Abstract
Chaperone mediated autophagy (CMA) targets soluble proteins for lysosomal degradation. Here we show that CMA is activated in T cells in response to T cell receptor (TCR) engagement, which induces the expression of the lysosomal CMA receptor, LAMP-2A. In activated T cells, CMA targets the ubiquitin ligase Itch and the calcineurin inhibitor Rcan-1 for degradation to maintain activation-induced responses. Consequently, deletion of Lamp2a in T cells causes deficient in vivo responses to immunization or Listeria infection. Impaired CMA activity also occurs in T cells with age, which negatively impacts their function. Restoration of LAMP-2A in aged T cells results in enhancement of activation-induced responses. Our findings define a role for CMA in the regulation of T cell activation through the targeted degradation of negative regulators of T cell activation.
Degradation of cellular components in the lysosomes through autophagy has a central role in essential cellular functions that range from maintenance of cell homeostasis, prevention of damaged proteins accumulation and amino acids recycling for new protein synthesis, to regulation of cell differentiation, protection against biological, physical or chemical stress or regulation of cell death and survival1,2. Three major forms of autophagy have been described in mammalian cells: macroautophagy (MA), microautophagy and chaperone mediated autophagy (CMA), which use different mechanisms to target substrates into the lysosome and are also differentially regulated. CMA is a selective form of autophagy that targets single soluble cytosolic proteins bearing a consensus targeting motif biochemically related to the KFERQ pentapeptide3. CMA substrate proteins are recognized by the cytosolic heat shock chaperone Hsc70 and its associated co-chaperones, which deliver the substrate proteins to the lysosomal membrane4. The mRNA message for the Lamp2 gene undergoes alternative splicing and generates three different single-span membrane proteins: LAMP-2A, LAMP-2B and LAMP-2C, which have different C-terminal domains. Only LAMP-2A is involved in CMA5. LAMP-2A is the lysosomal receptor for CMA and mediates the translocation of substrates into the lysosomal lumen assisted by a lumenal resident form of Hsc706,7. CMA activity is directly dependent on the amount of LAMP-2A at the lysosomal membrane, because the binding of substrate proteins to LAMP-2A is the limiting step in the CMA pathway5,8. Lysosomal amounts of LAMP-2A are usually regulated through changes in its turnover and intralysosomal distribution and do not usually involve de novo protein synthesis 8,9. However, under conditions requiring maximal activation of this autophagic process, such as in response to oxidative stress, activation of CMA occurs through upregulation of the expression of Lamp2a transcription and the synthesis of new protein10.
T cell activation requires a tight regulation of positive and negative modulators of signaling pathways downstream of the T cell receptor (TCR). The ability of CMA to selectively degrade specific proteins makes this type of autophagy a possible candidate to contribute to the regulation of the levels of different signaling intermediates during T cell activation. CMA contributes to the regulation of cellular quality control and the response to stress in several tissues and organs10-12, however there is yet very little evidence of what role, if any, CMA may play in the regulation of the adaptive immune system. In this study we present evidence that CMA is an essential regulator of T cell activation through the targeted degradation of the ubiquitin ligase Itch and the calcineurin inhibitor Rcan-1, two negative regulators of TCR-signaling. Consequently, activation of CMA in response to TCR engagement helps maintain activation-induced T cell responses. Furthermore, we show that an age-dependent decline of CMA activity in T cells appears to underlie the deficient T cell function associated to aging.
Results
TCR engagement induces CMA by upregulating LAMP-2A expression
Macroautophagy is induced in activated T cells, where it regulates cell metabolism, survival and proliferation13,14. To determine if CMA, the other inducible form of autophagy, plays any role in the modulation of T cell responses and is induced in response to T cell activation, we examined if expression of LAMP-2A changed in response to TCR engagement. Immunoblots of LAMP-2A protein showed a marked increase in the amount of LAMP-2A in total cell lysates in different subsets of CD4+ T cells, including naïve and effector cells, following activation with anti-CD3 and anti-CD28 antibodies (Fig.1a). An increase in LAMP-2A expression was seen in T cells activated with anti-CD3 in absence of anti-CD28, suggesting the process was not dependent on costimulation, although CD28 engagement appeared to accelerate the kinetics of LAMP-2A induction (Fig.1a). Increased LAMP-2A protein expression in activated T cells correlated with upregulation of Lamp2a mRNA expression (Fig.1b). Immunofluorescence analyses using anti-LAMP-2A antibodies also showed an increase in the number of LAMP-2A positive puncta, which distributed in a characteristic lysosomal pattern (Fig.1c). An increased number of lysosomes was also indicated by morphometric analysis of electron microscopy images, which showed that activated T cells presented an increased number of electrodense lysosomes per cell (Fig.1d). The increase in the number of lysosomes in activated T cells was however not dependent on LAMP-2A, as a similar increase was detected in LAMP-2A-deficient T cells using the lysosomal probe lysotracker (Supplementary Fig.1a). As the transcription factor TFEB has been identified as a key regulator of lysosomal biogenesis15 and its expression has been reported in activated T cells16, we investigated if induction of TFEB expression in activated T cells would account for the increased lysosomal biogenesis in these cells. Expression of TFEB was upregulated in activated CD4+ T cells (Supplementary Fig.1b). Knockdown of TFEB expression with specific siRNAs markedly reduced the increased in lysosomal numbers induced by TCR-CD28 engagement in CD4+ T cells, suggesting that, as it has been described for macroautophagy17, TFEB may, in some conditions, link lysosomal biogenesis and CMA activation (Supplementary Fig.1c-e).
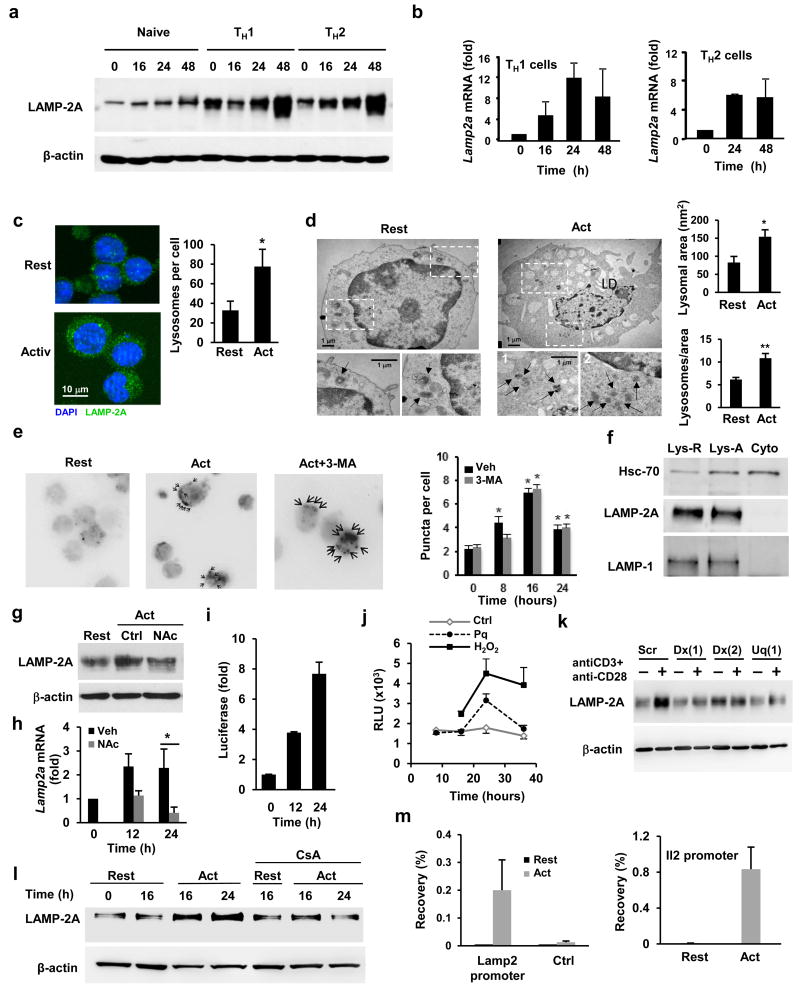
Figure 1. TCR engagement activates CMA.
(a-b) Immunoblot of LAMP-2A in total cell lysates (representative of 5 experiments) and quantification of Lamp2a gene expression by qPCR (relative to resting cells) in naïve CD4+ T cells and TH1 and TH2 cells following activation with anti-CD3 and anti-CD28 antibodies. (c) LAMP-2A (green) expression determined by immunofluorescence in resting and activated TH1 cells. Nuclei were stained with DAPI (blue). Original magnification 630x. Quantification of number of lysosomes (mean+s.e.m.) performed in 5 fields (20-30cells) from each of three different experiments (n=3; t test; *p=0.013). (d) Electron microscopy images of resting and 24 hours activated TH1 cells. Fields show representative cells and higher-magnification fields show individual lysosomes (black arrows denote lysosomes). Morphometric analysis performed in 8 micrographies (original magnification ×12,000), corresponding to cells from two different experiments. Values are mean+s.e.m.. (n=2; t test; *p =0.037; **p=0.034). (e) KFERQ–PA–mCherry1 distribution in TH1 cells transiently transfected with a CMA reporter vector, photoactivated and stimulated with anti-CD3+anti-CD28 in the presence or absence of 3-Methyladenine (3-MA). Images show representative cells (with arrows indicating mCherry positive puncta; original magnification of 630x). Quantification shown in graph (mean+s.e.m. of 3 different experiments with >50 cells counted per experiment) (n=3; ANOVA with Tukey post-test; *p<0.05). (f) Hsc70 detected by immunoblot in cytosolic (Cyto) and lysosomal fractions from resting (Lys-R) and stimulated (Lys-A) TH1 cells. LAMP-1 and LAMP-2A are used as controls. Blot is representative of 6 different experiments. (g) Immunoblot of LAMP-2A in resting TH1 cells and cells activated in the presence or absence of N-acetylcysteine (NAc) (representative of two different experiments). (h) Lamp2a gene expression quantified by qPCR in TH1 cells and cells activated in the presence or absence of NAc (values, fold increase relative to resting cells, are mean+s.e.m.; n= 4; t test; *p=0.007). (i-j) Relative luciferase activity (to resting cells) in TH1 cells transiently transfected with a vector containing a luciferase reporter under the control of the Lamp2 proximal promoter and stimulated with (i) plate-bound anti-CD3 and anti-CD28 or (j) 50 μM paraquat (Pq) or 1 mM H2O2 for for 16 hours. Results are mean+s.e.m. from (i) 4 or (j) 3 different experiments. (k) Immunoblot showing LAMP-2A protein in total cell lysates (representative blot from 2 similar experiments) of resting and activated (24 hours) TH1 cells transfected with a control non-targeting siRNA (Scr) or siRNAs specific for Duox1 (Dx) or Uqcrfs1 (Uq). (l) Immunoblot showing LAMP-2A in lysates from TH1 cells resting or activated in the presence or absence of cyclosporine A (CsA) (representative of 5 different experiments). (m) Chromatin immunoprecipitation showing NFAT1 recruitment to the Lamp2 gene promoter (% of input precipitated using anti-NFAT1 antibodies) in resting and activated TH1 cells. Binding to Adad1 (Ctrl), is used as negative control, while binding to the Il2 promoter is used as positive control. Graph shows mean+s.e.m. from 4 independent experiments.
To test if the increase in LAMP-2A amounts in lysosomes resulted in higher CMA activity, we transfected in vitro differentiated CD4+ TH1 cells with a vector that directs the expression of KFERQ-PA-Mcherry-1, a photoactivable CMA reporter18 that allows monitoring the CMA activity in living cells. In this experimental system, photoactivation of the KFERQ-mCherry-1 reporter allows detection of CMA activation as red puncta patterns that results from the redistribution of the fluorescent CMA substrate from the cytosol to the lysosomes membrane. T cell activation with anti-CD3 and anti-CD28 antibodies significantly increased the number of mCherry+ puncta per cell, further supporting that TCR engagement induces CMA activation (Fig.1e). Treatment with 3-methyladenine, an inhibitor of macroautophagy which has no effect on CMA19, did not affect mCherry cellular redistribution (Fig.1e). Furthermore, a control photoactivable mCherry-1 protein that did not express a functional CMA-targeting KFERQ motif did not undergo lysosomal redistribution in activated T cells (data not shown).
CMA occurs only at lysosomes that express the resident Hsc70 chaperone, which is required for the translocation of substrate proteins into lysosomes, in their lumen20. This subgroup of lysosomes usually expands in number when CMA is maximally activated. We then examined if Hsc70+ lysosomes were enriched in activated T cells. Immunoblotting of lysosomal fractions showed increased amounts of Hsc70 in the lysosomes of activated T cells compared to resting T cells (Fig.1f), suggesting an expansion of the CMA-active population of lysosomes following T cell activation. T cells expressing higher amounts of LAMP2A following activation (Fig.1a) also increased the size of their lysosomal compartment (Fig.1c) and as such the relative amount of LAMP-2A protein per lysosome did not change (Fig.1f). Collectively, these results indicate that TCR engagement activates CMA activity in CD4+ T cells through the upregulation of LAMP-2A expression.
Lamp2a expression is regulated by TCR-induced ROS production
Because mild oxidative stress activates CMA in fibroblasts through transcriptional upregulation of Lamp2a10 we next explored if reactive oxygen species (ROS) production following T cell activation was responsible for the increased LAMP-2A expression in T cells. T cells treated with N-acetylcysteine, a ROS scavenger, failed to increase LAMP-2A protein expression (Fig.1g) or induce Lamp2a mRNA expression (Fig.1h) following activation. A luciferase reporter vector controlled by the proximal promoter of the Lamp2 gene was sensitive to TCR triggering and showed an increase in luciferase activity with kinetics similar to the induction of the endogenous Lamp2a mRNA in activated T cells (Fig.1i). In addition, increased luciferase activity was also induced by the oxidative stress-inducing agent paraquat and by hydrogen peroxide treatment (Fig.1j), indicating the presence of ROS-responsive elements in the Lamp2 proximal promoter. Mitochondrial and cytosolic generated ROS are known to be required for efficient TCR engagement-induced store operated calcium signaling21,22. To further investigate the role of ROS in the TCR-dependent induction of Lamp2a transcription, we knocked down the expression of two enzymes, RISP (encoded by Uqcrfs) and Duox1, that regulate TCR signaling through the production of mitochondrial and cytosolic ROS respectively21,22 (Supplementary Fig.1f-g)27,28. Activated T cells expressing siRNAs specific for either RISP or Duox1 showed a marked reduction in the expression of LAMP-2A protein, compared to control cells transfected with non-targeting siRNAs (Fig.1k). One of the main signaling targets of store operated calcium entry is the calcineurin-mediated activation NFAT, which is decreased in the absence of TCR-induced generation of ROS in T cells22. We identified several putative NFAT binding sites in the Lamp2 proximal promoter, suggesting that ROS might induce Lamp2a expression through NFAT. TCR-induced expression of LAMP-2A was prevented by the calcineurin inhibitor cyclosporine A (Fig.1l) and specific binding of NFAT1, the most abundant NFAT protein in effector T helper cells23, was detected at the Lamp2 promoter by chromatin immunoprecipitation in activated TH1 cells (Fig.1m), suggesting a direct role of NFAT in the TCR-induced expression of Lamp2a. Binding of NFAT1 in the Lamp2 promoter was prevented by cyclosporine A (Supplementary Fig.1h). These results suggest that TCR-induced ROS production modulates Lamp2a expression in CD4+ T cells through activation of NFAT.
CMA activity maintains T cell activation
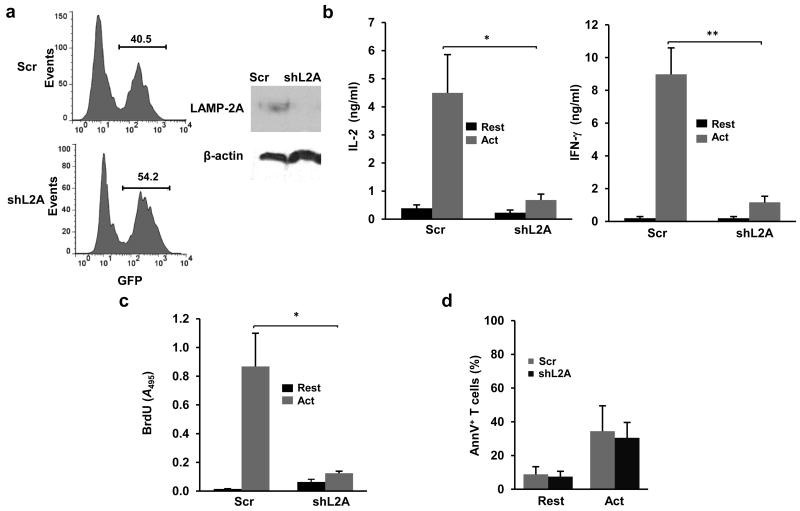
To determine the importance of CMA activation during T cell activation we analyzed the effects of silencing Lamp2a and, therefore, inhibiting CMA, in T cells. TH1 cells transduced with retroviruses expressing control or Lamp2a-specific shRNAs tagged by GFP expression were sorted based on GFP expression (Fig.2a). Silencing of Lamp2a expression significantly reduce both activation-induced cytokine production (Fig.2b) and cell proliferation (Fig.2c) in TH1 cells. The specificity of this effect was confirmed using three shRNAs targeting different regions of the Lamp2a mRNA (Supplementary Fig.2a-c). Furthermore, a vector expressing human LAMP2-A, which is not targeted by the shRNA we used, restored LAMP-2A expression in LAMP-2A silenced T cells and restored cytokine production and proliferative responses to levels similar to those of control cells (Supplementary Fig.2a-c). Impaired T cell responses caused by LAMP-2A silencing were not due to increased apoptosis, as there were no significant differences in annexin V binding between LAMP-2A silenced and control T cells (Fig.2d). These data indicate that LAMP-2A expression was required to maintain T cell activation.
Figure 2. Silencing of LAMP-2A expression impairs T cell activation.
(a) Immunoblot of LAMP-2A protein in sorted GFP+ T cells transduced with a lentivirus expressing GFP and a specific shRNA for Lamp2a (shL2A) or a non-targeting control shRNA (Scr). (b) IL-2 and INF-γ production measured by ELISA in resting and 24 hours stimulated (anti-CD3+anti-CD28) TH1 cells transduced with a lentivirus expressing shL2A or Scr. Results are mean+s.e.m. from 4 (IL-2) or 5 (IFN-γ) different experiments (t test; *p=0.031; **p=0.038). (c) T cell proliferation measured by BrdU incorporation in resting and 24 hours stimulated TH1 cells transduced with a lentivirus expressing shL2A or Scr. Results are mean+s.e.m. from 4 different experiments (t test *p=0.013). (d) Quantification of Annexin V+ cells (mean+s.e.m.) in resting and stimulated TH1 cells transduced with a lentivirus expressing shL2A or Scr (n=3).
Itch and Rcan-1 are degraded by CMA in activated T cells
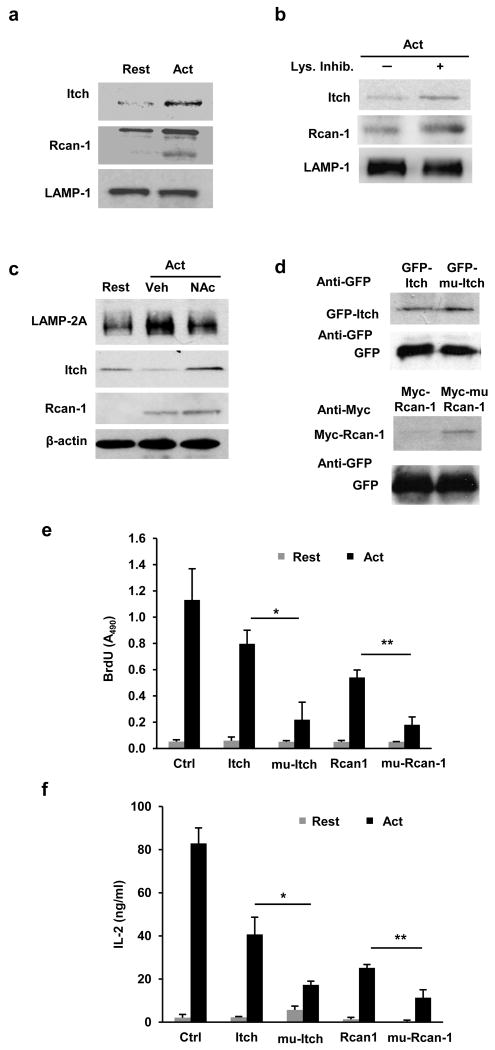
To investigate how CMA was required for T cell activation, we tested if CMA was required for the selective degradation of inhibitors of TCR signaling. CMA is a selective form of autophagy in which substrate specificity is conferred by the presence of a pentapeptide motif of biochemical properties comparable to those of KFERQ in the substrate protein, which allows recognition of the substrate by the cytosolic Hsc703. Candidate proteins with a previously defined negative regulatory effect on TCR signaling were screened for the presence of one or more CMA targeting motifs in their amino acid sequences. We identified the presence of three CMA targeting motifs in the E3 ubiquitin ligase Itch (data not shown) and the calcineurin inhibitor Rcan1, which was previously defined as a CMA substrates in neurons24. Both proteins were found enriched in the lysosomal fractions of activated T cells (Fig.3a), suggesting that they were being targeted to the lysosome for degradation following activation. A faster band specific for Rcan1, likely representing a smaller isoform of Rcan125, was also detected by immunoblot in lysosomes isolated from activated cells (Fig.3a). To determine if Itch and Rcan1 proteins were degraded in the lysosomes, we measured lysosomal amounts of Itch and Rcan-1 in T cells activated in the presence or absence of lysosomal protease inhibitors. Both Rcan-1 and Itch were further enriched in the lysosomal fractions of activated T cells in the presence of protease inhibitors as comopared to non-treated cells, suggesting lysosomal degradation in activated T cells (Fig.3b). Furthermore, N-acetylcystein, which prevented the upregulation of LAMP-2A expression, caused defective degradation of Itch in activated T cells (Fig.3c). Expression of Rcan-1 was previously reported to be upregulated in activated T cells26. Following this new synthesis (Supplementary Fig.3a), Rcan-1 was also degraded in TH1 cells (Supplementary Fig.3a), likely to maintain the cellular homeostasis of this protein. Treatment with N-acetylcysteine impaired Rcan-1 degradation, resulting in higher cellular amounts of Rcan-1 in activated T cells compared to non-treated cells (Fig.3c).
Figure 3. Degradation of Rcan-1 and Itch by CMA in activated T cells.
(a) Immunoblot showing enrichment of Itch and Rcan-1 in lysosomal fractions isolated from activated (24 hours antti-CD3+anti-CD28) TH1 cells compared to lysosomes obtained from resting cells. LAMP-1 is used as control. (b) Itch and Rcan-1 detected by immunoblot in lysosomal fractions from TH1 cells activated for 24 hours where lysosomal proteases had been inhibited or not for the last 3 hours with NH4Cl and leupeptin (Lys.Inhib.). LAMP-1 is used as control. (c) Immunoblot showing total cellular Itch and Rcan-1 protein in resting TH1 cells and in cells activated (24 hours) in the presence or absence of N-acetylcysteine (N-Ac). β-actin is used as control. (d) Immunoblot of cell lysates from TH1 cells transfected with plasmids expressing GFP or Myc tagged wild type or KFERQ-mutants (GFP-mu-Itch and Myc-mu-Rcan-1) Itch or Rcan-1, and activated for 24 hours with anti-CD3+anti-CD28. Efficiency of transfection is controlled in all cells with a co-transfected GFP expression plasmid. Blots are representative of 4 (a and b), 2 (c) and 3 (d) different experiments. (e-f) Cell proliferation and IL-2 production measured in TH1 cells transfected with plasmids expressing GFP (Ctrl), wild type Itch, mu-Itch, Rcan-1, mu-Rcan-1 and activated with anti-CD3+anti-CD28 for 24 (IL-2 production) or 48 (proliferation) hours Results are mean+s.e.m. from three different experiments (t test; (e) *p=0.019; **p=0.0034; (f) *p=0.046; **p=0.044).
To confirm that Itch and Rcan-1 were CMA substrates, we mutated all putative targeting CMA motifs in both proteins. TH1 cells were transfected with plasmids that expressed tagged wild-type (GFP-Itch or Myc-Rcan1) or mutant (GFP-mu-Itch or Myc-mu-Rcan1) Itch or Rcan1 and then activated them for 24 hours with anti-CD3 and anti-CD28 antibodies. Wild-type and mutant Itch and Rcan1 proteins expression was similar to those of the endogenous proteins also expressed in the transfected T cells (Supplementary Fig.3b-c). GFP-mu-Itch and Myc-mu-Rcan1 showed higher accumulation in activated T cells than GFP-Itch and Myc-Rcan1 (Fig.3d and Supplementary Fig.3b-e). This accumulation was not a consequence of differences in the efficiency of transfection, because similar expression of GFP , expressed from a co-transfected reporter vector, was detected in all conditions (Fig.3d). In addition, GFP-mu-Itch and Myc-mu-Rcan1 were not efficiently targeted to the lysosomal compartment (Supplementary Fig.3f-g). Transfection with GFP-mu-Itch and Myc-mu-Rcan1 suppressed activation-induced T cell proliferation (Fig.3e) and cytokine production compared to transfection of wild-type proteins (Fig.3f). These results indicate that degradation of Itch and Rcan-1 by CMA was required to maintain efficient T cell activation.
LAMP-2A deficiency diminishes adaptive immune responses in mice
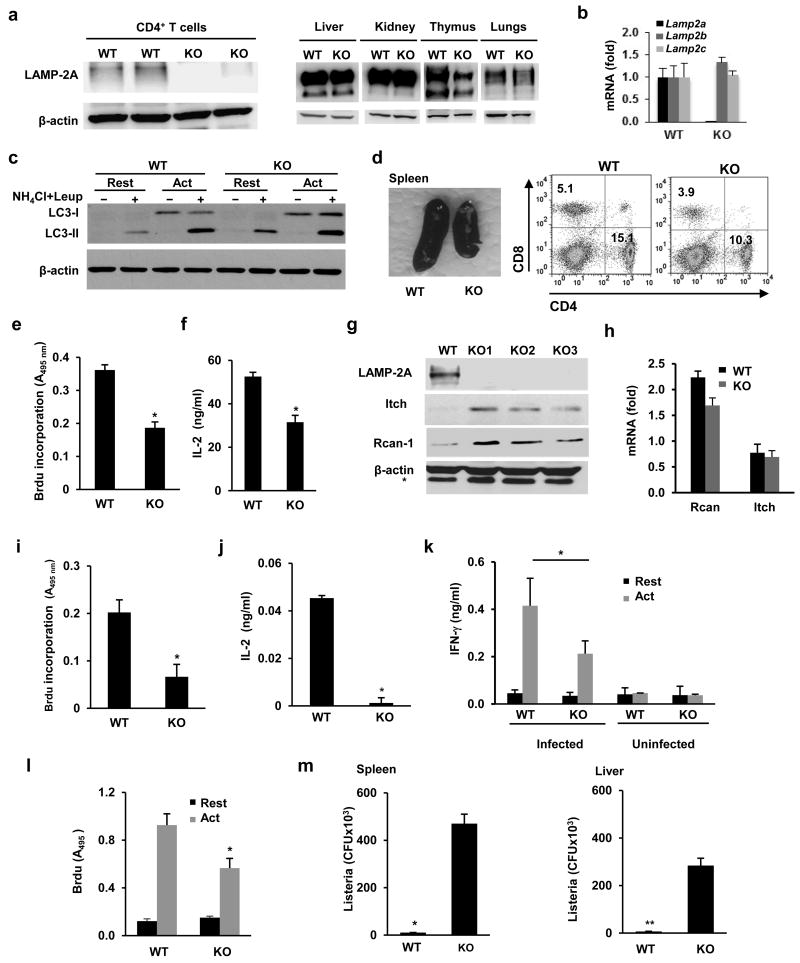
To analyze the function of CMA in T cells in vivo, we used a newly generated mouse model in which we specifically deleted Lamp2a in T cells. C57Bl/6 mice expressing exon 8a of the Lamp2 gene flanked by LoxP sites 27 were crossed with C57Bl/6 mice that expressed Cre recombinase under the control of the Lck promoter (Lck-cre). In these mice (called L2A-cKO here), deletion of exon 8a prevents the expression of Lamp2a (encoding for LAMP-2A, the only LAMP-2 isoform that acts as a receptor for CMA), while preserving the expression of the other two Lamp2 splicing variants, Lamp2b and Lamp2c. L2A-cKO mice showed specific loss of LAMP-2A expression in the T cell compartment, while expression was normal in other tissues (Fig.4a). Real time PCR analyses confirmed loss of expression of Lamp2a in T cells in the L2A-cKO mice, while Lamp2b and Lamp2c mRNA expression was comparable with control littermate mice (Fig.4b). Macroautophagy was not impaired in CD4+ T cells in these animals indicating the existence of a normal functional lysosomal compartment (Fig.4c). Basal macroautophagy activity was slightly elevated in LAMP-2A-deficient T cells (Fig.4c) , likely due to the previously described compensatory effect between these two forms of autophagy28, while activation-induced macroautophagy was similar in wild-type and LAMP-2A-deficient T cells (Fig.4c). Furthermore, the expression of lysosomal structural proteins (LAMP-1), lysosomal proteases and glycosidases (cathepsin D and β-hexosaminidase), as well as lysosomal stability were not decreased in LAMP-2A-deficient T cells (Supplementary Fig.4a-c)
Figure 4. Reduced T cell responses in CMA-deficient mice.
(a) Immunoblot of LAMP-2A in CD4+ T cells, liver, kidneys, thymus and lung of L2A-cKO (KO) mice or wild type littermates (WT). (b) Lamp2a, b and c mRNA expression (mean+s.e.m, expressed as relative to WT values) measured by qPCR in T cells from WT or L2A-cKO mice (n=3). (c) Macroautophagy flux (LC3 flux) measured in resting CD4+ T cells from WT and KO mice or cells activated with anti-CD3 and anti-CD218 for 24 hours (Act). (d) Representative image (from 6 different mice) of spleens from WT and L2A-cKO mice. Distribution of peripheral CD4+ and CD8+ T cells was determined by FACS. (e-f) Cell proliferation and IL-2 production measured in CD4+ T cells isolated from spleens of WT and KO mice following stimulation for 48 hours. Results are mean+s.e.m. of 4 (e) 6 (f) independent experiments (t test, (e) *p=0.0062; (f) *p=0.006). g. Immunoblot of Itch and Rcan-1 proteins in total lysates from activated (24 hours) CD4+ T cells of 3 different KO mice compared to WT littlermate controls. * Non-specific band. (h) Expression of Lamp2a mRNA assessed by qPCR in samples obtained from resting or activated for 24 hours CD4+ T cells isolated form WT or KO mice Data (mean+s.e.m. from 3 different experiments) is presented as fold induction of mRNA expression following activation. (i-j) Recall responses to OVA323-339 peptide (cell proliferation and IL-2 production) in draining lymph nodes isolated from OVA323-339-immunized KO mice and WT littermates. Results are mean+s.e.m. from 4 different experiments (t test; (i) *p=0.0054; (j) *p=0.0052). (k-l) Recall responses (cell proliferation and cytokine production) to Listeria in CD4+ T cells isolated from naïve or previously infected WT or KO mice. Graphs show mean+s.e.m. from 6 mice analyzed in two independent experiments (t test; (k) *p=0.032; (l) *p=0.02). (m) Listeria CFU in spleen and liver measured in previously immunized WT and KO mice after reinfection with a high dose of Listeria. Results show mean+s.e.m. from data obtained in triplicate from three different mice (t test; *p=0.008; **p=0.001).
LAMP-2A-deficient mice had a reduced spleen and thymus size, with decreased numbers of total thymocytes and a 20-30% reduction in the percentage of peripheral CD4+ and CD8+ T cells, but conserved numbers of CD4+CD25+ T cells (Fig.4d and Supplementary Fig.4). L2A-cKO mice also presented slightly higher percentages of CD4- CD8- (double negative) thymocytes, but no major differences in other thymic populations were apparent (Supplementary Fig.4d-f). Cell death in the spleen and thymus of L2A-cKO mice was comparable to control wild-type littermates (data not shown). Furthermore, peripheral LAMP-2A-deficient CD4+ T cells did not appear to have any defect in early signaling events downstream of the TCR (Supplementary Fig.5a-d).
As we saw in vitro, CD4+ T cells isolated from L2A-cKO mice showed impaired T cell responses, with a significant decrease in activation-induced proliferation and cytokine expression (Fig.4e-f). Expression of Itch and Rcan1 proteins was markedly higher in T cells from L2A-cKO mice than in T cells isolated from littermate controls (Fig.4g) and this was not a consequence of differences in expression of Rcan1 or Itch mRNA in LAMP-2A-deficient T cells (Fig.4h). Furthermore, we observed decreased amounts of Rcan-1 and Itch in the lysosomal compartment of LAMP-2A-deficient T cells, and whereas overexpression of Myc-mu-Rcan1 resulted in accumulation of the mutant Rcan1 protein in wild-type cells, Myc-Rcan1 and Myc-mu-Rcan1 accumulated similarly in LAMP-2A-deficient T cells (Supplementary Fig.3h and 5e-f).
To further analyze the role of CMA in the regulation of T cell function in vivo, we measured T cell responses in two different experimental models. First, mice were immunized with OVA323-339 peptide (two immunizations separated by one week). Seven days after the second immunization, draining lymph nodes were isolated and recall responses to OVA323-339 peptide was analyzed. Cell proliferation as well as cytokine production in response to re-challenge with OVA323-339 were significantly reduced in L2A-cKO mice compared to wild-type littermates (Fig.4i-j). Furthermore, anti-OVA323-339 specific immunoglobulin in serum was significantly lower in immunized L2A-cKO mice (Supplementary Fig.4g). In a second model, L2A-cKO mice and wild-type littermates were challenged intraperitoneally with attenuated Listeria monocytogenes. Seven days after injection, CD4+ T cells isolated from spleen of L2A-cKO mice proliferated less and showed significantly lower cytokine production than those isolated from control wild-type littemates mice after ex-vivo re-challenge with Listeria-infected antigen presenting cells (Fig.4k-l), while levels of specific anti-Listeria antibodies were lower in the serum of infected L2A-cKO mice compared to control littermates (Supplementary Fig.4h). Furthermore, when mice were re-challenged in vivo with a high dose of Listeria, L2A-cKO mice were less efficient in controlling this pathogen than wild-type littermates, which resulted in higher titers of Listeria in spleen and liver (Fig.4m).Together, these data indicate that CMA activity in T cells is required to generate effective in vivo T cell responses.
Deficient CMA contributes to age-associated decrease in T cell responses
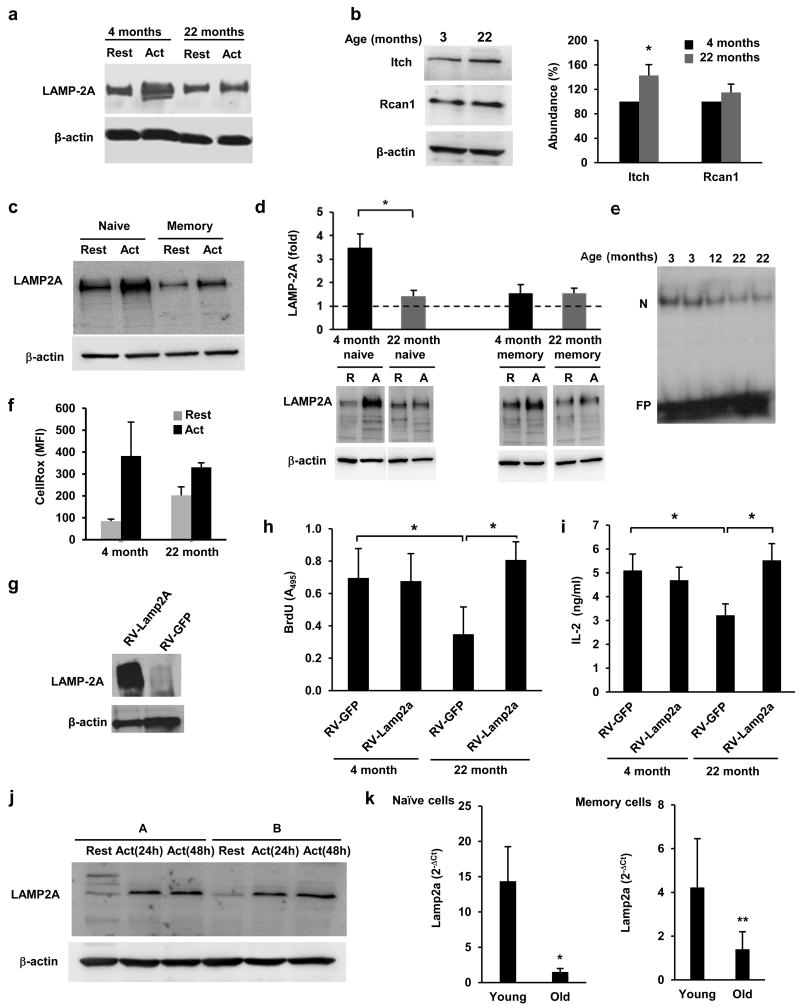
Because one of the best characterized physiological conditions where CMA activity is reduced is aging29, we investigated whether decreased CMA occurred in T cells with age and could account for the diminished T cell responses observed in aging organisms. Effector T cells differentiated from naïve cells isolated form aged mice failed to efficiently upregulate the expression of LAMP-2A in response to TCR engagement (Fig.5a). Consequently, higher expression of Itch (40%) and Rcan-1 (15%) protein was detected in activated T cells from 22-month old mice compared to T cells from 4 month old mice, though that difference was only statistically significant for Itch (Fig.5b). Freshly isolated naïve and memory CD4+ T cells isolated from young mice responded to TCR activation with increased LAMP-2A expression, though LAMP-2A upregulation was less pronounced in memory than in naive T cells (Fig.5c). Naïve CD4+ T cells from (22 month) old mice showed a significant impairment in their ability to upregulate LAMP-2A expression compared to young (4 month) mice, whereas this defect was not as evident in memory T cells (Fig.5d). Itch and Rcan1 accumulated in naïve T cells isolated from old mice (Supplementary Fig.5g), which also showed decreased activation-induced nuclear translocation of NFAT (Fig.5e). Furthermore, whereas as expected, naïve CD4+T cells isolated from 22-month old mice had higher basal ROS production, they were nonetheless less able to increase ROS expression upon TCR engagement (Fig.5g and Supplementary Fig.5h). Memory CD4+ T cells also showed some age-associated accumulation of Itch and Rcan-1, though the total cellular amounts of these proteins were much lower than those detected in naïve cells (Supplementary Fig.5g).
Figure 5. Deficient regulation of LAMP-2A expression in aged T cells.
(a) LAMP-2A protein detected by immunoblot in CD4+ T cells from 4-month and 22-month old mice that were left resting or stimulated with anti-CD3+anti-CD28 for 24 hours. Image representative of 3 different experiments. (b) Immunoblot showing Rcan-1 and Itch in CD4+ T cells from 4 and 22 month old mice stimulated for 24 hours. A quantification of blots from 3 different mice is shown (% of the protein content in 4 month old mice. mean+s.e.m.; t test; *p=0.044). (c-d) Immunoblot of LAMP-2A in total cell lysates prepared form resting and activated (24 hours) naive and memory CD4+ T cells (c). Similar experiments using naïve and memory CD4+ T cells from 4 month and 22 month old mice (d). A representative immunoblot and the quantification from blots of 3 different young and 3 old mice are shown (mean+s.e.m.; t test; *p=0.009). (e) EMSA showing NFAT binding using nuclear extracts from CD4+ T cells isolated from 3, 12 or 22 month old mice and activated for 4 hours. (N:NFAT; FP: free probe) (f) ROS production measured by FACS using CellRox-green in CD4+ T cells isolated from 4 or 22 month old mice and activated for 12 hours. Data (mean+s.e.m. from 3 different mice) is presented as mean fluorescence intensity (MFI). (g-i) Immunoblot for LAMP-2A (g), proliferation (h) and IL-2 production (i) measured in TH1 cells from 4 or 22 month old mice transduced with a retrovirus expressing human LAMP-2A and GFP (RV-Lamp2a) or just GFP (RV-GFP). Infected cells were sorted by GFP expression, cultured for 2 weeks and then stimulated for 24 to 48 hours. Results are mean+s.e.m. from 4 (h) and 5 (i) different experiments (ANOVA with Tukey post-test; *p<0.05). (j) Immunoblot showing LAMP-2A in total cell lysates obtained from resting and activated CD4+ T cells from two human donors (A and B). (k) Lamp2a mRNA expression measured by real time PCR in naïve and memory CD4+ T cells obtained from 3 young (26±2 years old) and 6 old (72±6 years old) human donors and activated for 24 hours. 2−ΔCt values (mean+s.e.m.) were calculated using actin as a reference gene (Mann-Whitney; *p=0.0033; **p=0.047).
To determine the functional relevance of decreased CMA activation in T cells with age, we analyzed the consequences of restoring the expression of LAMP-2A in aged T cells. T cells from old mice were less responsive to TCR engagement and showed decreased proliferation and cytokine expression compared to T cells from young mice (Fig.5h) However, activation-induced cell proliferation and cytokine production were both restored in naïve effector CD4+ T cells from 22 month old mice transduced with a LAMP-2A expressing retrovirus to levels similar to those detected in T cells from 4 month old mice (Fig.5g-i). The ability of CD4+ T cells to upregulate the expression of LAMP-2A in response to activation was also tested in human cells. CD4+ T cells isolated from leukopaks obtained from anonymous donors and activated with anti-CD3 and anti-CD28-coated beads for 24 or 48 hours also increased the expression of LAMP-2A upon activation (Fig.5j). We also analyzed the activation-induced Lamp2a expression in naïve and memory CD4+ T cells isolated from whole blood from young (26±2 years old) and old (78±6 years old) subjects and activated for 24 hours with anti-CD3 and anti-CD28-coated beads. Naive T cells from old subjects showed a significant defect in the induction of Lamp2a expression following activation compared to young T cells (Fig.5k). As seen in mice, memory human CD4+ T cells from young individuals also showed a less pronounced induction of Lamp2a compared to naïve cells. In addition, memory T cells from old subjects also showed a significant decreased in their capacity to upregulate Lamp2a expression compared to their young counterparts (Fig.5k). Together these data indicate that decreased CMA activity with age is a determinant factor of T cell immunosenescence.
Discussion
CMA contributes to the maintenance of the cellular homeostasis during prolonged starvation30 and to cellular quality control in response to different stressors such as oxidative stress10. The unique selectivity of this type of autophagy confers also important regulatory functions through the selective degradation of individual proteins in a tightly regulated manner31,32. Here we describe a regulatory role for CMA in T helper cells. Our data show that in response to TCR engagement, CD4+ T cells activate CMA through the upregulation of LAMP-2A expression. In activated T cells CMA directs the degradation of two negative regulators of TCR signaling, Itch and Rcan1, which allows the generation of effective T cell responses.
T cell activation leads to mitochondrial generation of ROS, which are required to ensure NFAT activation and IL-2 production22. Furthermore, the NADPH oxidase Duox1 couples H2O2 generation with TCR-downstream signaling events and contributes to store operated calcium entry and NFAT activation in CD4+ T cells21. Our data indicate that NFAT1 is recruited to the Lamp2 promoter in activated T cells and that Lamp2a upregulation in those cells can be prevented by the calcineurin inhibitor cyclosporine A or by blocking ROS production, which supports that ROS-activated calcium-calcineurin-mediated NFAT activation upregulates expression of Lamp2a and activates CMA following TCR engagement.
Basal macroautophagy maintains organelle homeostasis in T cells33,34. Macroautophagy is also activated in response to TCR engagement and regulates cell proliferation, energy metabolism and cell death13,14. Selective macroautophagy also regulates TCR signaling by targeting ubiquitinated Bcl-10, a component of the IKK-activating complex, for degradation35. The early kinetics of macroautophagy activation in T cells and the relatively late activation of CMA suggest that both processes may have distinct roles during T cell activation, as it has been shown in other systems36. Consistent with the late kinetics of CMA activation, we did not detect any defect in early signaling events in CMA-deficient T cells. Our data support that the maintenance of activation-induced T cell responses by CMA can be explained by the selective degradation of Itch and Rcan-1. These proteins have been described as negative regulators of TCR signaling, which would explain the need for a tight regulation of their cellular content to maintain T cell activation37. Mice deficient in Itch develop a systemic progressive autoimmune disease with hyperactive T cells38. Anergic T cells also upregulate the expression of Itch, which targets PLC-γ and PKC-θ for degradation39. Other proteins inactivated by Itch in T cells include JunB or the TCRζ chain, defining this E3 ubiquitin ligase as a negative regulator of TCR signaling38,40. Itch activity is regulated by tyrosine phosphorylation mediated by Fyn41. Though degradation as a regulatory mechanism of Itch activity has not been described in T cells, it has been proposed for other E3 ubiquitin ligases that regulate T cell activation, such as Cbl-b42. Rcan-1 is an inhibitor of calcineurin that contributes to setting the threshold for TCR activation and modulates calcineurin signaling-dependent gene expression43. Rcan-1 expression increases following TCR engagement26. A feed-back loop occurs in which NFAT2 activates Rcan-1 expression, which in turns modulates NFAT activation by controlling calcineurin activity44. Our results support a model in which CMA would participate in this loop through the regulated degradation of Rcan-1, preventing its accumulation, which otherwise could potentially lead to an uncontrolled inhibition of calcineurin activity.
Though the increased CMA activity that follows TCR engagement could account for the increased degradation of Itch and Rcan-1, we cannot exclude that posttranslational modifications may also contribute to this effect, either by creating new CMA motifs or by inducing conformational changes that may expose otherwise hidden CMA motifs45. It is also possible that CMA may regulate the degradation of other regulatory proteins in activated T cells. In this sense, CMA modulates cytosolic IκB content in response to nutritional stress in fibroblasts46.
CMA activity decreases with age in different tissues due to a gradual reduction of the lysosomal content of LAMP-2A, leading to the accumulation of damaged proteins and inefficient responses to stress8,29. In a murine transgenic model, restoration of LAMP-2A expression in liver in old mice resulted in decreased signs of cell damage and improved hepatic function12. In the aging immune system, the immunosenescent phenotype is particularly evident in the T cell compartment. Several mechanisms have been proposed to account for the decreased efficacy of T cell responses with age, from defects in signaling pathways and immunological synapse formation to the inability to maintain chromosomal integrity47-49. Our results show that old T cells also show reduced capacity to upregulate LAMP-2A expression and support that this dysfunction is not just a consequence of aging but contributes to the deficient T cell responses associated with age, as activation-induced responses are markedly improved when expression of LAMP-2A is restored in aged T cells. Decrease upregulation of TCR-induced LAMP-2A expression with age is very clear in naïve cells in mice and humans. However, while memory CD4+ T cells from aged mice show a similar capacity to increase LAMP-2A expression than cells from young mice, upregulation of LAMP-2A is clearly deficient in human old memory T cells. Although the reasons for this discrepancy between mouse and human remain to be determined, it might reflect the fact that memory T cell responses acquired early in life appeared to be conserved in mice49, while they are deficient in humans50. However, the fact that basal levels of Itch and Rcan-1 are much lower in memory than in naïve CD4+ T cell could indicate that in memory T cells CMA might be less important to eliminate those proteins, whose presence in the cell would be reduced during the generation of memory.
We propose a model in which TCR-induced ROS generation upregulates LAMP-2A expression, leading to the activation of CMA and the regulated degradation of TCR-signaling inhibitors, which is required to maintain T cell activation. Failure of this process with age would prevent the normal function of this regulatory mechanism, leading to decreased T cell responses. Modulation of CMA activity in T cells could, therefore, become a therapeutic tool to improve T cell function with age.
Methods
Human samples
Where indicated T cells were isolated from anonymous leukopaks obtained from the New York Blood Center. For the age-controlled studies samples were obtained from participants of the Einstein Longevity Study51. Informed written consent was obtained from participants. The protocol was approved under the guidelines of the Institutional Review Board of the Albert Einstein College of Medicine.
Mice
Six to eight-week-old C57BL/6 female mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and were maintained in pathogen-free conditions. To generate T cell-specific LAMP-2A deficient mice, Lamp2af/f mice, in which the LAMP-2A specific exon 8a of the Lamp2 gene is flanked by LoxP sites, were generated and crossed with mice expressing Cre recombinase under the control of an Lck promoter purchased from The Jackson Laboratory. Studies were performed in Lamp2af/f-Lck-Cre and littermate controls lacking the Cre transgene (Lamp2af/f). Deletion of Lamp2af/f was assessed by PCR and confirmed by immunoblot or real-time quantitative PCR. Where several animal treatments were compared, littermates were randomly assigned to each experimental group. All animal work was approved and performed according to the guidelines set by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee.
Cell culture
CD4+ T cells were isolated from lymph nodes and spleens of mice using anti-CD4-coupled magnetic beads (Life Technologies). Isolated T cells were stimulated with 0.5 μg/ml plate-bound anti-CD3 and 0.5 μg/ml anti-CD28 (clones 2C11 and 37.51; BD Biosciences) and in some cases differentiated to T helper 1 (TH1) or T helper 2 (TH2) for 6 d with IL-12 (10 ng/ml) (Cell Sciences), anti-IL-4 (10 μg/ml) (11B.11; NCI BRB Preclinical Repository) and 10 U/ml recombinant human IL-2 (NCI BRB Preclinical Repository), or IL-4 (10 ng/ml) and anti-IFNγ (10 μg/ml) (BD Biosciences) respectively. Where indicated, memory and naïve populations were separated using the CD4+CD62L+ T cell isolation Kit II (Miltenyi Biotec). CD4+ T cells and HEK293 and NIH-3T3 cell lines (American Type Culture Collection) were maintained in exponential growth in DMEM supplemented with 10% FCS, 2 mM L-glutamine, nonessential amino acids (Cambrex), essential vitamins (Cambrex), and 50 μM 2-mercaptoethanol. Where indicated, the ROS scavenger (10 μM) N-acetyl-cysteine and the pro-oxidants compounds (0.05-0.1 mM) Paraquat-methyl-dichloride hydrate and (1 mM) Hydrogen peroxide (Sigma-Aldrich) were added to the media. Human CD4+ T cells were isolated from PBMCs obtained from leukopaks or whole blood using a CD4+ T cell isolation Kit II (Miltenyi Biotec) and activated where indicated with Dynaneads Human T-activator CD3/CD28 beads (Life Technologies). Where indicated, further separation into naïve and memory populations was performed using a Memory CD4+ T cell Isolation Kit (Miltenyi Biotec). Human T cells were grown in RPMI supplemented with supplemented with 10% FCS and 2 mM L-glutamine.
ELISA
T cells (2.5-5×104) were stimulated with 0.5 μg/ml plate-bound anti-CD3 and anti-CD28 in 96-well plates for 24-48 hours. Supernatants were collected, and IL-2 and IFNγ levels were measured in a sandwich ELISA following the manufacturer's recommendations (BD Biosciences). Direct ELISA using OVA323-339 peptide precoated plates was used to detect anti-OVA antibodies in serum from immunized mice.
Proliferation assay
T cells (5×104) were stimulated with 0.5 μg/ml plate-bound anti-CD3 and anti-CD28 in 96-well plates. Forty eight hours later, BrdU was added for 12 h. Incorporation of BrdU was measured by ELISA according to the manufacturer's instructions (Roche).
Apoptosis assay
Apoptosis was determined with an Annexin V-PE apoptosis detection kit (BD Biosciences). Stained cells were acquired by FACS using an LSRII (Becton Dickinson) and data were analyzed with FlowJo software (Tree Star, Inc).
ROS production assessment
CD4+ T cells isolated from 3- or 22-month old mice were left resting or activated for 12 hours with anti-CD3 and anti-CD28 antibodies. 5 μM CellROX-green (Life Technologies) were added to the cell cultures for the last 30 minutes and cells were then analyzed by FACS.
Real-time PCR
cDNA was synthesized from total RNA, and gene expression was analyzed by real time PCR using SYBR Green in a Step One Plus Thermocycler (Applied Biosystems). Expression of each gene was normalized to actin. Primers used are shown in Supplementary Table 1.
Chromatin immunoprecipitation
Nuclear lysates from 106 to 107 paraformaldehyde fixed T cells were incubated overnight with anti-NFAT1 antibodies (clone 25A10, Thermo Scientific). Immune complexes were collected by Protein G Dynabeads (Life Technologies), and the recovered DNA fragments subjected to real time PCR. Specific primer pairs used are shown in Supplementary Table 1. Specific enrichments were calculated and expressed as percent recovery of inputs after subtracting background recovery obtained with non-specific isotype-matched antibodies.
Plasmid transfections and reporter assays
The murine proximal promoter of the Lamp2 gene was cloned in a luciferase reporter vector. TH1 cells were transfected by electroporation using a Nucleofector electroporator (Amaxa) following the manufacturer's recommendations with the firefly luciferase reporter plasmid under the control of a Lamp2 proximal promoter element and a renilla luciferase expression vector regulated by the pRL-TK promoter (ratio 20:1) as an internal control. Cells were stimulated with 0.5 μg/ml plate-bound anti-CD3 and 0.5 μg/ml anti-CD28 with or without ROS scavengers, 24 h after transfection. Cells were lysed and assayed for luciferase activity using the Promega Dual-Luciferase Reporter Assay System. To measure CMA using the photoactivable CMA reporter, TH1 cells were electroporated with KFERQ-PA-mCherry-1 19 and 24 hours post-transfection, the cells were exposed to a 405 nm light immediately following activation with 0.5 μg/ml plate-bound anti-CD3 and 0.5 μg/ml anti-CD28, T cells were collected at different times after activation, fixed and changes in the cellular pattern of the reporter were analyzed by fluorescence microscopy. Images were acquired with an Axiovert 2000 fluorescence microscope with apotome and quantification of images was done using ImageJ software (National Institutes of Health). Activation of CMA was measured as changes in the number of fluorescent puncta per cell.
PCR site-directed mutagenesis
CD4+ T cells were electroporated with a Nucleofector electroporator (Amaxa) following the manufacturer's recommendations with wild type Itch and Rcan-1 expressing plasmids, or vector expressing versions of those proteins containing mutated KFERQ motives. The QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Stratagene) was used to mutate the glutamines resides in all identified KFERQ motifs of Itch and Rcan-1 expression vectors (a gift from S. Ryeom, University of Pennsylvania). Primers used to create the mutant proteins are shown in Supplementary Table 1.
Lentiviral and retroviral infection of T cells
Knock down of Lamp2a was performed by transducing T cells with supernatants from HEK293 that were transfected with LV-IRES-GFP in which hairpin versions of one of three sequences targeting exon 8a of the Lamp2a gene 5′-GACTGCAGTGCAGATGAAG-3′, 5′-CTGCAATCTGATTGATTA-3′ or 5′-TAAACACTGCTTGACCACC-3′)28 were subcloned in the lentiviral vector. A control a LV-IRES-GFP expressing a scramble shRNA was also used Phoenix Ecotropic cells (a gift from G. Nolan, Stanford University) were transfected with retroviral vectors RV-IRES-GFP and RV-LAMP2-A-IRES-GFP developed as it is described above. Supernatants were collected 48 h after transfection, supplemented with 8 μg/ml polybrene, and used to infect CD4+ T cells 24 h after stimulation. Infected cells were sorted for GFP expression. LAMP-2A expression was assessed by immunoblot using an anti-human LAMP2 antibody (H4B4; Developmental Studies Hybridoma Bank).
siRNA transfections
5 million T cells were transfected with 15 μg of siRNA specific for Tfeb (5′- CAAGAAGGAUCUGGACUUA or 5′- AUGGCCAUGCUACAUAUCA); Duox1 (5′-CUGAAGAUGUGGAUGCACU or 5′-CACUUACAGCCGGGACAGA); Uqcrfs1 (5′- CAAGUUGUCUGAUAUCCCU or 5′-CUCAGAAGAGUGGAGAGUA) or a control non-targeting siRNA, by electroporation using a Nucleofector (Amaxa) After 24 hours mRNA levels were measured by RT-PCR or protein by immunoblot. T cells were then stimulated for 24 hours before quantifying lysosome numbers, using lysotracker Red (Life Technologies) or LAMP-2A levels, by immunoblot.
In vivo immunizations
Lamp2aF/F-Lck-Cre and littermate controls lacking the Cre transgene (Lamp2aF/F) were immunized with two rounds of subcutaneous injections of OVA323-339 peptide and Freud's adjuvant (Sigma-Aldrich) separated 1 week from each other. One week after the last immunization the mice were sacrificed and draining lymph nodes were isolated to analyze recall responses in vitro to the OVA peptide by ELISA and proliferation assay. Serum was also obtained from those mice to quantify anti-OVA peptide antibodies by ELISA.
Immunizations with Listeria monocytogenes
Mice were immunized with an attenuated Listeria monocytogenes as described previously52. Briefly, Lamp2aF/F-Lck-Cre and littermate controls were immunized intraperitoneally once with 107 CFU of Listeria. One week later, both groups of mice were euthanized and CD4+ T cells were isolated from spleen. To analyze T cell responses, 2.5×105 T cell-depleted splenocytes from C57Bl/6 mice were incubated with 2.5×103 Listeria CFU. After 1 hour, gentamycin was added to the media for an additional hour at 50 μg/ml to kill extracellular Listeria. Purified CD4+ T cells from naïve and Listeria infected Lamp2aF/F-Lck-Cre or littermate were then added add to the splenocytes and incubated for 48h. Cell proliferation and the cytokine expression were measured as described above. Where indicated, Listeria CFU were isolated from spleen and liver as described previously53. Lamp2aF/F-Lck-Cre and littermate wild type mice were injected with 0.5×107 CFU of Listeria on day one, and received a second dose of 5×108 Listeria CFU on day 8. Two days later, spleen and liver were dissected and homogenized, plated on agar. Listeria colonies were counted after 24 hours. The number of Listeria CFU was calculated per gram of tissue.
Immunoblotting
Total cellular lysates were prepared using RIPA buffer (1% Triton-X 100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate, pH 7.2). Fractions enriched in lysosomes active for CMA were isolated from cultured cells (0.5-1 ×108) after disruption of the plasma membrane by nitrogen cavitation through differential centrifugation and flotation in percoll/metrizamide discontinuous density gradients54. Mouse spleen lysosome-enriched fractions were isolated in a discontinuous metrizamide density gradient as described20. Stability of lysosomes at the moment of the isolation was measured by β-hexosaminidase latency54. Primary antibodies used are shown in Supplementary table 2.
LC3 Flux
Macroautophagy activity was quantified as the degradation of the autophagosome-associated protein LC3. Cells were left resting or activated for 20 hours and then treated with or without with 20 mM NH4Cl and 100 μM leupeptin for 4 hrs. Immunoblot against LC3 was performed to detect autophagosome-associated LC3 (LC3-II). Flux was calculated as the ratio of treated to untreated lines after normalization to actin55.
Electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared from T cells stimulated for 4 hours with anti-CD3 and anti-CD28 antibodies using the NE-PER system (Pierce). NFAT-specific oligonucleotides (5′-AGCTAGCTAGGAATATTCCTGGATGATC) were labeled with [32P] ATP and incubated with nuclear extracts (3-5μg) in a buffer containing 10 mM HEPES, pH 7.0, 125 mM NaCl, 10% glycerol, 0.25 mM DTT and 100ng/μl poly (dI-dC) for 20 min at 4 °C. DNA-protein complexes were resolved in polyacrylamide gel electrophoresis on a 4% non-denaturing gel.
Fluorescence microscopy
T cells or NIH3T3 cells were fixed with 4% paraformaldehyde, permeabilized, blocked and incubated with antibodies against LAMP-2A, LAMP-1 (The Developmental Studies Hybridoma Bank), Rcan1, Itch or Myc. Images were taken in a Zeiss Axiovert inverted microscope using an Apotome optical-sectioning module. Where indicated t cells were activated with anti-CD3 and anti-CD28 antibodies and NIH3T3 starved form serum for 24 hours to activate CMA. Particle measurements on immunofluorescence fields were performed using Image J software (National Institutes of Health).
Electron microscopy
Cells were fixed in 2.5% glutaraldehyde in 100 mM sodium cacodylate, pH 7.43, and postfixed in 1% osmium tetroxide in 100 mM sodium cacodylate, pH 7.43, followed by 1% uranyl acetate20. After ethanol dehydration and embedding in LX112 resin (LADD Research Industries, Williston, VT), ultrathin sections were cut on a Reichert Ultracut E and stained with uranyl acetate followed by lead citrate. All grids were viewed on a JEOL 100CX II transmission electron microscope at 80 kV. Morphometric analysis was performed using ImageJ software (National Institutes of Health) in 15–20 different micrographs for each condition after thresholding.
Statistical analysis
Analyses were performed using GraphPad Instat (GraphPad Software). Differences between multiple groups were analyzed by one-way ANOVA with a Tukey post-test. Comparisons between data pairs were analyzed using a t test or a Mann-Whitney test as indicated. For the human studies, power calculations were performed post-hoc after collecting data from 2 samples to determine the sample size required to obtain significance (defined as p<0.05) with a minimum 85% power. In vivo experiments using mice were performed with a minimum of 3 mice analyzed in at least two different days. All animals experiments were performed with samples unblinded to the investigator but analyses of human samples were blinded during the experiment and the assessment of results.
Supplementary Material
Acknowledgments
We would like to thank S. Kaushik for help in the preparation of lysosomal fractions and S. Bandyopadhyay for assistance in chromatin immunoprecipitation experiments. This work was supported by National Institutes of Health grant AG031782 and a Glenn Foundation Award (to F.M. and A.M.C). The use of specific research cores was supported by the Einstein Nathan Shock Center in Basic Research in Aging (AG038072).
Footnotes
Author contribution. R.V. designed and did experiments, analyzed data and wrote manuscript. Y.B., E.M., I.G-R., D.C. and H.K. did experiments and contributed to data interpretation. C.G. designed experiments and interpreted data. A.M.C. designed project and interpreted data. F.M. conceived and directed project, interpreted data and wrote the manuscript.
None of the authors has any no competing financial interest.
References
- 1.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM, Macian F. Autophagy, nutrition and immunology. Mol Aspects Med. 2012;33:2–13. doi: 10.1016/j.mam.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 4.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 5.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113 Pt 24:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 6.Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 8.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, et al. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 16.Huan C, Kelly ML, Steele R, Shapira I, Gottesman SR, et al. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat Immunol. 2006;7:1082–1091. doi: 10.1038/ni1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga H, Martinez-Vicente M, Arias E, Kaushik S, Sulzer D, et al. Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J Neurosci. 2011;31:18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 21.Kwon J, Shatynski KE, Chen H, Morand S, de Deken X, et al. The nonphagocytic NADPH oxidase Duox1 mediates a positive feedback loop during T cell receptor signaling. Sci Signal. 2010;3:ra59. doi: 10.1126/scisignal.2000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macian F, Garcia-Cozar F, Im SH, Horton HF, Byrne MC, et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009;23:3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- 25.Fuentes JJ, Pritchard MA, Estivill X. Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics. 1997;44:358–361. doi: 10.1006/geno.1997.4866. [DOI] [PubMed] [Google Scholar]
- 26.Narayan AV, Stadel R, Hahn AB, Bhoiwala DL, Cornielle G, et al. Redox response of the endogenous calcineurin inhibitor Adapt 78. Free Radic Biol Med. 2005;39:719–727. doi: 10.1016/j.freeradbiomed.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 27.Schneider JL, Suh Y, Cuervo AM. Deficient Chaperone-Mediated Autophagy in Liver Leads to Metabolic Dysregulation. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 30.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 31.Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q, She H, Gearing M, Colla E, Lee M, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 34.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard VM, Valdor R, Macian F, Cuervo AM. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology. 2012;13:21–35. doi: 10.1007/s10522-011-9331-x. [DOI] [PubMed] [Google Scholar]
- 37.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 38.Fang D, Elly C, Gao B, Fang N, Altman Y, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 39.Heissmeyer V, Macian F, Im SH, Varma R, Feske S, et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Jeon MS, Liao L, Yang C, Elly C, et al. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C, Zhou W, Jeon MS, Demydenko D, Harada Y, et al. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol Cell. 2006;21:135–141. doi: 10.1016/j.molcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Bardos T, Li D, Gal I, Vermes C, et al. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J Immunol. 2002;169:2236–2240. doi: 10.4049/jimmunol.169.5.2236. [DOI] [PubMed] [Google Scholar]
- 43.Ryeom S, Greenwald RJ, Sharpe AH, McKeon F. The threshold pattern of calcineurin-dependent gene expression is altered by loss of the endogenous inhibitor calcipressin. Nat Immunol. 2003;4:874–881. doi: 10.1038/ni966. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya S, Deb J, Patra AK, Thuy Pham DA, Chen W, et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin--NFAT signaling network. J Exp Med. 2011;208:823–839. doi: 10.1084/jem.20100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuervo AM, Hu W, Lim B, Dice JF. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou JP, Effros RB. T cell Replicative Senescence in Human Aging. Curr Pharm Des. 2012;19:1680–1698. doi: 10.2174/138161213805219711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenbraun MD, Tamir A, Miller RA. Altered composition of the immunological synapse in an anergic, age-dependent memory T cell subset. J Immunol. 2000;164:6105–6112. doi: 10.4049/jimmunol.164.12.6105. [DOI] [PubMed] [Google Scholar]
- 49.Maue AC, Yager EJ, Swain SL, Woodland DL, Blackman MA, et al. T-cell immunosenescence: lessons learned from mouse models of aging. Trends Immunol. 2009;30:301–305. doi: 10.1016/j.it.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goronzy JJ, Weyand CM. Understanding immunosenescence to improve responses to vaccines. Nat Immunol. 2013;14:428–436. doi: 10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 52.Kim SH, Castro F, Gonzalez D, Maciag PC, Paterson Y, et al. Mage-b vaccine delivered by recombinant Listeria monocytogenes is highly effective against breast cancer metastases. Br J Cancer. 2008;99:741–749. doi: 10.1038/sj.bjc.6604526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandra D, Jahangir A, Quispe-Tintaya W, Einstein MH, Gravekamp C. Myeloid-derived suppressor cells have a central role in attenuated Listeria monocytogenes-based immunotherapy against metastatic breast cancer in young and old mice. Br J Cancer. 2013;108:2281–2290. doi: 10.1038/bjc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storrie B, Madden EA. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 55.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.