm liver with a smooth edge but coarse in echotexture, no biliary dilation, patent portal vein.
Upper gastrointestinal endoscopy—grade I oesophageal varix plus portal hypertensive gastropathy.
Genetic analysis of the E. coli—strains found to be of the ST131 multilocus sequence type. A number of resistance genes were present (figure 1), including the plasmidic AmpC, CMY-2. Several known virulence factors were also identified in the isolates, including fimbrial and other adhesins (papI, sfaC, dra, fim, nfa), genes encoding for iron uptake (chu, ent, fep), cellular invasion (ompA, traJ) and serum resistance (TraT).
Differential diagnosis
Necrotising fasciitis
Non-necrotising soft tissue infection
Treatment
The patient was resuscitated with intravenous fluids in the emergency department but his blood pressure remained low. He received empiric antimicrobial therapy in accordance with local guidelines for presumed necrotising fasciitis, namely coamoxiclav 1.2 g intravenously three times daily plus clindamycin 900 mg intravenously four times daily plus single dose gentamicin (5 mg/kg) intravenously.
Surgical exploration revealed no evidence of necrosis, with all muscle tissue appearing healthy. Fasciotomies were carried out to mitigate against possible future swelling and samples were taken for microbiological testing. The patient was transferred to intensive care for inotropic support due to his resistant hypotension. At this time, antibiotic therapy was changed to empiric therapy for a non-necrotising soft tissue infection: flucloxacillin 2 g intravenously four times daily plus a vancomycin infusion, the intention being to stop vancomycin once methicillin-resistant Staphylococcus aureus carriage was excluded on screening swabs.
The unexpected discovery of a Gram-negative bacillus in blood cultures late on the first day of his admission prompted a reassessment of his antibiotic therapy. The decision was made to broaden therapy back to the initial regimen of coamoxiclav and clindamycin pending further information.
The following day, the organism had been identified as E. coli and initial disc diffusion susceptibility testing suggested resistance to both coamoxiclav and gentamicin, but susceptibility to piperacillin-tazobactam and meropenem. A cautious approach was adopted given the atypical microbiology of the case, and broad-spectrum antibiotic therapy with meropenem 500 mg intravenously three times daily was administered pending confirmation of antimicrobial susceptibilities. At this time the patient remained in the ICU but was alert and not intubated. The E. coli was subsequently identified as ESBL-positive by the BD Phoenix automated system and the decision was made to continue antibiotic therapy with ertapenem 1 g intravenously once daily.
The patient improved and was discharged to the cardiac unit, and subsequently to the plastic surgery ward. His antibiotic therapy was converted to oral ciprofloxacin 500 mg twice daily to complete 3 weeks of therapy.
He required two further visits to theatre for wound exploration, where very small areas of superficial fat necrosis were debrided. He ultimately underwent a skin graft to repair the defects in his lower leg and was discharged from hospital with a vacuum dressing in situ.
Outcome and follow-up
The patient made a full recovery from the acute infective episode and was discharged from hospital with follow-up for wound and skin graft care. Additionally, arrangements were made for ongoing follow-up with the hepatologists for monitoring of his newly diagnosed portal hypertension.
Discussion
While the majority of spontaneous skin and soft tissue infections are due to Gram-positive organisms, patients with impaired immunity are at risk of infection with less common pathogens. E. coli is an uncommon but recognised cause of soft tissue infections in the immunocompromised. It has been reported in solid organ transplant recipients1 patients with haematological malignancies2 on haemodialysis3 and with cirrhosis.4 It is also well recognised in children with nephrotic syndrome.5
In this case the patient suffered from a complex E. coli soft tissue infection of the leg with a secondary bacteraemia but was not known to be immunocompromised. Testing for HIV was negative. Investigations during his admission revealed previously undiagnosed liver disease, likely due to a combination of excess alcohol and congestion due to his cardiac failure; he tested negative for hepatitis viruses. Peripheral oedema likely also contributed to his condition, and he had scratched and breached the skin on his thigh, possibly providing a portal of entry for the E. coli, which may have been inadvertently spread from the groin/perineal region. Empiric therapy for a soft tissue infection covering only Gram-positive organisms resulted in a delay to effective therapy, known to be associated with increased mortality. This case shows that broader empiric therapy is often necessary in immunocompromised patients; it also highlights the importance of investigating possible underlying immunosuppression in patients whose infections are found to be due to atypical pathogens.
WGS has been shown to be sensitive and specific in the prediction of antimicrobial resistance,6 7 and here determined the underlying genetic mechanisms of resistance. The disc diffusion susceptibility testing had failed to pick-up on third generation cephalosporin resistance, and the BD Phoenix misclassified the organism as an ESBL-producer. Both phenotypic methods identified the organism as piperacillin-tazobactam susceptible, when this would probably be inadequate for treatment given the presence of CMY-2.8
Isolates were ST131, a commonly identified global clinical lineage, frequently associated with antimicrobial resistance (although more commonly with CTX-M-15 and fluoroquinolone resistance), invasive infections and severe disease.9 The genotypic virulence profile was consistent with this lineage, with a large number of virulence factors identified.10
Learning points.
Common infectious syndromes may be caused by less common aetiological pathogens in immunocompromised hosts—the presence of such a pathogen in a ‘normal’ host should trigger investigation for causes of immunosuppression.
It is reasonable to give broad empirical antibiotic cover in severe infections in a debilitated host even when a particular group of organisms is most likely.
Whole genome sequencing can comprehensively characterise pathogens, including their virulence factors and resistance genes, and represents a more reliable ‘one-stop’ test which could be used ‘in real time’ for both clinical diagnostics and epidemiology. However, further large-scale genome-wide association studies would be needed to correlate clinical phenotypes with genotype.
Acknowledgments
Many thanks to Robert Newnham for his work preparing the isolates for sequencing; thanks also to Adam Giess for his assistance with genome sequence data analysis.
Footnotes
Contributors: RB wrote the clinical portion of the manuscript including performing the necessary literature searches. NS reviewed the whole genome sequence data and wrote the scientific portion of the manuscript. DC and ICJWB provided advice throughout the process and edited the manuscript prior to publication.
Competing interests: None.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Janny S, Bert F, Dondero F, et al. Fatal Escherichia coli skin and soft tissue infections in liver transplant recipients; a report of three cases. Transpl Infect Dis 2013;15:E49–53 [DOI] [PubMed] [Google Scholar]
- 2.Sunder S, Haguenoer E, Bouvet D, et al. Life threatening Escherichia coli cellulitis in patients with haematological malignancies. J Med Microbiol 2012;61:1324–7 [DOI] [PubMed] [Google Scholar]
- 3.Liu CT, Chen YC, Chen TH, et al. Necrotizing fasciitis of thigh associated with Escherichia coli bacteraemia in a patient on chronic hemodialysis. Hemodial Int 2012;16:564–7 [DOI] [PubMed] [Google Scholar]
- 4.Levy V, Reed C, Abbott SL, et al. Escherichia coli myonecrosis in alcoholic cirrhosis. J Clin Gastroenterol 2003;36:443–5 [DOI] [PubMed] [Google Scholar]
- 5.Asmar BI, Bashour BN, Fleischmann LE, et al. Escherichia coli cellulitis in children with idiopathic nephrotic syndrome. Clin Pediatr (Phila) 1987;26:592–4 [DOI] [PubMed] [Google Scholar]
- 6.Gordon NC, Price JR, Cole K, et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 2014;52:1182–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoesser N, Batty EM, Eyre DW, et al. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data .J Antimicrob Chemother 2013;68:2234–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009;22:161–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price LB, Johnson JR, Aziz M, et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 2013;4:e00377–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco J, Mora A, Mamani R, et al. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 2013;51:3358–67 [DOI] [PMC free article] [PubMed] [Google Scholar]


