Abstract
Artificial microRNA (amiRNA) technology offers highly specific and versatile gene silencing in diverse plant species. The principal challenge in amiRNA application is to select potent amiRNAs from hundreds of bioinformatically designed candidates to enable maximal target gene silencing at the protein level. To address this issue we developed the epitope-tagged protein-based amiRNA (ETPamir) screens, in which single or multiple target genes encoding epitope-tagged proteins are constitutively or inducibly co-expressed with individual amiRNA candidates in plant protoplasts. Accumulation of tagged proteins, detected by immunoblotting with a commercial tag antibody, inversely and quantitatively reflects amiRNA efficacy in vivo. The core procedure, from protoplast isolation to identification of optimal amiRNA, can be completed in 2-3 days. The ETPamir screens circumvent the widespread shortage of plant antibodies and the complexity of plant amiRNA silencing at target mRNA or/and protein levels. This method can be extended to verify predicted endogenous target genes for plant natural miRNAs.
Keywords: Plant artificial microRNA, Gene silencing, Plant microRNA, Target gene validation
INTRODUCTION
The rapidly expanding genomic information across the plant kingdom stresses an urgent need for reliable and versatile tools to decipher the functions of newly discovered genes and their regulatory networks. Determination of gene functions often requires examination of loss-of-function phenotypes. In the model plant Arabidopsis thaliana, T-DNA insertion lines represent the most important resource for loss-of-function mutants. Targeted genome editing tools, including zinc finger nucleases1, transcription activator-like effector nucleases2,3, and RNA-guided Cas9 endonucleases4–6, have recently opened up promising new avenues for generating targeted loss-of-function mutants for Arabidopsis genes lacking T-DNA insertion mutants and for genes in other plant species. However, lethality and complex long-term physiological and developmental consequences associated with stable mutants have imposed limitations in functional characterization of most genes essential for plant growth and reproduction. It is also more challenging to use T-DNA insertion mutants to study functionally redundant and physically linked genes in plant genomes7. The artificial microRNA (amiRNA)-based method for targeted gene silencing provides an invaluable alternative approach for conditional, reversible and multiplex control of gene activities for systematic functional genomic analyses in plants.
Targeted gene silencing in plant research has been obtained mostly by hair-pin RNAs (hpRNAs), amiRNAs and virus-induced gene silencing (VIGS). The amiRNA technology exploits the biogenesis and silencing machineries of natural miRNAs for silencing one or multiple genes of interest. A desired amiRNA can be easily generated using a native miRNA precursor (pre-miRNA) backbone by replacing its original mature miRNA sequence with a custom sequence that base-pairs with and triggers cleavage, decay or/and translational inhibition of target mRNAs of interest8–13. The homogeneity of a single silencing amiRNA produced by a pre-amiRNA and the prerequisite of a near-perfect complementarity between plant amiRNAs and target mRNAs ensure the superb silencing specificity of plant amiRNAs8–13, whereas hpRNAs and VIGS often exhibit off-target effects due to the unpredictable heterogeneity of the siRNAs produced. In addition, the amiRNA-targeted genes can be easily modified to resist amiRNA activities and then used for functional complementation in transgenic mutant plants with amiRNA-mediated gene silencing, to establish a solid genotype-phenotype correlation9,10.
Although manual design of plant amiRNAs is feasible14, the resourceful web-based miRNA designer (WMD) facilitates an automatic design of gene-specific amiRNA candidates for over 100 plant species with fully sequenced genomes or extensive databases of ESTs10. However, the in vivo silencing efficacy of individual amiRNA candidates can be highly variable10,11,15–18. This is largely owing to unpredictable factors, such as amiRNA expression and processing, target mRNA structure and accessibility, and effects of potential target mRNA binding proteins11,18,19. Therefore, optimal amiRNAs for gene silencing are not readily recognizable among dozens to hundreds of candidates in the WMD prediction list. Without rapid in vivo screen and quantitative evaluation of the performance of selected amiRNA candidates, tremendous time and labor investment in generating and screening amiRNA-expressing transgenic plants could lead to ineffective or partial rather than complete silencing of the target gene(s) at the protein level. Therefore, a facile and robust method for identifying optimal amiRNAs in a broad range of plant species will facilitate highly efficient gene silencing in plants and promote scientific advances and discoveries in plant research.
Development of the ETPamir screens
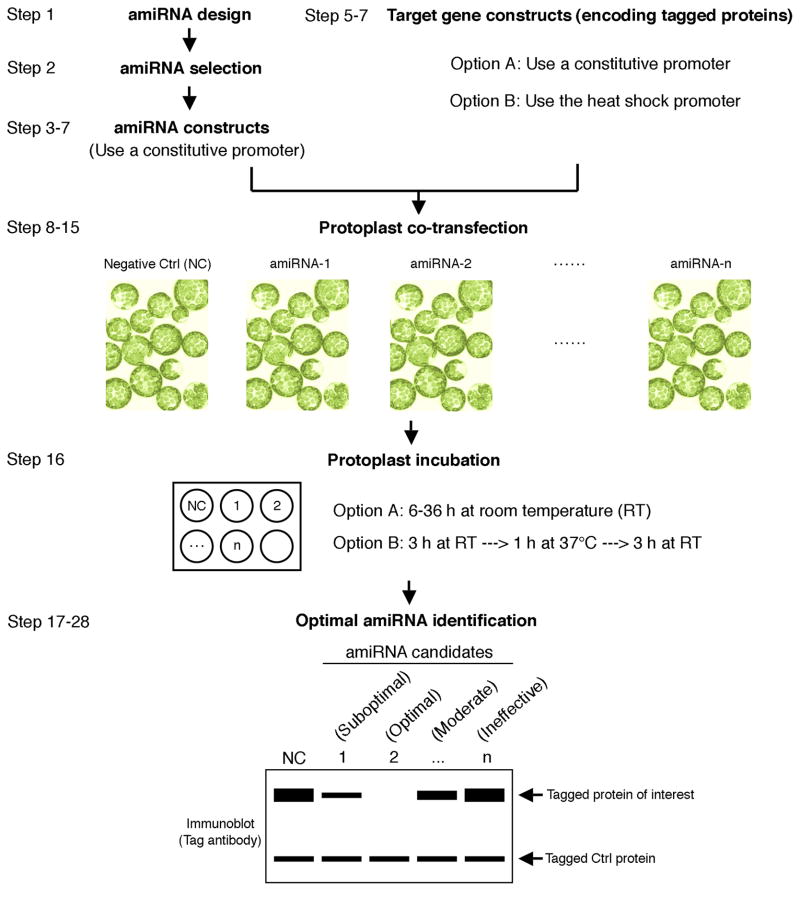
To pinpoint the most potent amiRNAs from bioinformatically designed candidates for silencing single or multiple target genes, we have developed a straightforward and widely adaptable method, the epitope-tagged protein-based amiRNA (ETPamir) screen11. Our strategy is to constitutively or inducibly co-express full-length target genes encoding epitope-tagged proteins with individual amiRNA candidates in plant mesophyll protoplasts, which are freshly isolated leaf cells lacking cell walls that support highly efficient DNA transfection20. Transfected protoplasts are incubated for sufficient time to allow each amiRNA to accumulate and exert its inhibitory effect on target mRNAs, through a combination of cellular mechanisms, to suppress the production of tagged proteins. This suppression is quantified by immunoblotting with the suitable tag antibody. One option for co-expression of amiRNA and its target gene(s) is to use a constitutive promoter to drive the expression of both. This option requires longer protoplast incubation time (e.g., 36 h) to determine the amiRNA efficacy, considering the turn-over time of the tagged proteins synthesized from escaped target mRNAs at the beginning of co-expression (i.e., in the absence of sufficient amiRNA activity). An alternative option is to allow sufficient amiRNAs to be produced under a constitutive promoter for 3 h before a 1 h heat induction of target mRNA expression, which is driven by the heat shock promoter. The amiRNA efficacy is then distinguishable after another 3 h of protoplast incubation. Using either option, the accumulation of tagged proteins from target mRNAs quantified by immunoblotting is inversely correlated with the in vivo silencing efficacy of each amiRNA. We have observed excellent consistency between the amiRNA efficacy determined by the ETPamir screen in protoplasts and its corresponding silencing phenotypes in transgenic plants11. The protocol presented here is a streamlined procedure covering steps from the selection of computationally designed amiRNA candidates to the identification of an optimal amiRNA for a single target gene (Fig. 1).
Figure 1.
Flow chart of the ETPamir screens for identifying optimal amiRNAs. Co-expression of the target gene encoding epitope-tagged proteins with different amiRNAs in plant protoplasts and subsequent immunoblot analysis of target protein accumulation using tag antibodies facilitate a quick and reliable discrimination of potent, moderate and ineffective amiRNAs from computationally designed candidates. Two co-expression strategies, Option A and Option B, are provided each with particular advantages. The protoplast incubation time in Option A depends on the target protein stability - unstable target proteins require a shorter incubation time (e.g., 6-12 h). An untargeted control gene is co-expressed in every transfection experiment as an indicator of equal transfection efficiency and absence of side effects of amiRNA expression.
Applications of the ETPamir screens
Our protocol for the ETPamir screens can be used to identify optimal amiRNAs for silencing single or multiple target genes in Arabidopsis and other plant species listed in Table 1, all of which have established protocols for protoplast-based transient gene expression and have been included in the WMD genome database for computational amiRNA design. If amiRNA candidates are manually designed according to the procedure of Eamens and co-workers14, our protocol in principle can be adapted to any plant species amenable to protoplast isolation and DNA transfection. The protocol can also be used to screen potent amiRNAs for the silencing of viral mRNAs to confer enhanced viral resistance in transgenic plants expressing these amiRNAs13. By replacing amiRNA candidates with hpRNA or trans-acting small interfering RNA (tasiRNA)21 candidates, this protocol can also be used to rapidly evaluate the in vivo efficiency of other post-transcriptional gene silencing techniques. The key concept of the ETPamir screen can be further extended to validate in silico predicted target genes for natural miRNAs from plants or interacting organisms including fungal pathogens and pests22 (Fig. 2; this procedure is described in Box 1). In addition, this protocol can be used to determine the silencing specificity of amiRNAs or other gene silencing methods, and the fates of target mRNAs in plant cells by parallel quantification of proteins by immunoblotting and of mRNAs by RT-qPCR (see ref. 11).
TABLE 1.
Plant species in WMD genome database with established protoplast transient assay.
| Plant Latin name | Common name | Group | Reference |
|---|---|---|---|
| Actinidia deliciosa | Kiwifruit | Dicot | 29 |
| Arabidopsis thaliana | Dicot | 20 | |
| Arachis hypogaea | Peanut | Dicot | 30 |
| Avena sativa | Oat | Monocot | 31 |
| Brassica napus | Rapeseed | Dicot | 32 |
| Brassica oleracea | Dicot | 33 | |
| Capsicum annuum | Pepper | Dicot | 34 |
| Carica papaya | Papaya | Dicot | 35 |
| Catharanthus roseus | Dicot | 11 | |
| Chlamydomonas reinhardtii* | Alga | 36 | |
| Citrus sinensis | Sweet orange | Dicot | 37 |
| Cucumis sativus | Cucumber | Dicot | 38 |
| Festuca arundinacea | Tall fescue | Monocot | 39 |
| Glycine max | Soybean | Dicot | 40 |
| Gossypium hirsutum | Cotton | Dicot | 41 |
| Helianthus annuus | Sunflower | Dicot | 11 |
| Hordeum vulgare | Barley | Monocot | 42 |
| Lactuca sativa | Lettuce | Dicot | 43 |
| Medicago sativa | Alfalfa | Dicot | 44 |
| Nicotiana benthamiana | Dicot | 11 | |
| Nicotiana sylvestris | Dicot | 45 | |
| Nicotiana tabacum | Tobacco | Dicot | 46 |
| Oryza sativa | Rice | Monocot | 47 |
| Panicum virgatum | Switchgrass | Monocot | 48 |
| Petunia hybrida | Dicot | 49 | |
| Phaseolus vulgaris | Bean | Dicot | 50 |
| Physcomitrella patens | Bryophyte | 51 | |
| Pinus pinaster | Maritime pine | Pinophyta | 52 |
| Pisum sativum | Pea | Dicot | 53 |
| Populus tremula × alba | Poplar | Dicot | 54 |
| Saccharum officinarum | Sugarcane | Monocot | 55 |
| Selaginella moellendorffii | Lycophyte | 56 | |
| Solanum lycopersicum | Tomato | Dicot | 11 |
| Solanum tuberosum | Potato | Dicot | 57 |
| Taraxacum officinale | Dandelion | Dicot | 58 |
| Triticum aestivum | Wheat | Monocot | 59 |
| Vigna unguiculata | Cowpea | Dicot | 60 |
| Vitis vinifera | Grapevine | Dicot | 61 |
| Zea mays | Maize | Monocot | 62 |
Chlamydomonas reinhardtii is transformed by the glass-bead method36 instead of protoplast transfection.
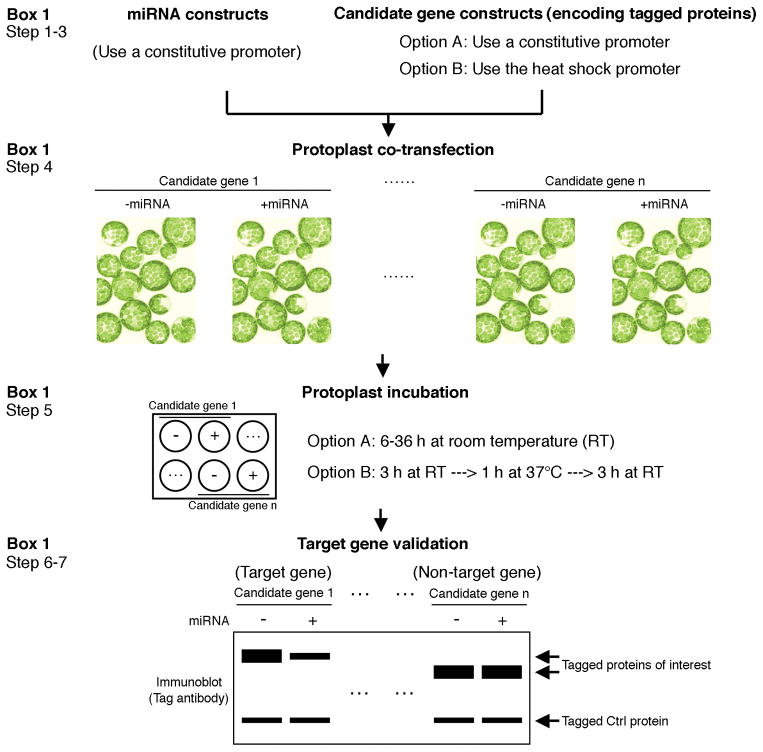
Figure 2.
Flow chart of the protein-based validation of predicted target genes for plant natural miRNAs. Co-expression of predicted candidate genes encoding epitope-tagged proteins with the miRNA of interest in plant protoplasts and subsequent immunoblot analysis of candidate protein accumulation by tag antibodies allow an easy and robust identification of authentic miRNA target genes. Two co-expression strategies, Option A and Option B, are provided each with particular advantages. The protoplast incubation time in Option A depends on the candidate protein stability. An untargeted control gene is co-expressed in every transfection experiment as an indicator of equal transfection efficiency and absence of side effects of miRNA expression.
BOX 1. PROTEIN-BASED VALIDATION OF PREDICTED TARGET GENES OF ENDOGENOUS PLANT MIRNAS.
The key strategy of the ETPamir screen can be extended to validate computationally predicted target genes of endogenous plant miRNAs (Fig. 2).
PROCEDURE
-
1|
Input the sequence of the miRNA of interest on the “Target Search” webpage of the WMD website (http://wmd3.weigelworld.org) to predict its endogenous target genes.
-
2|
Clone the miRNA and its individual target candidate genes according to Step 5 of the main Procedure.
-
3|
Extract plasmid DNA according to Steps 6 and 7 of the main Procedure.
-
4|
Co-transfect protoplasts with the miRNA and individual candidate gene constructs expressing epitope-tagged proteins, as described in Steps 8-15 of the main Procedure. For each target candidate gene, set up a negative control, in which the miRNA construct is replaced by empty vector as described in Step 9 of the main Procedure.
-
5|
Co-express the miRNA and individual target candidate genes in protoplasts using either Option A or Option B according to Step 16 of the main Procedure.
-
6|
Monitor candidate protein accumulation by SDS-PAGE and immunoblotting, as described in Steps 17-28 of the main Procedure.
-
7|
Identify authentic target genes whose expression is reduced in the presence of the miRNA.
? TROUBLESHOOTING
Comparison with other methods
Current routinely used methods for evaluating the efficacy of plant amiRNAs or miRNAs include RT-qPCR and RNA blot analyses for monitoring target transcript levels8,9, and RNA ligase mediated-5′ rapid amplification of cDNA ends (RLM-RACE) for detecting products of amiRNA/miRNA-mediated target mRNA cleavage23. However, the results of both methods do not reflect the amiRNA/miRNA action at the protein level and may lead to misinterpretation of amiRNA/miRNA activities given the complexity of the potential silencing mechanisms11,24,25. The ETPamir screen directly examines the ultimate outcome of gene silencing at the protein level, bypassing the complexity that amiRNA/miRNA can mediate gene silencing at the target mRNA or/and protein level11,24,25. The use of epitope tags and tag antibodies in the screens not only circumvents the technical obstacle of plant antibody paucity but also offers enhanced sensitivity and flexibility. Although translational repression has been analyzed by co-expression of plant miRNA and the GFP fusion to a specific target gene through agroinfiltration of Nicotiana benthamiana leaves and microscopic visualization26, our protoplast-based ETPamir screen offers four advantages over that method. First, the leaf agroinfiltration-mediated transient assay is only amenable in several plant species, whereas the protoplast transient expression system renders the ETPamir screen applicable in a broad range of plant species (Table 1), thus offering higher possibility to evaluate amiRNA/miRNA activities in near-native cellular contexts in the plant species of interest. Second, leaf agroinfiltration has relatively lower efficiency and higher variability in DNA co-delivery than the protoplast transient assay20. Third, GFP visualization is not as sensitive and quantitative as protein blot analyses. Fourth, the large size of the GFP protein may interfere with the stability, function and regulation of target proteins.
Experimental design
Proper amiRNA expression backbone and experimental controls are key for identifying optimal amiRNAs in a conclusive and reliable manner. An appropriate endogenous miRNA backbone from the plant species of interest or its close relatives should be used to express amiRNA precursors to avoid potential problems associated with amiRNA expression and processing. Table 2 summarizes miRNA backbones that have been proven useful for amiRNA expression in dicot, monocot, tree or alga species. If a native or species-related miRNA backbone is not readily available, the Arabidopsis miR319a (ath-miR319a) backbone or the rice miR528 (osa-miR528) backbone can be used as an alternative for amiRNA expression in dicots and monocots, respectively (see many examples in Table 2). In the ETPamir screen, a negative control with the target gene expression alone should be conducted in parallel with other amiRNA screens to monitor target protein accumulation without amiRNA co-expression. An untargeted control gene (e.g., GFP) should be co-expressed with the target gene in every transfection experiment (including in the negative control) to indicate comparable transfection efficiencies between samples, as well as the absence of nonspecific silencing effects of amiRNA expression. The protein products of the untargeted control gene should be clearly distinguishable in size from the proteins of interest. On the user’s first attempt of the ETPamir screen, we recommend that a positive control (i.e, co-expression of a target gene with its verified optimal amiRNA) may be conducted to ensure the ETPamir screen procedure is working properly in user’s own experimental conditions (target genes and their verified optimal amiRNA constructs are available from the authors). Regarding the target gene validation for endogenous plant miRNAs, the miRNA expression backbone is not an issue because the endogenous pre-miRNAs of interest will be expressed. However, the same requirements on the control setup should be followed.
TABLE 2.
Reported amiRNA backbones in diverse plant species.
| Plant name | Common name | Group | amiRNA backbone | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | Dicot | ath-miR159a | 13 | |
| ath-miR164 | 8 | |||
| ath-miR169d | 63 | |||
| ath-miR172a | 9 | |||
| ath-miR319a | 9, 11 | |||
| Catharanthus roseus | Dicot | ath-miR319a | 11 | |
| Chlamydomonas | Alga | cre-miR1162 | 64 | |
| reinhardtii | cre-miR1157 | 65 | ||
| Glycine max | Soybean | Dicot | ath-miR319a | 66 |
| Gossypium hirsutum | Cotton | Dicot | ghi-miR169a | 67 |
| Helianthus annuus | Sunflower | Dicot | ath-miR319a | 11 |
| Medicago sativa | Alfalfa | Dicot | ath-miR319a | 68 |
| Medicago truncatula | Dicot | mtr-miR159b | 69 | |
| Nicotiana benthamiana | Dicot | ath-miR319a | 11 | |
| Nicotiana tabacum | Tobacco | Dicot | ath-miR164b | 8 |
| Oryza sativa | Rice | Monocot | osa-miR528 | 11, 15 |
| Physcomitrella patens | Bryophyte | ath-miR319a | 70 | |
| Populus tremula × alba | Poplar | Dicot | ptc-miR408 | 71 |
| Solanum lycopersicum | Tomato | Dicot | ath-miR319a | 11, 72 |
| ath-miR164 | 8 | |||
| Solanum melongena L. | Eggplant | Dicot | ath-miR319a | 73 |
| Solanum tuberosum | Potato | Dicot | ath-miR168a | 18 |
| Triticum aestivum | Wheat | Monocot | osa-miR395 | 74 |
| Vitis vinifera | Grapevine | Dicot | vvi-miR166f | 75 |
| Zea mays | Maize | Monocot | zma-miR396 | 76 |
| ath-miR319a | 11 |
For protoplast incubation in the ETPamir screens, users can choose Option A (i.e., constitutive co-expression of amiRNA and target mRNAs) if less hands-on manipulation is preferred or if the protein products of the target gene are relatively unstable. Alternatively, users can choose Option B (i.e., constitutive expression of amiRNA but inducible expression of target mRNAs) if a quicker identification of optimal amiRNAs is desired. Accordingly, target gene and untargeted control gene should be expressed using a constitutive promoter for Option A, or using the heat shock promoter for Option B. In Option A, 36 h of co-expression is empirically considered optimal for clearly discriminating potent, moderate and ineffective amiRNAs for most target genes, while shorter co-expression time (e.g., 6-12 h) is required for target genes encoding unstable proteins. For example, the Arabidopsis ZAT6 (Zinc Finger of Arabidopsis thaliana 6) protein has a short half-life around 10 min. The optimal amiRNA for the ZAT6 gene was found to completely block ZAT6-FLAG protein accumulation within 6 h co-expression11.
The procedure presented in this protocol is specific for identifying an optimal amiRNA for a single target gene. When applying the ETPamir screen to identifying a single optimal amiRNA for multiple target genes, one can conduct the co-expression of each target gene with each amiRNA candidate in a pairwise manner and determine the optimal amiRNA that is able to potently silence all the target genes. Alternatively, one can co-express all the target genes together plus individual amiRNA candidates. In the latter case, to monitor different silencing profiles of individual target genes, one can employ the same tag for all the target genes if their proteins are well distinguishable by size, or use different tags for different target genes if the proteins migrate too closely in SDS-PAGE.
Limitations
In the ETPamir screens, optimal amiRNAs are identified based on a transient assay. Therefore, we cannot completely rule out the possibility that reduction of the endogenous target gene expression by these optimal amiRNAs in transgenic plants can trigger enhanced target gene transcription to counterbalance the silencing effects, as some gene expression is controlled by transcriptional regulatory loops in planta10. In those cases, more potent amiRNA may be required. In terms of target gene validation of endogenous plant miRNAs by the ETPamir screen, one needs to be aware that target validation in this assay is conducted in conditions of miRNA over-expression in mesophyll protoplasts.
MATERIALS
REAGENTS
4-morpholineethanesulfonic acid (MES, Sigma, cat. no. M3671)
Mannitol (MP Biomedicals, cat. no. 102248)
MgCl2 (Sigma, cat. no. M9272) KCl (Sigma, cat. no. P3911)
NaCl (Sigma, cat. no. S9888)
CaCl2 (Sigma, cat. no. C7902)
CsCl (American Bioanalytical, cat. no. AB00300)
Polyethylene glycol 4000 (PEG4000, Sigma, cat. no. 81240)
Tween-20 (Sigma, cat. no. P7949)
Phusion DNA polymerase (NEB, cat. no. M0535)
PVDF membrane (Immobilon, cat. no. IPVH304F0)
Nonfat dry milk (Santa Cruz Biotechology, cat. no. sc-2325)
Bovine calf serum (HyClone, cat. no. SH30072.03)
HA HRP-conjugated antibody (Roche, cat. no. 12013819001)
SuperSignal West Pico chemiluminescent kit (Thermo Scientific, cat. no. 34080)
SuperSignal West Femto chemiluminescent kit (Thermo Scientific, cat. no. 34095)
10% precast polyacrylamide gel (Bio-Rad, cat. no. 456-1034)
Terrific broth (American Bioanalytical, cat. no. AB01966)
Custom oligo primers for PCR generating amiRNA precursors (Oligo synthesis service provider)
pHBT-HA constitutive expression vector (Available from the authors upon request)
pHSP-HA heat shock inducible expression vector (Available from the authors upon request)
pHBT-ath-miR319a constitutive expression plasmid (Available from the authors upon request)
EQUIPMENT
CL2 clinical centrifuge (Thermo Scientific, cat. no. 004260F)
Mini-PROTEAN Tetra system (Bio-Rad, cat. no. 165-8006)
Trans-Blot SD semi-dry electrophoretic transfer cell (Bio-Rad, cat. no. 170-3940)
Fisher Scientific Isotemp heating block (Fisher Scientific, cat. no. 11-715-305Q)
2-ml round-bottom microcentrifuge tubes (USA Scientific, cat. no. 1620-2700)
1.5-ml microcentrifuge tubes (USA Scientific, cat. no. 1615-5500)
6-well culture plates (Falcon, cat. no. 3046)
1000-ml storage bottle with 0.22 μm vacuum filter (Corning, cat. no. 430517)
REAGENT SETUP
Mannitol, 0.8 M stock
Dissolve 146 g mannitol in Milli-Q water to a final volume of 1 L. This solution can be stored at 25°C for 6 months.
NaCl, 5 M stock
Dissolve 292.2 g NaCl in Milli-Q water to a final volume of 1 L. This solution can be stored at 25°C for 12 months.
CaCl2, 1 M stock
Dissolve 111 g CaCl2 in Milli-Q water to a final volume of 1 L. This solution can be stored at 25°C for 6 months.
KCl, 2 M stock
Dissolve 149.1 g KCl in Milli-Q water to a final volume of 1 L. This solution can be stored at 25°C for 6 months.
MgCl2, 2 M stock
Dissolve 190.4 g MgCl2 in Milli-Q water to a final volume of 1 L. This solution can be stored at 25°C for 6 months.
MES, 0.2 M stock (pH 5.7)
Dissolve 39 g MES in 700 ml Milli-Q water, adjust to pH 5.7 with KOH, and finalize the volume to 1 L with Milli-Q water. This solution can be stored at 4°C for 12 months.
Tris-HCl, 1.5 M stock (pH 6.8)
Dissolve 181.65 g Tris base in 700 ml Milli-Q water, adjust to pH 6.8 with 120 ml concentrated HCl. This solution can be stored at 25°C for 12 months.
Calf serum, 5% (vol/vol)
Mix 25 ml bovine calf serum with 475 ml sterile Milli-Q water. This solution can be stored at 4°C for up to 6 months.
MMg solution
Mix 250 ml 0.8 M mannitol stock solution, 3.75 ml 2 M MgCl2 stock solution and 10 ml 0.2 M MES stock solution, and finalize the volume to 500 ml with Milli-Q water. Sterilize by passing through a 0.22 μm filter and collect the flow-through into a storage bottle. This solution can be stored at 4°C for 6 months.
WI solution
Mix 312.5 ml 0.8 M mannitol stock solution, 5 ml 2 M KCl stock solution and 10 ml 0.2 M MES stock solution. and finalize the volume to 500 ml with Milli-Q water. Sterilize by passing through a 0.22 μm filter and collect the flow-through into a storage bottle. This solution can be stored at 4°C for 6 months.
W5 solution
Mix 15.4 ml 5 M NaCl stock solution, 62.5 ml 1 M CaCl2 stock solution, 1.25 ml 2 M KCl stock solution and 5 ml 0.2 M MES stock solution. Finalize the volume to 500 ml with Milli-Q water. Sterilize by passing through a 0.22 μm filter and collect the flow-through into a storage bottle. This solution can be stored at 4°C for 6 months.
PEG solution
To make 10 ml PEG solution, dissolve 4 g PEG4000 in a mixture of 3 ml water, 2.5 ml 0.8 M mannitol stock solution and 1 ml 1 M CaCl2 stock solution. This solution should be freshly made before use.
SDS-PAGE loading buffer (4×)
Dissolve 0.8 g SDS, 2 mg bromophenol blue in a mixture of 1.7 ml 1.5 M Tris-HCl, pH 6.8, 4 ml glycerol and 0.8 ml β-mecaptomethanol. Finalize the volume to 10 ml with Milli-Q water. This solution can be stored at 25°C for up to 6 months.
Tris-Glycine-SDS Running buffer
To make a 10× stock solution, dissolve 30.3 g Tris base, 144 g glycine, 10 g SDS in Milli-Q water to a final volume of 1 L. This stock solution can be stored at 25°C for 12 months and can be diluted to 1× before use.
Transfer buffer
Dissolve 3 g Tris base, 14.4 g glycine in 800 ml Milli-Q water, add 100 ml methanol, and finalize the volume to 1 L with Milli-Q water. This solution can be stored at 25°C for 3 months.
TBST buffer
Dissolve 6.1 g Tris base, 8.8 g NaCl in 800 ml Milli-Q water, adjust to pH 7.4 with HCl. Add 0.5 ml Tween-20 and finalize the volume to 1 L with Milli-Q water. This solution can be stored at 25°C for up to 6 months.
Blocking buffer
Dissolve 5 g nonfat dry milk in 100 ml TBST buffer. This solution should be freshly made before use.
PROCEDURE
Design and selection of amiRNAs ● TIMING 1-2 days
-
1|
Follow the detailed instruction on the WMD website (http://wmd3.weigelworld.org) to obtain a list of predicted, gene-specific amiRNA candidates for the gene(s) of interest. In the “Designer” webpage of WMD, the user can either input the gene identification number or the gene sequence in the fasta format as “Target genes”, and select the intended plant genome from the WMD genome database as “Genome”, and input “0” as “Accepted off-targets” to ensure that the designed amiRNA candidates are specific to the gene(s) of interest.
-
2|
Select 3-4 amiRNA candidates satisfying all the criteria in Table 3.
CRITICAL STEP WMD ranks amiRNA candidates based on sequence complementarity and small RNA properties10. The amiRNA ranking on the WMD prediction list may or may not be correlated with its experimentally determined efficacy11. However, it is convenient that the search of suitable amiRNA candidates starts from the top candidate on the list. By clicking into each amiRNA candidate on the list, the user can access detailed characteristics about the candidate, including the target site location, mismatch number and position, hybridization energy and potential off-targets. It should be noted that potential off-targets are different from the “defined” off-targets excluded in Step 1, as the formers may have considerable sequence complementarity with a given amiRNA but the mismatch positions or/and hybridization energy parameters prohibit the WMD algorithm from making a clear judgment.
? TROUBLESHOOTING
-
3|
Input individual selected amiRNA sequences on the “Oligo” webpage of WMD to design primers for generating amiRNA precursors (pre-amiRNAs) by PCR.
-
4|
Assemble individual pre-amiRNAs using an appropriate endogenous miRNA backbone (see Experimental design and Table 2) by overlapping PCR according to the detailed instructions on the WMD website.
TABLE 3.
Criteria for selecting amiRNA candidates from the WMD prediction list.
| Number | Criterion |
|---|---|
| 1 | Target site within the 5′ 200 nucleotides (nt) of the coding sequence (CDS) |
| 2 | No identical or overlapping target sequence with other selected amiRNA candidates |
| 3 | Less than 2 mismatches between the amiRNA candidate and its target mRNA |
| 4 | Mismatch can be acceptable only at the position 1 or 15-21 of an amiRNA candidate |
| 5 | Hybridization energy between the amiRNA candidate and its target sequence should be above 80% of that between the amiRNA and a perfect complement |
| 6 | No potential off-target is predicted by WMD |
The criteria were empirically determined based on the evaluation of 79 amiRNA-target mRNA interactions in Arabidopsis mesophyll protoplasts11.
Generation of amiRNA and target gene constructs ● TIMING 1-2 weeks
-
5|
Clone individual pre-amiRNAs into a transient expression plasmid (e.g., the pHBT-ath-miR319a plasmid) containing a constitutive and strong promoter and the NOS terminator. Meanwhile, clone the target gene of interest or an untargeted control gene (see Experimental design) into a transient expression plasmid encoding hemagglutinin (HA)-tagged proteins under a constitutive and strong promoter (e.g., the pHBT-HA plasmid; for Option A in Step 16) or under the heat shock promoter11 (e.g., the pHSP-HA plasmid; for Option B in Step 16).
CRITICAL STEP The HA tag (YPYDVPDYA) and FLAG tag (DYKDDDDK) are highly recommended due to their small sizes and excellent antibody resources. Their 27-bp and 24-bp coding sequences, respectively, can be easily fused with the target gene coding sequence as part of the primer sequence through PCR. Other epitope tags and fluorescent proteins (e.g., GFP) with commercial antibodies available can also be used. A binary plasmid can also be used instead of the transient expression plasmid but may lead to reduced protoplast transfection efficiency.
-
6|
Transform E. coli and grow a single colony in 200 ml Terrific broth with appropriate antibotics at 37°C for 16 h.
-
7|
Purify the DNA of the plasmids expressing amiRNAs and target genes.
CRITICAL STEP Obtaining high-quality and concentrated (2 μg/μl) plasmid DNA is crucial for high transfection efficiency in protoplasts, and we highly recommend using CsCl gradient ultracentrifugation for this purpose (its protocol is provided on the Sheen lab website: http://molbio.mgh.harvard.edu/sheenweb/protocols_reg.html). DNA preparation by homemade silica resin27 or by commercial DNA maxiprep kits is acceptable. The commercial DNA maxiprep kits are more convenient but expensive, and their plasmid DNA yields in general result in lower protoplast transfection efficiency.
PAUSE POINT Purified DNA can be stored at −20°C until use.
Protoplast isolation ● TIMING 3-4 h
-
8|
Follow the detailed protocol20 for isolating mesophyll protoplasts from 4-week-old Arabidopsis plants. We used this protocol successfully, with no modification, to isolate protoplasts from, but not limited to, 4-week-old tobacco, 3-week-old Catharanthus roseus, and 2-week-old tomato or sunflower11.
CRITICAL STEP Using healthy plants is very critical for achieving high quality protoplasts that allow efficient DNA transfection and protein expression, and maintain cell integrity during prolonged (e.g., >24 h) incubation.
Co-transfection of amiRNA and target gene constructs ● TIMING 15 min for 5 samples
-
9|
For each co-transfection, mix DNA of the following three plasmids in a 2-ml round-bottom microcentrifuge tube to generate a 21 μl DNA cocktail (2 μg DNA/μl): 16 μl (32 μg) of the amiRNA construct, 4 μl (8 μg) of the target gene-HA tag construct, and 1 μl (2 μg) of control gene (e.g., GFP)-HA tag construct. For a negative control, prepare an additional DNA cocktail replacing the amiRNA construct with empty vector to monitor target protein accumulation without amiRNA co-expression.
-
10|
Add 200 μl protoplasts (2 × 105 cells/ml in MMg solution) to each tube.
-
11|
Add 220 μl PEG solution to each tube and mix well by gently tapping on the tube bottom 15 times.
-
12|
Incubate the samples at room temperature for 5 min.
-
13|
Quench the transfection by adding 800 μl W5 solution and inverting the tube twice.
-
14|
Pellet the protoplasts by centrifugation at 100 × g for 2 min at room temperature using a CL2 clinical centrifuge, and remove the supernatant.
CRITICAL STEP The supernatant (~1.2 ml) should be pipetted out using a 1 ml pipette with caution. To avoid disturbing the protoplast pellet at the tube bottom, there is no need to completely remove the supernatant and 20-30 μl of supernatant can be left in the tube.
-
15|
Resuspend the transfected protoplasts with 100 μl W5 solution per sample and transfer cells to 1 ml WI solution in a 6-well culture plate pre-coated with 5% calf serum, and mix well.
Protoplast incubation ● TIMING 6-36 hr
-
16|
Use either Option A or Option B for protoplast incubation, each with particular advantages (see Experimental design). Accordingly, construct target gene and untargeted control gene expression plasmids using a constitutive promoter for Option A, or the heat shock promoter for Option B (see Step 5).
-
Constitutive co-expression of amiRNA and target mRNAs:
-
Incubate the transfected protoplasts under normal plant growth conditions for 6-36 h (see Experimental design). Normal plant growth conditions are photoperiods of 12 h light (75 μmolm−2s−1) at 23°C and 12 h dark at 20°C20.
? TROUBLESHOOTING
-
-
Constitutive expression of amiRNA but inducible expression of target mRNAs:
Incubate the transfected protoplasts under normal plant growth conditions for 3 h.
Incubate protoplasts at 37°C for 1 h.
Incubate protoplasts under normal plant growth conditions for another 3 h.
-
Identification of optimal amiRNAs ● TIMING 6 h
-
17|
Resuspend the protoplasts by gently swirling the 6-well plate and transfer cells to 1.5 ml microcentrifuge tubes.
-
18|
Pellet the protoplasts by centrifugation at 100 × g for 2 min at room temperature using the CL2 clinical centrifuge.
-
19|
Remove most of the supernatant and leave ~30 μl WI solution and the pellet at the bottom intact.
-
20|
Add 10 μl 4 × SDS-PAGE loading buffer to each tube, briefly vortex, and boil the samples at 95°C for 5 min.
PAUSE POINT Protein samples can be stored at −20°C until further analysis.
-
21|
Resolve all protein samples (~40 μl each) in a 10% precast polyacrylamide gel until the dye is running out.
-
22|
Transfer the proteins from the gel to a PVDF membrane.
-
23|
Incubate the membrane with the blocking buffer under gentle (70 rpm) shaking at room temperature for 30 min.
-
24|
Incubate the membrane with the blocking buffer containing HA HRP-conjugated antibodies (1:10,000 dilution) under gentle shaking at room temperature for 2 h.
-
25|
Wash the membrane three times (10 min each time) with the TBST buffer under gentle shaking.
-
26|
Detect tagged proteins with the SuperSignal West Pico chemiluminescent kit.
? TROUBLESHOOTING
-
27|
Quantify the immunoblot signals by densitometric analysis using the Image J program (the program can be downloaded at http://rsbweb.nih.gov/ij/download.html).
-
28|
Identify the optimal amiRNA(s) whose co-expression completely blocks or leads to minimal target protein accumulation relative to the negative control.
? TROUBLESHOOTING
TIMING
Step 1-4, design and selection of amiRNAs: 1-2 days
Step 5-7, generation of amiRNA and target gene constructs: 1-2 weeks
Step 8, protoplast isolation: 3-4 h
Step 9-15, co-transfection of amiRNA and target gene constructs: 15 min for 5 samples (4 amiRNA samples plus 1 negative control)
Step 16, protoplast incubation: 6-36 h
Step 17-28, identification of optimal amiRNAs: 6 h
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 4.
TABLE 4.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 2 | WMD cannot design any single amiRNA to target the multiple gene targets Insufficient or no amiRNA candidates fulfill all the criteria in Table 3 |
Target genes do not share sufficient sequence identity Target gene has a limited number of designed amiRNA candidates targeting its 5′ 200 nt of CDS |
Reduce target gene number or use multiple amiRNAs to taregt these genes Relax the amiRNA target site requirement to include the entire CDS |
| 16A(i) | Bacteria are detected at the end of the incubation step | Experimental environment is not hygienic | Add 200 μg/ml (final) carbenicillin to WI solution in Step 15 |
| 26 and Box 1, step 7 | No target protein is detectable (even in the negative control) | Target gene is expressed low due to inherent reason (e.g., large protein size or codon usage bias) Target protein is unstable |
Use the SuperSignal West Femto chemiluminescent kit to enhance immunoblot signals Use Option A in step 16 with shorter co-expression time (e.g., 6-12 h) |
| 27 | No amiRNA candidate can sufficiently suppress target gene expression | Tested amiRNAs are not potent Target gene is highly expressed and its protein products are very stable |
Return to Step 2 to select 3-4 additional amiRNA candidates and repeat the ETPamir screen Use Option B in Step 16 |
ANTICIPATED RESULTS
A typical result of the ETPamir screen is shown at the bottom of Figure 1. In general, at least one optimal amiRNA can be identified from 3-4 selected amiRNA candidates for a single target gene by following this protocol. The optimal amiRNAs should be able to reduce the target protein accumulation by over 90% compared to the negative control, given that the expression of the untargeted control gene is comparable between samples. Although constitutive expression of moderate to suboptimal amiRNAs can generate target gene knockdown phenotypes, constitutive expression of those optimal amiRNAs would very likely lead to “functional knockout” of target gene expression, conferring silencing phenotypes resembling genetic null mutants11. Optimal amiRNAs can also be expressed using a chemically inducible promoter or a tissue-specific promoter in transgenic plants to gain tight temporal and spatial control of target gene activity during the functional study.
Using the key strategy of the ETPamir screen, bioinformatically predicted target genes for a given endogenous plant miRNA can be experimentally validated as illustrated in Figure 2, where the protein products of an authentic target gene will be reduced in the presence of miRNA, while those of a false target gene will not be affected. Even if the validated target gene and the miRNA were not co-expressed in planta26, the results of this assay may still be biologically relevant considering the possibility of intercellular movement of many plant natural miRNAs28.
Acknowledgments
The authors thank the Weigel lab for developing the versatile WMD platform and members in the Sheen lab for their efforts to test and improve the ETPamir screens. This work has been supported by Massachusetts General Hospital ECOR Postdoctoral Fellowship for Medical Discovery to J.F.L. and the National Science Foundation Grant IOS-0843244 and the National Institutes of Health Grants R01 GM60493 and R01 GM70567 to J.S.
Footnotes
Contributor Information
Jian-Feng Li, Email: jli@molbio.mgh.harvard.edu.
Dandan Zhang, Email: ddzhang@molbio.mgh.harvard.edu.
Jen Sheen, Email: sheen@molbio.mgh.harvard.edu.
References
- 1.Zhang F, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 3.Christian M, Qi Y, Zhang Y, Voytas DF. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3. 2013;3:1697–1705. doi: 10.1534/g3.113.007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JF, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 6.Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Modes of gene duplication contribute differently to genetic novelty and redundancy but show parallels across divergent angiosperms. PLoS ONE. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez JP, et al. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 11.Li JF, et al. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell. 2013;25:1507–1522. doi: 10.1105/tpc.113.112235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu QW, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 14.Eamens AL, McHale M, Waterhouse PM. The use of artificial microRNA technology to control gene expression in Arabidopsis thaliana. Methods Mol Biol. 2014;1062:211–224. doi: 10.1007/978-1-62703-580-4_11. [DOI] [PubMed] [Google Scholar]
- 15.Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park W, Zhai J, Lee JY. Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Rep. 2009;28:469–480. doi: 10.1007/s00299-008-0651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deveson I, Li J, Millar AA. MicroRNAs with analogous target complementarities perform with highly variable efficacies in Arabidopsis. FEBS lett. 2013;587:3703–3708. doi: 10.1016/j.febslet.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Bhagwat B, et al. An in vivo transient expression system can be applied for rapid and effective selection of artificial microRNA constructs for plant stable genetic transformation. J Genet Genomics. 2013;40:261–270. doi: 10.1016/j.jgg.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 20.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 21.Felippes FF, Wang JW, Weigel D. MIGS: miRNA-induced gene silencing. Plant J. 2012;70:541–547. doi: 10.1111/j.1365-313X.2011.04896.x. [DOI] [PubMed] [Google Scholar]
- 22.Weiberg A, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 24.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 25.Iwakawa HO, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 26.Palatnik JF, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Li JF, Li L, Sheen J. A rapid and economical procedure for purification of plasmid or plant DNA with diverse applications in plant biology. Plant Methods. 2010;6:1. doi: 10.1186/1746-4811-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gursanscky NR, Searle IR, Carroll BJ. Mobile microRNAs hit the target. Traffic. 12:1475–1482. doi: 10.1111/j.1600-0854.2011.01253.x. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira MM, Barroso J, Pais MS. Direct gene transfer into Actinidia deliciosa protoplasts: analysis of transient expression of the CAT gene using TLC autoradiography and a GC-MS-based method. Plant Mol Biol. 1991;17:235–242. doi: 10.1007/BF00039498. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Cheng M, Demski JW, Jarret RL. Improved electroporation buffer enhances transient gene expression in Arachis hypogaea protoplasts. Genome. 1995;38:858–863. doi: 10.1139/g95-113. [DOI] [PubMed] [Google Scholar]
- 31.Huttly AK, Baulcombe DC. A wheat alpha-Amy2 promoter is regulated by gibberellin in transformed oat aleurone protoplasts. EMBO J. 1989;8:1907–1913. doi: 10.1002/j.1460-2075.1989.tb03593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauls PK, Kunert K, Huttner E, Grandbastien MA. Expression of the tobacco Tnt1 retrotransposon promoter in heterologous species. Plant Mol Biol. 1994;26:393–402. doi: 10.1007/BF00039548. [DOI] [PubMed] [Google Scholar]
- 33.Eimert K, Siegemund F. Transformation of cauliflower (Brassica oleracea L. var. botrytis)-an experimental survey. Plant Mol Biol. 1992;19:485–490. doi: 10.1007/BF00023396. [DOI] [PubMed] [Google Scholar]
- 34.Chung E, et al. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta. 2004;220:286–295. doi: 10.1007/s00425-004-1372-9. [DOI] [PubMed] [Google Scholar]
- 35.Jiang L, Pan LJ. Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol Biol Rep. 2012;39:7105–7115. doi: 10.1007/s11033-012-1542-y. [DOI] [PubMed] [Google Scholar]
- 36.Kindle KL. High-efficiency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niedz RP, McKendree WL, Shatters RC., Jr Electroporation of embryogenic protoplasts of sweet orange (Citrus sinensis L. Osbeck) and regeneration of transformed plants. In Vitro Cell Dev Biol – Plant. 2003;39:586–594. [Google Scholar]
- 38.Graham IA, Baker CJ, Leaver CJ. Analysis of the cucumber malate synthase gene promoter by transient expression and gel retardation assays. Plant J. 1994;6:893–902. doi: 10.1046/j.1365-313x.1994.6060893.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZY, et al. Transgenic plants of tall fescue (Festuca arundinacea Schreb.) obtained by direct gene transfer to protoplasts. Biotechnology. 1992;10:691–696. doi: 10.1038/nbt0692-691. [DOI] [PubMed] [Google Scholar]
- 40.Lin W, Odell JT, Schreiner RM. Soybean protoplast culture and direct gene uptake and expression by cultured soybean protoplasts. Plant Physiol. 1987;84:856–861. doi: 10.1104/pp.84.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao X, et al. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011;66:293–305. doi: 10.1111/j.1365-313X.2011.04491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gopalakrishnan B, Sonthayanon B, Rahmatullah R, Muthukrishnan S. Barley aleurone layer cell protoplasts as a transient expression system. Plant Mol Biol. 1991;16:463–467. doi: 10.1007/BF00023996. [DOI] [PubMed] [Google Scholar]
- 43.Czarnecka E, Verner FL, Gurley WB. A strategy for building an amplified transcriptional switch to detect bacterial contamination of plants. Plant Mol Biol. 2012;78:59–75. doi: 10.1007/s11103-011-9845-2. [DOI] [PubMed] [Google Scholar]
- 44.Loake GJ, et al. Phenylpropanoid pathway intermediates regulate transient expression of a chalcone synthase gene promoter. Plant Cell. 1991;3:829–840. doi: 10.1105/tpc.3.8.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi Y, Dokiya Y, Sugiura M, Niwa Y, Sugita M. Genomic organization and organ-specific expression of a nuclear gene encoding phage-type RNA polymerase in Nicotiana sylvestris. Gene. 2001;279:33–40. doi: 10.1016/s0378-1119(01)00729-6. [DOI] [PubMed] [Google Scholar]
- 46.Ranjan R, et al. Development and functional analysis of novel genetic promoters using DNA shuffling, hybridization and a combination thereof. PLoS ONE. 2012;7:e31931. doi: 10.1371/journal.pone.0031931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazarei M, Al-Ahmad H, Rudis MR, Stewart CN., Jr Protoplast isolation and transient gene expression in switchgrass, Panicum virgatum L. Biotechnol J. 2008;3:354–359. doi: 10.1002/biot.200700189. [DOI] [PubMed] [Google Scholar]
- 49.de Lange P, de Boer GJ, Mol JN, Kooter JM. Conditional inhibition of beta-glucuronidase expression by antisense gene fragments in petunia protoplasts. Plant Mol Biol. 1993;23:45–55. doi: 10.1007/BF00021418. [DOI] [PubMed] [Google Scholar]
- 50.Roby D, Broglie K, Gaynor J, Broglie R. Regulation of a chitinase gene promoter by ethylene and elicitors in bean protoplasts. Plant Physiol. 1991;97:433–439. doi: 10.1104/pp.97.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thevenin J, et al. A new system for fast and quantitative analysis of heterologous gene expression in plants. New Phytol. 2012;193:504–512. doi: 10.1111/j.1469-8137.2011.03936.x. [DOI] [PubMed] [Google Scholar]
- 52.Gomez-Maldonado J, Crespillo R, Avila C, Canovas FM. Efficient preparation of maritime pine (Pinus pinaster) protoplasts suitable for transgene expression analysis. Plant Mol Biol Rep. 2001;19:361–366. [Google Scholar]
- 53.Ballas N, Wong LM, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) J Mol Biol. 1993;233:580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- 54.Guo J, et al. Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS ONE. 2012;7:e44908. doi: 10.1371/journal.pone.0044908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao SJ, et al. Enhanced transgene expression in sugarcane by co-expression of virus-encoded RNA silencing suppressors. PLoS ONE. 2013;8:e66046. doi: 10.1371/journal.pone.0066046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin C, Richter U, Borner T, Weihe A. Evolution of phage-type RNA polymerases in higher plants: characterization of the single phage-type RNA polymerase gene from Selaginella moellendorffii. J Mol Evol. 2009;68:528–538. doi: 10.1007/s00239-009-9229-2. [DOI] [PubMed] [Google Scholar]
- 57.Feltkamp D, Masterson R, Starke J, Rosahl S. Analysis of the involvement of ocs-like bZip-binding elements in the differential strength of the bidirectional mas1’2’ promoter. Plant Physiol. 1994;105:259–268. doi: 10.1104/pp.105.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahler D, et al. Polyphenoloxidase silencing affects latex coagulation in Taraxacum species. Plant Physiol. 2009;151:334–346. doi: 10.1104/pp.109.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee B, Murdoch K, Topping J, Kreis M, Jones MG. Transient gene expression in aleurone protoplasts isolated from developing caryopses of barley and wheat. Plant Mol Biol. 1989;13:21–29. doi: 10.1007/BF00027332. [DOI] [PubMed] [Google Scholar]
- 60.Mirabella R, Franken C, van der Krogt GN, Bisseling T, Geurts R. Use of the fluorescent timer DsRED-E5 as reporter to monitor dynamics of gene activity in plants. Plant Physiol. 2004;135:1879–1887. doi: 10.1104/pp.103.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchive C, et al. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE. 2013;8:e54185. doi: 10.1371/journal.pone.0054185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheen J. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell. 1991;3:225–245. doi: 10.1105/tpc.3.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu C, et al. A simple artificial microRNA vector based on ath-miR169d precursor from Arabidopsis. Mol Biol Rep. 2009;37:256–262. doi: 10.1007/s11033-009-9713-1. [DOI] [PubMed] [Google Scholar]
- 64.Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
- 65.Molnar A, et al. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- 66.Melito S, et al. A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biol. 2010;10:104. doi: 10.1186/1471-2229-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ali I, Amin I, Briddon RW, Mansoor S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol J. 2013;10:231. doi: 10.1186/1743-422X-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verdonk JC, Sullivan ML. Artificial microRNA (amiRNA) induced gene silencing in alfalfa (Medicago sativa) Botany. 2013;91:117–122. [Google Scholar]
- 69.Devers EA, Teply J, Reinert A, Gaude N, Krajinski F. An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol. 2013;13:82. doi: 10.1186/1471-2229-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khraiwesh B, Ossowski S, Weigel D, Reski R, Frank W. Specific gene silencing by artificial microRNAs in Physcomitrella patens: an alternative to targeted gene knockouts. Plant Physiol. 2008;148:684–693. doi: 10.1104/pp.108.128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi R, Yang C, Lu S, Sederoff R, Chiang VL. Specific down-regulation of PAL genes by artificial microRNAs in Populus trichocarpa. Planta. 2010;232:1281–1288. doi: 10.1007/s00425-010-1253-3. [DOI] [PubMed] [Google Scholar]
- 72.Fernandez AI, et al. Flexible tools for gene expression and silencing in tomato. Plant Physiol. 2009;151:1729–1740. doi: 10.1104/pp.109.147546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toppino L, et al. Reversible male sterility in eggplant (Solanum melongena L.) by artificial microRNA-mediated silencing of general transcription factor genes. Plant Biotechnol J. 2011;9:684–692. doi: 10.1111/j.1467-7652.2010.00567.x. [DOI] [PubMed] [Google Scholar]
- 74.Fahim M, Millar AA, Wood CC, Larkin PJ. Resistance to wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol J. 2012;10:150–163. doi: 10.1111/j.1467-7652.2011.00647.x. [DOI] [PubMed] [Google Scholar]
- 75.Roumi V, et al. Transient expression of artificial microRNAs confers resistance to grapevine virus A in nicotiana benthamiana. J Plant Pathol. 2012;94:643–649. [Google Scholar]
- 76.Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012;8:e1002419. doi: 10.1371/journal.pgen.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]