Key Points
Loss of Bim contributes to adaptive rather than intrinsic bortezomib resistance in multiple myeloma.
A Bim-targeting strategy combining an HDACI with a BH3 mimetic overcomes such resistance through a new link between autophagy and apoptosis.
Abstract
Bim contributes to resistance to various standard and novel agents. Here we demonstrate that Bim plays a functional role in bortezomib resistance in multiple myeloma (MM) cells and that targeting Bim by combining histone deacetylase inhibitors (HDACIs) with BH3 mimetics (eg, ABT-737) overcomes bortezomib resistance. BH3-only protein profiling revealed high Bim levels (Bimhi) in most MM cell lines and primary CD138+ MM samples. Whereas short hairpin RNA Bim knockdown conferred bortezomib resistance in Bimhi cells, adaptive bortezomib-resistant cells displayed marked Bim downregulation. HDACI upregulated Bim and, when combined with ABT-737, which released Bim from Bcl-2/Bcl-xL, potently killed bortezomib-resistant cells. These events were correlated with Bim-associated autophagy attenuation, whereas Bim knockdown sharply increased autophagy in Bimhi cells. In Bimlow cells, autophagy disruption by chloroquine (CQ) was required for HDACI/ABT-737 to induce Bim expression and lethality. CQ also further enhanced HDACI/ABT-737 lethality in bortezomib-resistant cells. Finally, HDACI failed to diminish autophagy or potentiate ABT-737–induced apoptosis in bim−/− mouse embryonic fibroblasts. Thus, Bim deficiency represents a novel mechanism of adaptive bortezomib resistance in MM cells, and Bim-targeting strategies combining HDACIs (which upregulate Bim) and BH3 mimetics (which unleash Bim from antiapoptotic proteins) overcomes such resistance, in part by disabling cytoprotective autophagy.
Introduction
Multiple myeloma (MM) is an accumulative disorder of mature plasma cells. Recent treatment advances, including proteasome inhibitors (eg, bortezomib, carfilzomib) and immunomodulatory agents have significantly improved MM patient outcomes.1 However, relapse and drug resistance occur in virtually all responding patients.2 Like many malignancies, MM is characterized by dysregulation of the Bcl-2 family,3 divided into pro- and antiapoptotic groups. The former consists of multidomain (eg, Bak and Bax) and BH3-only proteins (eg, Bim, Bid, Puma, Noxa, Bad, Bik, Bmf, and Hrk), and the latter include multidomain proteins (eg, Bcl-2, Bcl-xL, Mcl-1).4 Whereas Bax and Bak are absolutely required for apoptosis, BH3-only proteins include activators (eg, Bim) and sensitizers or derepressors (eg, Noxa, Bik).5 Attention has focused on Bim because it determines the activity of diverse agents targeting oncogene-driven pathways.6,7 Bim is upregulated by inhibition of pathways (eg, MEK/ERK and PI3K/AKT) that repress expression through transcriptional regulation and/or posttranslational modifications, particularly phosphorylation.8 Bim phosphorylation promotes ubiquitination and proteasomal degradation.9,10 Notably, proteasome inhibitors (eg, bortezomib) block the latter process that results in Bim accumulation, which represents a mechanism of action (MOA) of these agents.11 However, not all MM patients respond to bortezomib (intrinsic resistance), and initial responders eventually relapse (adaptive or acquired resistance),12 thus prompting efforts to understand and overcome these events.
BH3 mimetics such as ABT-737 bind and inactivate antiapoptotic Bcl-2 family proteins, which induces apoptosis in MM cells.3,13 ABT-737 has two clinical analogs: ABT-263 (Navitoclax) and the newer-generation ABT-199, both of which target Bcl-2 and show promising activity in certain cancers,14 including hematopoietic malignancies.15 Mechanistically, Bim release from Bcl-2/Bcl-xL represents a major ABT-737 MOA.16 Notably, BH3 mimetics also induce autophagy by releasing Beclin-1 from Bcl-2/Bcl-xL.17 In contrast to apoptosis, autophagy is generally a cytoprotective mechanism that maintains intracellular homeostasis by removing harmful mal-folded proteins, protein aggregates, and damaged organelles,18 whereas autophagy inhibition promotes BH3-mimetic lethality.19 Importantly, a recent study demonstrated that Bim inhibits autophagy by sequestering Beclin-1 at microtubules.20 Conversely, histone deacetylase inhibitors (HDACIs) upregulate Bim in tumor cells, including MM cells.21,22 Among HDACIs, romidepsin and vorinostat have been approved for use in cutaneous T-cell lymphoma and peripheral T-cell lymphoma.23 HDACI lethality involves multiple mechanisms, including oxidative injury, death receptor upregulation, antiapoptotic protein downregulation, Bim upregulation, and disabling of chaperone and DNA repair proteins, among others.24 Notably, HDACIs also modulate autophagy.25-28
Currently, the role of Bim in resistance to proteasome inhibitors such as bortezomib is largely unknown. Here we report that Bim is widely expressed in MM cells, and although basal Bim levels do not correlate with intrinsic bortezomib resistance, Bim downregulation confers adaptive bortezomib resistance in Bimhi MM cells. Furthermore, HDACIs prime bortezomib-resistant cells that display Bim downregulation to BH3-mimetic lethality by increasing Bim expression. Mechanistically, Bim upregulation by HDACIs disables cytoprotective autophagic responses to BH3 mimetics. Finally, in Bimlow MM cells, which display minimal Bim upregulation in response to HDACIs, autophagy disruption (eg, by chloroquine [CQ]) is required for full response to this strategy. Collectively, these findings provide proof of principle for a Bim-targeting strategy in which HDACIs, which upregulate Bim, are combined with BH3 mimetics (eg, ABT-737), which unleash Bim from antiapoptotic Bcl-2 family proteins in bortezomib-resistant MM.
Materials and methods
Cells and reagents
Human MM cell lines,29,30 primary MM samples, and Bim knockout mouse embryonic fibroblasts (MEFs)31 are described in supplemental Materials, available on the Blood Web site. To establish human MM cells adaptively resistant to bortezomib, RPMI8226 and U266 cells were continuously cultured in gradually increasing concentrations of bortezomib (initially 0.5 nM and increasing in stepwise increments of 0.2 nM) to 10 nM or 15 nM, respectively.
The Bcl-2/Bcl-xL/Bcl-w antagonist ABT-737 and ABT-199 were kindly provided by Abbott Laboratories (Abbott Park, IL).32 Suberoyl bishydroxamic acid (SBHA)33 and CQ were purchased from Calbiochem (San Diego, CA) and Sigma-Aldrich (St. Louis, MO), respectively. Drugs were dissolved in sterile dimethylsulfoxide (final concentration <0.1%), prepared into aliquots, and stored at −20°C.
Autophagy analysis
LC3 (microtubule-associated protein 1A/1B-light chain 3) puncta were visualized after stable transfection with pE green fluorescent protein (GFP)-LC3B (plasmid 11546; Addgene, Cambridge, MA)34 by using a Zeiss LSM 700 confocal microscope. LC3 processing from LC3-I to LC3-II was monitored by immunoblotting analysis using anti-LC3 antibody (Novus, Littleton, CO).35,36 MEFs were stained with acridine orange (Sigma-Aldrich) to detect acidic vesicular organelles, indicating autophagy.37
Other procedures
See the supplemental Methods.
Statistical analysis
Values represent the means ± standard deviation for ≥3 independent experiments performed in triplicate. The significance of differences between experimental variables was determined by using 1-way analysis of variance with Tukey-Kramer multiple comparisons test and Student t test. P < .05 was considered significant.
Results
Bim downregulation confers adaptive rather than intrinsic bortezomib resistance in Bimhi MM cells
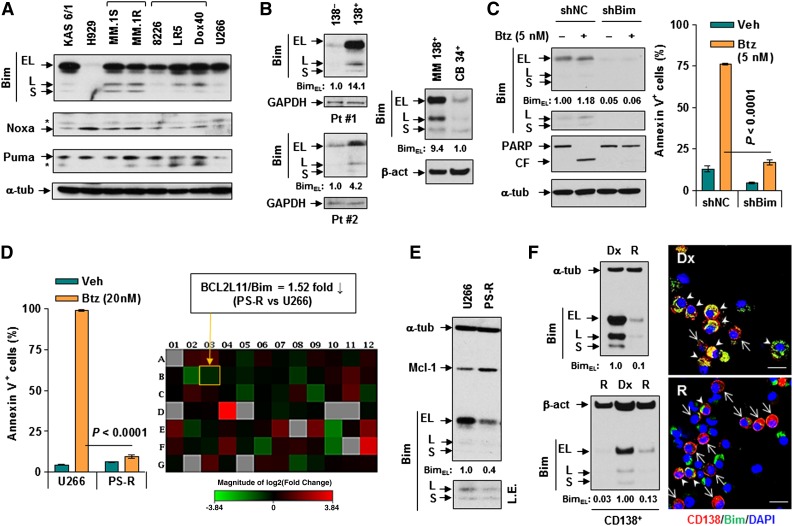
To examine basal levels of BH3-only proteins, 8 human MM cell lines were subjected to immunoblotting profiling. All untreated human MM cell lines analyzed displayed high Bim protein levels (Bimhi), with one exception: H929 cells (Bimlow) (Figure 1A). In contrast, basal Noxa or Puma expression was relatively low and varied between lines. Similarly, Bim was clearly expressed in primary CD138+ MM cells but was minimally expressed in their CD138– counterparts or normal cord blood CD34+ cells (Figure 1B and supplemental Figure 1A). Interestingly, H929 cells were equally if not more susceptible to bortezomib compared with other lines (eg, U266 and MM.1S; supplemental Figure 1B), suggesting lack of correlation between basal Bim levels and intrinsic bortezomib sensitivity. However, in Bimhi U266 cells, short hairpin RNA (shRNA) Bim knockdown sharply diminished poly adenosine diphosphate (ADP)-ribose polymerase (PARP) cleavage and reduced apoptosis (P < .0001 compared with negative controls [shNCs]) induced by bortezomib (5 nM, 24 hours; Figure 1C and supplemental Figure 1C), indicating a significant functional role for Bim in bortezomib sensitivity of Bimhi MM cells.
Figure 1.
Loss of Bim is associated with adaptive bortezomib resistance in Bimhi MM cells. (A) Immunoblotting analysis was performed to profile basal levels of the BH3-only proteins Bim (including EL, L, and S isoforms), Noxa, and/or Puma in untreated human MM cell lines. *Indicates nonspecific bands. (B) Three primary samples obtained from 2 patients with MM as well as normal CD34+ cells isolated from cord blood (CB) were subjected to immunoblotting analysis for basal levels of Bim. Blots for BimEL were quantified by using ImageJ software (available online). Values indicate fold-increase after normalization to loading controls (eg, glyceraldehyde-3-phosphate dehydrogenase [GAPDH] or β-actin [β-act]). (C) Bimhi U266 cells were stably transfected with constructs encoding shRNA-targeting Bim (shBim) or scrambled sequence as a negative control (shNC). Cells were then exposed to 5 nM bortezomib (Btz) for 24 hours, followed by immunoblotting for expression of Bim and cleavage of PARP (left panels) or flow cytometry to determine the percentage of apoptotic (annexin V+) cells (right panel). (D) U266 cells were continuously cultured with gradually increasing concentrations of bortezomib up to 15 nM to generate a subline (PS-R) that acquired marked resistance to bortezomib, as determined by flow cytometry (left panel). A Human Apoptosis RT2 Profiler Array Kit was then used to compare differences in the expression (messenger RNA [mRNA]) of key genes involved in apoptosis between PS-R and U266 cells. The heat map showed genes that were up- or downregulated in PS-R cells compared with U266 cells (right panel). (E) In parallel, immunoblotting analysis was conducted to monitor protein levels of Bim and/or Mcl-1 in untreated parental U266 cells and their bortezomib-resistant counterparts (PS-R). (F) Primary CD138+ cells were isolated from newly diagnosed (Dx; n = 2) or relapsed (R; n = 3, who had received prior bortezomib) MM patients, after which untreated cells were subjected to immunoblotting analysis (left) or immunofluorescence staining for CD138 (phycoerythrin, red) or Bim (AlexaFluor 488, green). Arrows and arrowheads indicate CD138+ cells displaying low and high Bim positivity, respectively. Scale bar = 10 μm; magnification ×40. α-tub, alpha tubulin; CF, cleaved fragment; DAPI, 4′,6 diamidino-2-phenylindole; L.E., long exposure, Veh, vehicle.
To assess Bim functional significance in adaptive bortezomib resistance, U266 cells were cultured in progressively higher bortezomib concentrations up to 15 nM, generating a highly resistant subline (PS-R). Whereas 20 nM bortezomib (24 hours) killed almost 100% of bortezomib-naïve U266 cells, PS-R cells displayed virtually complete resistance (Figure 1D, left, and supplemental Figure 1D). To identify candidate gene(s) responsible for resistance, a profiling array for 84 key apoptosis pathway genes compared PS-R cells to parental U266 cells (Figure 1D, right). The heat map identified the gene hit BCL2L11 (messenger RNA 1.52-fold lower in PS-R than in U266 cells), which encodes Bim, a finding validated by quantitative polymerase chain reaction (supplemental Figure 1E). PS-R cells consistently displayed sharply reduced Bim protein levels, particularly the EL isoform and, to a lesser extent, L and S isoforms, accompanied by modest increases in Mcl-1 expression (Figure 1E). Bim localized primarily to the mitochondria-enriched fraction38 where loss of Bim was also observed (supplemental Figure 1F). Bim expression was also clearly downregulated in bortezomib-resistant 8226/VR cells, which exhibited little change in Bcl-2, Bcl-xL, or Mcl-1 expression (supplemental Figure 1G). Notably, CD138+ cells isolated from newly diagnosed MM patients displayed higher basal Bim levels compared with those from relapsed patients who had received prior bortezomib (Figure 1F and supplemental Figure 1H). PS-R cells displayed little or no change in expression of other Bcl-2 family proteins, including antiapoptotic (eg, Bcl-2, Bcl-xL, Bfl1/A1, or Bcl-w) or other proapoptotic BH3-only proteins (eg, Puma, Bid, Bad, Bmf, and Bik; not shown). Interestingly, modest Noxa increases, but declines in Bfl1/A1 and Hrk expression were observed in PS-R cells (not shown). Together, these findings argue that whereas basal Bim expression per se does not predict bortezomib responsiveness, Bim downregulation represents a mechanism underlying adaptive forms of bortezomib resistance in MM cells initially expressing high Bim (ie, Bimhi).
Bim plays a significant functional role in BH3-mimetic sensitivity in MM cells
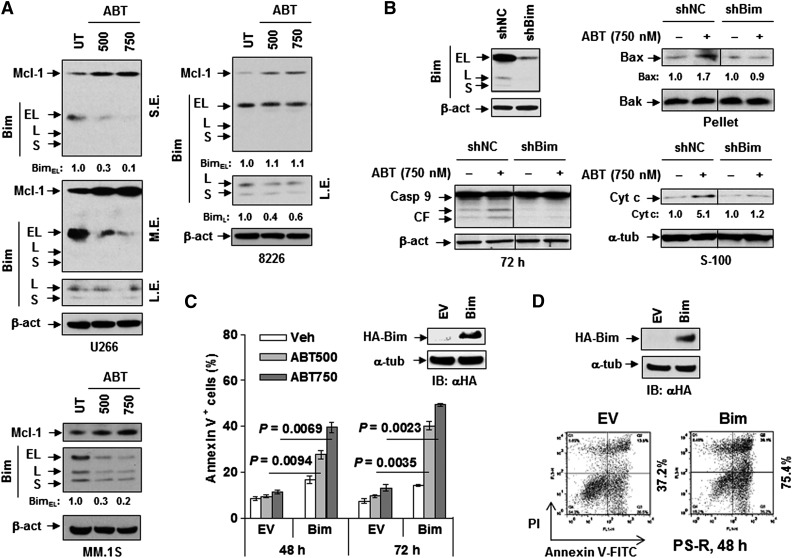
BH3 mimetics (eg, ABT-737) activate Bim by releasing it from antiapoptotic proteins (eg, Bcl-2, Bcl-xL), and BH3 peptides have been used to predict tumor cell susceptibility to anticancer therapies.39 Notably, exposure of various MM cells to ABT-737 clearly downregulated Bim, principally the EL isoform (Figure 2A). ABT-737 exposure also induced Mcl-1 upregulation, consistent with reports in lymphoma cells.40 These events presumably represent compensatory responses to disruption of Bim and Bcl-2/Bcl-xL interactions (supplemental Figure 2A). The functional role of Bim in ABT-737 lethality in MM cells was then assessed. Following exposure to 750 nM ABT-737 for 48 to 72 hours, shRNA knockdown of Bim diminished Bax mitochondrial translocation, cytosolic cytochrome c release, and caspase-9 cleavage (Figure 2B), as well as apoptosis (supplemental Figure 2B). Conversely, stable expression of hemagglutinin (HA)-tagged Bim (Figure 2C, inset) significantly sensitized MM cells to ABT-737 compared with empty-vector controls (P < .01 in each case; Figure 2C). Moreover, transient transfection of bortezomib-resistant PS-R cells with HA-tagged Bim resulted in robust apoptosis (Figure 2D), whereas ABT-737 failed to further increase lethality in these cells (supplemental Figure 2C). These findings suggest that BH3 mimetics elicit Bim downregulation and Mcl-1 upregulation as compensatory responses to disabling of Bcl-2/Bcl-xL and raise the possibility that a Bim-targeting strategy might be effective in bortezomib-resistant MM cells displaying Bim downregulation.
Figure 2.
Bim acts as a determinant of ABT-737 sensitivity in Bimhi MM cells. (A) U266 (upper left), RPMI8226 (upper right), and MM.1S (lower) were treated with the indicated concentrations (nM) of ABT-737 for 24 or 48 hours, after which expression of Bim and Mcl-1 was monitored by immunoblotting analysis. (B) U266 cells stably transfected with Bim shRNA (left upper panels) were treated with 750 nM ABT-737 for 72 hours, followed by immunoblotting analysis for monitoring caspase-9 (Casp-9) cleavage (left lower panels), Bax translocation (to mitochondria, right upper panels), and cytochrome c (Cyt c) release (to cytosol, right lower panels). Bak was probed for loading control of the mitochondria-enriched fraction. The vertical lines indicate the splice site in the composite image derived from a single blot. (C) RPMI8226 cells were stably transfected with HA-tagged human full-length Bim (inset, using an anti-HA antibody [αHA]) or an empty vector (EV). Cells were then exposed to 500 to 750 nM ABT-737 for 48 hours and 72 hours, after which the percentage of apoptotic cells was determined by flow cytometry. Numbers indicate P values. (D) Bortezomib-resistant U266 cells (PS-R) were transiently transfected with an empty vector or HA-tagged Bim construct. After 48 hours, untreated cells were lysed and subjected to immunoblotting analysis for HA-Bim (inset, using αHA) or flow cytometry to determine the percentage of apoptotic (annexin V+) cells. IB, immunoblot; FITC, fluorescein isothiocyanate; L.E., long exposure; M.E., medium exposure; S.E., short exposure; UT, untreated.
Bim upregulation primes bortezomib-resistant MM cells to BH3-mimetic lethality
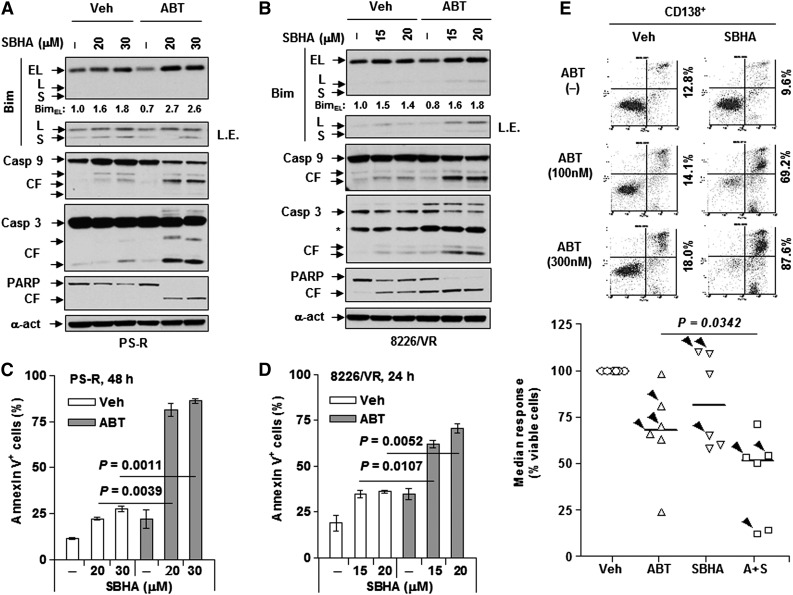
Treating drug-resistant tumor cells with BH3 (particularly Bim) peptides lowers the death threshold and increases sensitivity to anticancer agents (eg, ABT-737),41 that is, priming for cell death.42 Therefore, attempts were made to test whether Bim upregulation could mimic BH3 peptides in priming bortezomib-resistant MM cells that displayed loss of Bim to ABT-737–induced death by using HDACIs, which are known to upregulate Bim in myeloma cells.21,22 Exposure of either PS-R (Figure 3A) or 8226/VR cells (Figure 3B) to the HDACI SBHA (particularly in combination with ABT-737) clearly upregulated Bim (principally the EL isoform) and was accompanied by markedly increased caspase-3, caspase-9, and PARP cleavage and pronounced increases in apoptosis (P < .01 vs SBHA alone; Figure 3C-D). Similar results were obtained when the clinically relevant BH3 mimetic ABT-19914 was used (supplemental Figure 2D).
Figure 3.
Upregulation of Bim by SBHA potentiates ABT-737 lethality in bortezomib-resistant MM cells. (A-D) Bortezomib-resistant PS-R or 8226/VR cells were treated with ABT-737 (PS-R, 500 nM; 8226/VR, 300 nM) with or without the indicated concentrations (μM) of SBHA for 48 hours or 24 hours, respectively. After drug treatment, immunoblotting analysis and flow cytometry were performed to determine (A,B) levels of Bim and cleavage of caspases and PARP or (C,D) percentage of apoptosis. (E) Primary CD138+ cells were isolated from a bone marrow sample from a patient with relapsed MM. Cells were then treated with the indicated concentrations of ABT-737 (nM) with or without SBHA (μM) for 24 hours, after which flow cytometry was conducted to monitor percentage of early (annexin V+/PI–) and late (annexin V+/PI+) apoptosis. Values indicate the percentage of total annexin V+ cells. Parallel experiments were performed in additional primary samples derived from newly diagnosed MM patients or relapsed patients after treatment with bortezomib (arrowhead) (n = 6; 3 each for diagnosis and R patients). P < .05 for combination treatment vs each single agent. A+S, ABT-737 + SBHA; PI, propidium iodide.
To extend findings to primary bortezomib-resistant MM specimens, CD138+ cells were isolated from the bone marrow of a patient with relapsed MM after treatment with bortezomib. These cells exhibited resistance to bortezomib ex vivo (supplemental Figure 2E). Notably, whereas ABT-737 or SBHA administered individually induced little or no cell death, combined treatment strikingly increased apoptosis (70% to 90%) of CD138+ cells (Figure 3E, upper panel). In marked contrast, agents alone or in combination displayed little or no toxicity toward bone marrow nonmyeloma CD138– cells (supplemental Figure 2F). Similar results were observed in 6 additional primary samples, including 3 derived from relapsed patients (arrowhead; Figure 3E, lower panel). These findings argue that a dual Bim-targeting strategy combining HDACIs that upregulate Bim with BH3 mimetics that unleash Bim from antiapoptotic proteins (eg, Bcl-2/Bcl-xL) may be effective against adaptively bortezomib-resistant MM cells and raise the possibility that this strategy may preferentially target primary MM cells.
Autophagy disruption contributes functionally to HDACI/BH3-mimetic lethality
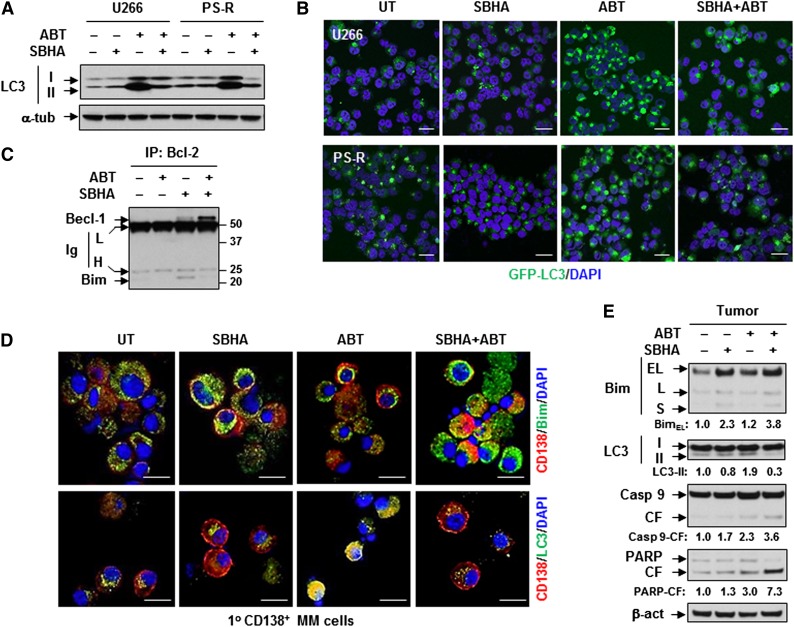
Recent findings indicate that in addition to triggering apoptosis,43,44 Bim also inhibits autophagy by localizing Beclin-1 to microtubules.20 Notably, BH3 mimetics such as ABT-737 potently induce autophagy by dissociating Beclin-1 from Bcl-2.17 Moreover, HDACIs also modulate autophagic responses.25-27,45 ABT-737 markedly increased levels of lipidated LC3-II, an event largely blocked by SBHA in bortezomib-naïve U266 cells and bortezomib-resistant PS-R cells (Figure 4A), findings confirmed by analysis of autophagic flux using bafilomycin A136 (supplemental Figure 3A). Consistent with these results, ABT-737 robustly increased the number of GFP-LC3 puncta, whereas SBHA alone had little effect. However, in both lines, SBHA sharply reduced ABT-737–induced GFP-LC3 puncta formation (Figure 4B). Interestingly, untreated PS-R cells exhibited modest increases in basal autophagy, reflected by both increased LC3-II expression and GFP-LC3 puncta compared with parental U266 cells (Figure 4A-B). Moreover, CD138+ cells derived from relapsed MM patients also exhibited relatively higher basal levels of autophagy compared with bortezomib-naïve patients (supplemental Figure 3B). However, clear changes in the expression of key proteins (eg, ULK1, Beclin-1, and ATG5) involved in autophagy initiation18 were not observed (supplemental Figure 3C).
Figure 4.
Bim upregulated by SBHA attenuates ABT-737–induced autophagy. (A) Drug-naïve (U266) and bortezomib-resistant (PS-R) cells were treated with 500 nM ABT-737 with or without 20 μM SBHA for 16 hours, followed by immunoblotting analysis to monitor LC3 processing. (B) U266 and PS-R cells stably expressing GFP-LC3 were treated (6 hours) as described in (A), and then analyzed for GFP-LC3 puncta by confocal microscopy. Scale bar = 10 mm (magnification × 20). (C) U266 cells were treated as described in (A), after which cells were lysed in 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) buffer and subjected to immunoprecipitation (IP) with anti-Bcl-2 antibody and subsequent immunoblotting analyses using anti–Beclin-1 (Becl-1) and anti-Bim antibodies. (D) Primary CD138+ cells derived from a patient with relapsed MM were treated with 100 nM ABT-737 with or without 10 μM SBHA for 16 hours, after which immunofluorescence staining was performed to monitor expression of Bim or LC3 (both AlexaFluor 488, green) in CD138-phycoerythrin-positive cells. Scale bar = 10 μm; magnification ×40. (E) Tumor tissues obtained from PS-R cell mouse xenografts following the indicated treatments were subjected to immunoblotting analysis to monitor expression of Bim and LC3, as well as cleavage of caspase-9 and PARP. H, heavy chain; Ig, immunoglobulin; L, light chain.
Although Bim has been reported to interact directly with and sequester Beclin-1,20 co-immunoprecipitation (co-IP) with either anti-Bim or anti-Beclin-1 antibodies did not detect direct Bim/Beclin-1 associations in either parental U266 cells (not shown) or bortezomib-resistant PS-R cells with or without drug treatment (supplemental Figure 3D). In contrast, marked Bim/Bcl-2 binding was observed on the same membrane, an event dramatically blocked by ABT-737 (supplemental Figure 3D). Notably, co-IP with anti-Bcl-2 antibody revealed that SBHA alone increased Bim/Bcl-2 binding, presumably secondary to Bim upregulation, whereas ABT-737 clearly released Bim from binding to Bcl-2 (Figure 4C), as previously reported.16 Interestingly, whereas SBHA alone modestly increased Beclin-1/Bcl-2 binding, coadministration of SBHA with ABT-737 induced a marked increase in Beclin-1/Bcl-2 association (Figure 4C), which prevents Beclin-1–induced autophagy.46 Similar results were observed in PS-R cells (not shown). These findings raise the possibility that release of Bcl-2 from Bim may contribute to increased Bcl-2/Beclin-1 binding. Notably, SBHA coadministration also upregulated Bim and diminished ABT-737–induced autophagy in primary CD138+ MM cells derived from relapsed MM patients, as shown by the representative images obtained from one patient (Figure 4D), as well as in tumor tissues obtained from bortezomib-resistant MM cell xenografts (Figure 4E).
Bim plays a functional role in autophagy disruption
The role of Bim in regulation of autophagy was then assessed. SBHA clearly reduced ABT-737–induced LC3-II expression in shNC cells (Figure 5A), consistent with previous results involving untransfected U266 cells. Notably, shBim cells, with or without drug treatment, exhibited striking increases in LC3-II levels and marked reductions in caspase-3 and PARP cleavage induced by SBHA with or without ABT-737. Similar results were obtained with tunicamycin, a classic autophagy inducer (supplemental Figure 3E).
Figure 5.
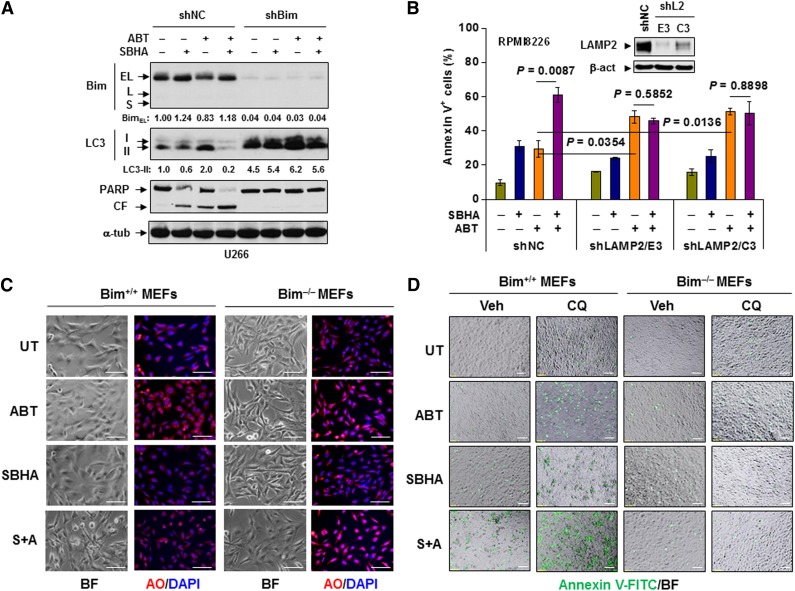
Bim is required for disruption of autophagy and induction of apoptosis induced by ABT-737. (A) U266 cells stably transfected with Bim or scrambled sequence shRNA were incubated with 300 nM ABT-737 with or without 20 μM SBHA for 24 hours. Following treatment, immunoblotting analysis was performed to monitor Bim expression, LC3 processing, and PARP cleavage. (B) RPMI8226 cells stably transfected with LAMP2 (two subclones designated E3 and C3) or scrambled sequence shRNA (inset) were exposed to 300 nM ABT-737 with or without 20 μM SBHA for 24 hours followed by flow cytometry to determine the percentage of apoptotic cells. (C) MEFs derived from wild-type (Bim+/+) or bim knockout (Bim−/−) mice were treated with 500 nM ABT-737 with or without 20 μM SBHA for 16 hours, after which cells were stained with acridine orange (AO). (D) MEFs were exposed to 500 nM ABT-737 with or without 20 μM SBHA in the presence or absence of 50 μM CQ for 24 hours, after which cells were stained with annexin V-FITC. For both 5C and 5D images were captured by inverted fluorescence microscopy. BF, bright field. Scale bar = 10 μm; magnification ×10. S+A, SBHA + ABT-737; shL2, shLAMP2.
To define the functional significance of autophagy disruption, RPMI8226 cells with stable shRNA knockdown of LAMP2, a key protein required for autophagosome maturation and lysosome fusion,47 were used. Two separate shLAMP2 clones (designated E3 and C3; Figure 5B inset) displayed significantly increased ABT-737 sensitivity (for each clone, P < .05 vs shNC cells; Figure 5B). These findings are consistent with previous results demonstrating the potentiation of ABT-737 lethality by CQ, which disrupts autophagosome maturation through inhibition of lysosomal acidification48 in lung cancer cells.19 However, whereas SBHA significantly increased ABT-737 lethality in shNC cells (P < .01 vs ABT-737 alone), it failed to potentiate ABT-737 lethality in shLAMP2 cells (P > .05 vs ABT-737 alone), presumably because autophagy had already been disabled in these cells. These findings argue that HDACI-induced Bim negatively regulates ABT-737–induced autophagy and that Bim-mediated suppression of a cytoprotective autophagic response plays a significant functional role in potentiation of BH3-mimetic lethality by HDACIs in both bortezomib-naïve and botezomib-resistant MM cells.
To define requirements for Bim in HDACI–BH3-mimetic interactions, bim gene knockout (Bim−/−) MEFs were used. Following ABT-737 exposure, wild-type (Bim+/+) MEFs stained with acridine orange displayed a clear increase in acidic vesicular organelles (orange or red; Figure 5C), including autophagosomes and lysosomes, that reflect enhanced autophagy.37 SBHA clearly inhibited ABT-737–induced autophagy. In sharp contrast, SBHA failed to attenuate ABT-737–induced autophagy in Bim−/− MEFs (Figure 5C). Moreover, ABT-737/SBHA markedly induced apoptosis in Bim+/+ MEFs, a phenomenon enhanced by CQ. However, bim gene knockout (Bim−/−) MEFs displayed sharply diminished ABT-737/SBHA-mediated apoptosis, even in the presence of CQ (Figure 5D). These findings argue that the presence of the bim gene is required for Bim-mediated inhibition of cytoprotective autophagy as well as the effectiveness of a strategy combining HDACIs with BH3 mimetics.
Autophagy disruption is required for enhanced HDACI/BH3-mimetic lethality in Bimlow MM cells
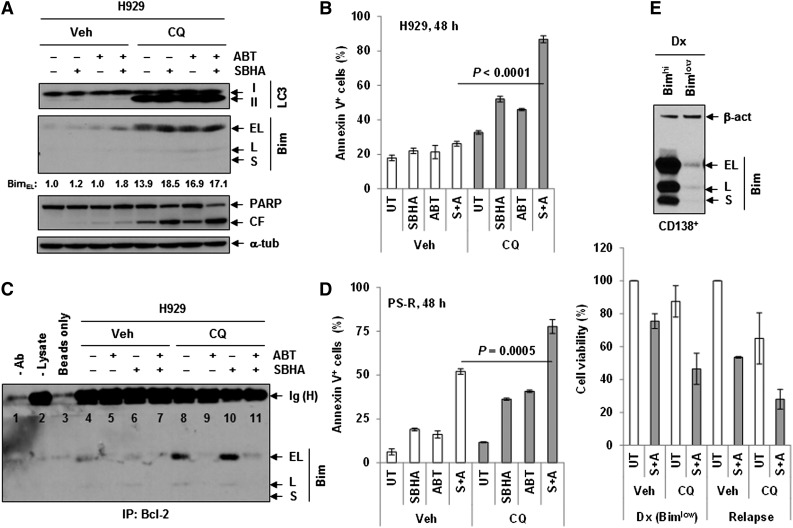
To determine whether analogous events also occur in Bimlow MM cells, H929 cells that intrinsically express low levels of Bim (Figure 1A) were used. In contrast to Bimhi cells, SBHA failed to upregulate Bim in H929 cells, whereas SBHA/ABT-737 coexposure induced very modest increases in Bim expression and PARP cleavage in these Bimlow cells (Figure 6A). Consistent with these results, SBHA/ABT-737 minimally induced apoptosis in H929 cells (Figure 6B). However, CQ disrupted autophagosome maturation, reflected by LC3-II accumulation and sharply upregulated Bim in H929 cells (Figure 6A), presumably because of interference with autophagic degradation of ubiquitinated proteins49 including Bim.9,10 Interestingly, in the presence of CQ, SBHA with or without ABT-737 treatment further increased Bim expression (Figure 6A). Moreover, co-IP revealed that Bim upregulation by CQ alone (lane 8) or SBHA plus CQ (lane 10) (Figure 6C) induced increased binding of Bim to Bcl-2, an established mechanism of Bim neutralization.50 Notably, such binding was essentially abrogated by ABT-737 (lanes 9 and 11; Figure 6C) thereby unleashing and activating Bim.16 These events were associated with pronounced potentiation of SBHA/ABT-737–mediated apoptosis by CQ (P < .0001 vs SBHA/ABT-737 treatment without CQ; Figure 6B) and PARP cleavage (Figure 6A). In contrast to H929 cells, in adaptively bortezomib-resistant cells (eg, PS-R) characterized by acquired Bim loss, SBHA clearly upregulated Bim and significantly increased ABT-737–mediated apoptosis (Figure 3A,C). However, CQ coadministration induced a pronounced further increase in cell death in PS-R cells (P < .01 vs SBHA/ABT-737 treatment without CQ; Figure 6D) but not in parental U266 cells (not shown). Moreover, CQ also enhanced Bim upregulation induced by SBHA with or without ABT-737 in PS-R cells (not shown), whereas ABT-737 diminished Bim/Bcl-2 associations (supplemental Figure 3D). Similar results were obtained in primary CD138+ MM cells displaying low basal Bim or derived from relapsed patients (Figure 6E). Together, these findings indicate that disrupting autophagy (eg, by CQ) plays a critical role in potentiating HDACI/BH3-mimetic lethality in Bimlow MM cells displaying minimal Bim upregulation after exposure to HDACIs, but it also significantly enhances anti-MM activity of this strategy in cells with acquired bortezomib resistance.
Figure 6.
CQ disrupts autophagy and sensitizes Bimlow MM cells to the HDACI/BH3 mimetic regimen. (A) H929 cells were exposed to 300 nM ABT-737 with or without 30 μM SBHA in the presence or absence of 50 μM CQ, after which immunoblotting analysis was performed to monitor LC3 processing, Bim expression, and PARP cleavage. (B) In parallel, flow cytometry was performed to determine the percentage of apoptotic H929 cells. (C) Alternatively, cells were lysed in CHAPS buffer and subjected to co-IP (IP with anti-Bcl-2, immunoblotting with anti-Bim). IP without anti-Bcl-2 (lane 1), cell lysate (lane 2), or both (lane 3) was performed as controls. (D) Similarly, PS-R cells were incubated with relatively lower concentrations of ABT-737 (300 nM) with or without SBHA (15 μM) with or without 50 μM CQ for 48 hours, followed by flow cytometry to monitor apoptosis. (E) Primary CD138+ cells derived from newly diagnosed (displaying low basal Bim levels, inset) or relapsed MM patients (n = 4; 2 samples each for Dx and R) were treated with 100 nM ABT-737 plus 10 μM SBHA in the presence or absence of 10 μM CQ for 24 hours, after which flow cytometry was performed to determine the percentage of viable cells (7-aminoactinomycin D [7AAD] staining-negative).
The HDACI/BH3-mimetic regimen is active in vivo but is inactivated by Bim shRNA knockdown
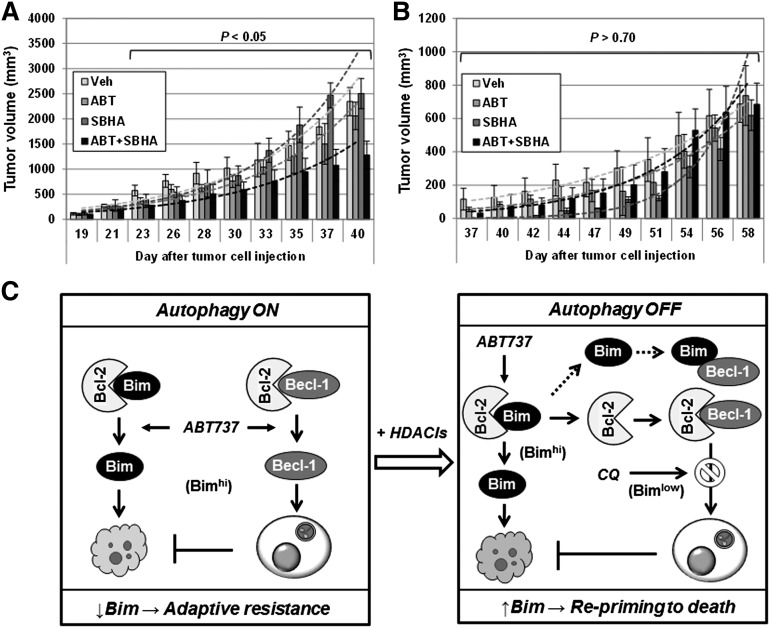
Finally, the in vivo activity of this regimen was examined in mouse xenograft models. Coadministration of ABT-737 with SBHA significantly reduced tumor burden (Figure 7A) and prolonged animal survival (not shown) in a bortezomib-resistant (PS-R) MM cell xenograft. In contrast, the same regimen was ineffective in a U266/shBim cell xenograft (Figure 7B), in which SBHA failed to upregulate Bim (supplemental Figure 3F). These findings suggest that Bim upregulation also plays a functional role in the in vivo activity of the HDACI/BH3-mimetic regimen.
Figure 7.
The HDACI/BH3-mimetic regimen displays activity in vivo which is blocked by Bim shRNA knockdown. (A-B) NOD/SCID-γ mice were subcutaneously inoculated in the flank with (A) 5 × 106 PS-R or (B) U266/shBim cells. Doses of 100 mg/kg ABT-737 and 200 mg/kg SBHA were administrated intraperitoneally individually or in combination (n = 5 per group) 3 days per week. Control animals were administered equal volumes of vehicle. Tumor size was measured by caliper, and volumes were calculated by using the formula (length × width2)/2. (C) A mechanistic model for the roles of Bim in adaptive drug resistance and priming resistant MM cells toward death. Left: BH3 mimetics (eg, ABT-737) simultaneously activate apoptosis and autophagy by releasing Bim and Beclin-1 from Bcl-2, respectively. Whereas the former action is therapeutically beneficial, it is opposed by the latter cytoprotective response (autophagy ON), which raises the cell death threshold and promotes drug resistance. In this setting, Bim downregulation is associated with adaptive resistance to targeted agents such as bortezomib. Right: HDACIs upregulate Bim in MM cells, including those resistant to bortezomib due to Bim downregulation, and thereby reprime them to BH3-mimetic–induced apoptosis. The latter event is related, at least in part, to disruption of cytoprotective autophagy (autophagy OFF) through release of Bcl-2 from Bim and resulting enhanced sequestration of Beclin-1 by Bcl-2. Notably, inhibition of autophagy (eg, by CQ) significantly increases HDACI/BH3 mimetic regimen lethality, particularly in Bimlow MM cells. Thus, loss of Bim expression can contribute to an adaptive form of bortezomib resistance, and a Bim-targeting strategy combining HDACIs, which upregulate Bim, with BH3 mimetics, which release Bim from antiapoptotic Bcl-2 family proteins, may represent an effective approach to this problem.
Discussion
Despite the introduction of effective therapies in MM, including front-line treatment with bortezomib, nearly all patients eventually relapse.2 One approach to this problem involves the use of second-generation proteasome inhibitors (eg, carfilzomib), which are effective in some bortezomib-refractory patients.12,51 However, there is a pressing need to understand mechanisms of resistance to proteasome inhibitors such as bortezomib and to develop strategies active against such resistant cells. Previous efforts have highlighted the contribution of the antiapoptotic Bcl-2 family protein Mcl-1 to MM cell survival and bortezomib resistance,52 but recent attention has focused on the role of proapoptotic BH3-only proteins (eg, Bim) in MM responsiveness to bortezomib.53 Bim downregulation correlates with poor prognosis in MM,54 but the precise role that Bim plays in bortezomib resistance has not yet been defined. Current studies emphasize an important contribution of Bim to adaptive (or acquired) rather than intrinsic forms of bortezomib resistance in MM. They also describe a Bim-targeting strategy that combines HDACIs that upregulate Bim21,22 with BH3 mimetics that unleash Bim from antiapoptotic proteins (eg, Bcl-2, Bcl-xL),16 which may overcome such forms of bortezomib resistance.
Our results suggest that Bim downregulation contributes functionally to adaptive (but not intrinsic) bortezomib resistance in MM cells. Recent gene expression profiling data reveal that BCL2L11 (Bim) is consistently and highly expressed in 4 MM subtypes (ie, HY, CCND1, MEF, and MMSET).55 Consistent with these findings, most of the MM cell lines and primary MM samples tested displayed high basal Bim levels (ie, Bimhi), suggesting that MM cells are primed by BH3-only proteins (eg, Bim) for cell death.41 Interestingly, MM cells (eg, H929) that exhibit low basal levels of Bim (Bimlow) were fully sensitive to bortezomib, arguing against a role for Bim in intrinsic bortezomib resistance. However, in Bimhi MM cells, shRNA knockdown of Bim dramatically reduced bortezomib sensitivity. Significantly, apoptosis pathway–targeted gene expression profiling identified BCL2L11 (Bim) as one of the genes downregulated in bortezomib-resistant MM cells, supported by clearly diminished Bim protein levels in these cells. Collectively, these observations argue that Bim downregulation is primarily involved in adaptive bortezomib resistance in which it raises the death threshold (ie, loss of priming).50 In addition, perturbations in the expression of other Bcl-2 family members (eg, Mcl-1, Hrk) may also contribute to or cooperate with Bim downregulation in conferring bortezomib resistance. For example, interactions between Bim and Mcl-1 may regulate MM cell survival56 or determine sensitivity to anti-MM agents (eg, melphalan).57 Efforts to test these hypotheses are underway.
A corollary of these findings is that agents that upregulate Bim may re-prime bortezomib-resistant MM cells toward death. Bim BH3 peptides prime neoplastic cells (including MM cells) for apoptosis induced by ABT-737,41 but the therapeutic use of peptides presents a challenge. Alternatively, HDACIs upregulate Bim in tumor cells, including MM cells,21,22 whereas BH3 mimetics (eg, ABT-737) unleash Bim from Bcl-2/Bcl-xL.16 Interestingly, ABT-737 induced a modest but discernible decline in Bim levels, accompanied by increased Mcl-1 expression,40 presumably representing compensatory responses to Bcl-2/Bcl-xL inhibition. Moreover, shRNA Bim knockdown markedly diminished ABT-737 lethality, whereas Bim overexpression increased ABT-737 sensitivity, arguing that Bim is critical for ABT-737 sensitivity.58 Notably, BH3 mimetics are effective against certain molecular MM subtypes that harbor a t(11;14) and a Bcl-2hi/Mcl-1low profile.59 Thus, the MOAs of HDACIs (eg, Bim upregulation) and BH3 mimetics (eg, unleashing Bim from Bcl-2/Bcl-xL) provide a specific rationale for combining these two classes of agents. Although we and other groups have described synergistic interactions between HDACIs and BH3 mimetics in various hematopoietic malignant cells,29,60-62 the ability of such a Bim-targeting strategy to re-prime bortezomib-resistant MM cells toward death has not previously been examined. Significantly, HDACIs, particularly when combined with BH3 mimetics, increased Bim expression and sharply increased BH3-mimetic lethality in bortezomib-resistant cells. Collectively, these findings indicate that, as described in the case of Bim peptides,41 an alternative Bim-targeting strategy that combines HDACIs and BH3 mimetics may be feasible and effective against certain adaptive forms of proteasome inhibitor resistance.
Apoptosis (type I) and autophagy (type II) represent two major forms of programmed cell death.63 Although excessive autophagy has been implicated in cell death in some settings, autophagy represents a cytoprotective mechanism conferring drug resistance in most circumstances.18 In this context, BH3 mimetics (including ABT-737) trigger autophagy by unleashing Beclin-1 from its inhibitory association with Bcl-2,17 whereas disruption of this event enhances BH3-mimetic lethality.19 Analogously, HDACIs induce autophagy in certain tumor cell types such as glioblastoma cells25 and breast cancer cells.26 Moreover, interference with autophagy (eg, by CQ or 3-MA (3-methyladenine), which disrupts autophagosome maturation64) enhances HDACI lethality.27 However, HDACIs may also inhibit autophagy under some conditions.46,65 Notably, in this study, HDACI coadministration predominantly attenuated BH3 mimetic–induced autophagy in MM cells.
Our results argue that the mechanism by which HDACIs disrupt autophagy in this setting involves Bim upregulation. A recent study demonstrated that Bim bridges the Beclin-1/LC8 interaction and thereby inhibits authophagy.20 Thus, HDACIs may inhibit BH3 mimetic–induced autophagy by upregulating Bim. Indeed, shRNA Bim knockdown induced a pronounced increase in autophagy and reduced the ability of SBHA to attenuate ABT-737–mediated autophagy in MM cells. SBHA also failed to prevent ABT-737–induced autophagy in Bim knockout MEFs. Furthermore, genetic disruption of autophagy (eg, by LAMP2 shRNA) significantly sensitized MM cells to ABT-737 but diminished potentiation of ABT-737 lethality by SBHA, presumably because autophagy had already been disrupted. Interestingly, although SBHA minimally upregulated Bim and increased ABT-737 lethality in Bimlow MM cells (eg, H929), disruption of autophagy by CQ strikingly increased apoptosis in association with pronounced Bim upregulation. Bim is a substrate for ubiquitination and degradation via the proteasome66 and autophagy.67 CQ also further increased HDACI/BH3-mimetic activity in bortezomib-resistant MM cells. In marked contrast, bim gene knockout (Bim−/−) MEFs were strikingly resistant to this regimen, even with CQ, indicating that the presence of the bim gene is required for this Bim-targeting strategy. Collectively, these findings argue that potentiation of BH3-mimetic lethality by HDACIs stems, at least in part, from impairment in a cytoprotective autophagic response secondary to Bim upregulation in MM cells.
In summary, our findings underscore the importance of Bim as a determinant of adaptive bortezomib resistance in MM cells and describe a Bim-targeting strategy designed to overcome such bortezomib resistance through attenuation of Bim-mediated autophagy. A model illustrating these concepts is provided in Figure 7C. In this model, BH3 mimetics (eg, ABT-737) release Bim from Bcl-2, which favors apoptosis, an event attenuated by autophagy induction (ie, autophagy ON) secondary to release of Beclin-1 from Bcl-2. HDACIs upregulate Bim which, in addition to potentiating apoptosis, attenuates autophagy (autophagy OFF). Alternatively, upregulated Bim may directly associate with and inactivate Beclin-1.20 A corollary of these finding is that this Bim-targeting strategy may be particularly effective against Bimhi MM cells, including those exhibiting adaptive bortezomib resistance stemming from loss of Bim. Finally, in Bimlow MM cells displaying impairment in HDACI-mediated Bim upregulation, disruption of autophagy (eg, by CQ) may be required to induce Bim expression and apoptosis by this regimen. Consequently, efforts to develop this Bim-targeting strategy further in bortezomib-resistant MM are currently underway.
Acknowledgments
The authors thank Dr Karla Kirkegaard (Stanford University) for pEGFP-LC3 plasmid, Dr Hisashi Harada (Virginia Commonwealth University) for wild-type and bim knockout MEFs, and the Tissue and Data Acquisition Core of Virginia Commonwealth University Department of Pathology for primary MM samples and corresponding information.
This work was supported by grants P50 CA142509-01 (S.G., R.Z.O., and Y.D.), P50 CA130805 (S.G.), CA100866 (S.G. and Y.D.), CA167708 (S.G.), and CA93738 (S.G. and Y.D.) from the National Institutes of Health (NIH), and LLSA #6181 from the Leukemia and Lymphoma Society of America. Plasmid preparation was performed at the Virginia Commonwealth University (VCU) Macromolecule Core Facility, supported in part with funding from NIH-National Cancer Institute Cancer Center Core Grant 5P30CA016059-29. Confocal microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported in part with funding from NIH-National Institute of Neurological Disorders and Stroke Center Core Grant 5P30NS047463.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.C. and Y.Z. designed and performed research, and analyzed data; L.Z., Y.L., H.L., M.K., X.-Y.P., and R.J. performed research; R.Z.O. helped design research; and Y.D. and S.G. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven Grant, Division of Hematology/Oncology, PO Box 980035, Virginia Commonwealth University, Richmond, VA 23298; e-mail: stgrant@vcu.edu; and Yun Dai, Virginia Commonwealth University and the Massey Cancer Center, Room 234, Goodwin Research Building, 401 College St, Richmond, VA 23298; e-mail: ydai@vcu.edu.
References
- 1.Munshi NC, Anderson KC. New strategies in the treatment of multiple myeloma. Clin Cancer Res. 2013;19(13):3337–3344. doi: 10.1158/1078-0432.CCR-12-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar SK, Lee JH, Lahuerta JJ, et al. International Myeloma Working Group. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study. Leukemia. 2012;26(1):149–157. doi: 10.1038/leu.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauhan D, Velankar M, Brahmandam M, et al. A novel Bcl-2/Bcl-X(L)/Bcl-w inhibitor ABT-737 as therapy in multiple myeloma. Oncogene. 2007;26(16):2374–2380. doi: 10.1038/sj.onc.1210028. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkholi R, Floros KV, Chipuk JE. The Role of BH3-Only Proteins in Tumor Cell Development, Signaling, and Treatment. Genes Cancer. 2011;2(5):523–537. doi: 10.1177/1947601911417177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda J, Taniwaki M. Involvement of BH3-only proteins in hematologic malignancies. Crit Rev Oncol Hematol. 2009;71(2):89–101. doi: 10.1016/j.critrevonc.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J. 2009;276(21):6050–6062. doi: 10.1111/j.1742-4658.2009.07329.x. [DOI] [PubMed] [Google Scholar]
- 8.Hübner A, Barrett T, Flavell RA, Davis RJ. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell. 2008;30(4):415–425. doi: 10.1016/j.molcel.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278(21):18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 10.Meller R, Cameron JA, Torrey DJ, et al. Rapid degradation of Bim by the ubiquitin-proteasome pathway mediates short-term ischemic tolerance in cultured neurons. J Biol Chem. 2006;281(11):7429–7436. doi: 10.1074/jbc.M512138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;8(12):3173–3180. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- 12.Kortuem KM, Stewart AK. Carfilzomib. Blood. 2013;121(6):893–897. doi: 10.1182/blood-2012-10-459883. [DOI] [PubMed] [Google Scholar]
- 13.Kline MP, Rajkumar SV, Timm MM, et al. ABT-737, an inhibitor of Bcl-2 family proteins, is a potent inducer of apoptosis in multiple myeloma cells. Leukemia. 2007;21(7):1549–1560. doi: 10.1038/sj.leu.2404719. [DOI] [PubMed] [Google Scholar]
- 14.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 15.Pan R, Hogdal LJ, Benito JM, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4(3):362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117(1):112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8(9):528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 19.Zinn RL, Gardner EE, Dobromilskaya I, et al. Combination treatment with ABT-737 and chloroquine in preclinical models of small cell lung cancer. Mol Cancer. 2013;12:16. doi: 10.1186/1476-4598-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo S, Garcia-Arencibia M, Zhao R, et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell. 2012;47(3):359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fandy TE, Shankar S, Ross DD, Sausville E, Srivastava RK. Interactive effects of HDAC inhibitors and TRAIL on apoptosis are associated with changes in mitochondrial functions and expressions of cell cycle regulatory genes in multiple myeloma. Neoplasia. 2005;7(7):646–657. doi: 10.1593/neo.04655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Bruyne E, Bos TJ, Schuit F, et al. IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms. Blood. 2010;115(12):2430–2440. doi: 10.1182/blood-2009-07-232801. [DOI] [PubMed] [Google Scholar]
- 23.Prince HM, Dickinson M. Romidepsin for cutaneous T-cell lymphoma. Clin Cancer Res. 2012;18(13):3509–3515. doi: 10.1158/1078-0432.CCR-11-3144. [DOI] [PubMed] [Google Scholar]
- 24.Khan O, La Thangue NB. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol Cell Biol. 2012;90(1):85–94. doi: 10.1038/icb.2011.100. [DOI] [PubMed] [Google Scholar]
- 25.Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA. 2012;109(17):6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao R, Balusu R, Fiskus W, et al. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012;11(4):973–983. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- 27.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torgersen ML, Engedal N, Bøe SO, Hokland P, Simonsen A. Targeting autophagy potentiates the apoptotic effect of histone deacetylase inhibitors in t(8;21) AML cells. Blood. 2013;122(14):2467–2476. doi: 10.1182/blood-2013-05-500629. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Dai Y, Pei XY, Grant S. Bim upregulation by histone deacetylase inhibitors mediates interactions with the Bcl-2 antagonist ABT-737: evidence for distinct roles for Bcl-2, Bcl-xL, and Mcl-1. Mol Cell Biol. 2009;29(23):6149–6169. doi: 10.1128/MCB.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai Y, Landowski TH, Rosen ST, Dent P, Grant S. Combined treatment with the checkpoint abrogator UCN-01 and MEK1/2 inhibitors potently induces apoptosis in drug-sensitive and -resistant myeloma cells through an IL-6-independent mechanism. Blood. 2002;100(9):3333–3343. doi: 10.1182/blood-2002-03-0940. [DOI] [PubMed] [Google Scholar]
- 31.Miller AV, Hicks MA, Nakajima W, Richardson AC, Windle JJ, Harada H. Paclitaxel-induced apoptosis is BAK-dependent, but BAX and BIM-independent in breast tumor. PLoS ONE. 2013;8(4):e60685. doi: 10.1371/journal.pone.0060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 33.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92(15):1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 34.Jackson WT, Giddings TH, Jr, Taylor MP, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, White EJ, Conrad C, Gomez-Manzano C, Fueyo J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009;453:273–286. doi: 10.1016/S0076-6879(08)04013-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Swanson BJ, Wang M, et al. Constitutive association of the proapoptotic protein Bim with Bcl-2-related proteins on mitochondria in T cells. Proc Natl Acad Sci USA. 2004;101(20):7681–7686. doi: 10.1073/pnas.0402293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12(2):171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115(16):3304–3313. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni Chonghaile T, Sarosiek KA, Vo TT, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334(6059):1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed JC. Cancer. Priming cancer cells for death. Science. 2011;334(6059):1075–1076. doi: 10.1126/science.1215568. [DOI] [PubMed] [Google Scholar]
- 43.Kuwana T, Bouchier-Hayes L, Chipuk JE, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17(4):525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Ren D, Tu HC, Kim H, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330(6009):1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Khoury V, Pierson S, Szwarcbart E, et al. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia. 2014;28(8):1636–1646. doi: 10.1038/leu.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Saftig P, Beertsen W, Eskelinen EL. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy. 2008;4(4):510–512. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 48.Bonapace L, Bornhauser BC, Schmitz M, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010;120(4):1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 50.Sarosiek KA, Ni Chonghaile T, Letai A. Mitochondria: gatekeepers of response to chemotherapy. Trends Cell Biol. 2013;23(12):612–619. doi: 10.1016/j.tcb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herndon TM, Deisseroth A, Kaminskas E, et al. U.S. Food and Drug Administration approval: carfilzomib for the treatment of multiple myeloma. Clin Cancer Res. 2013;19(17):4559–4563. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- 52.Podar K, Gouill SL, Zhang J, et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27(6):721–731. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 53.Chauhan D, Singh AV, Ciccarelli B, Richardson PG, Palladino MA, Anderson KC. Combination of novel proteasome inhibitor NPI-0052 and lenalidomide trigger in vitro and in vivo synergistic cytotoxicity in multiple myeloma. Blood. 2010;115(4):834–845. doi: 10.1182/blood-2009-03-213009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romagnoli M, Séveno C, Wuillème-Toumi S, et al. The imbalance between Survivin and Bim mediates tumour growth and correlates with poor survival in patients with multiple myeloma. Br J Haematol. 2009;145(2):180–189. doi: 10.1111/j.1365-2141.2009.07608.x. [DOI] [PubMed] [Google Scholar]
- 55.Gomez-Bougie P, Amiot M. Apoptotic machinery diversity in multiple myeloma molecular subtypes. Front Immunol. 2013;4:467. doi: 10.3389/fimmu.2013.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez-Bougie P, Bataille R, Amiot M. The imbalance between Bim and Mcl-1 expression controls the survival of human myeloma cells. Eur J Immunol. 2004;34(11):3156–3164. doi: 10.1002/eji.200424981. [DOI] [PubMed] [Google Scholar]
- 57.Gomez-Bougie P, Oliver L, Le Gouill S, Bataille R, Amiot M. Melphalan-induced apoptosis in multiple myeloma cells is associated with a cleavage of Mcl-1 and Bim and a decrease in the Mcl-1/Bim complex. Oncogene. 2005;24(54):8076–8079. doi: 10.1038/sj.onc.1208949. [DOI] [PubMed] [Google Scholar]
- 58.Morales AA, Kurtoglu M, Matulis SM, et al. Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. Blood. 2011;118(5):1329–1339. doi: 10.1182/blood-2011-01-327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodet L, Gomez-Bougie P, Touzeau C, et al. ABT-737 is highly effective against molecular subgroups of multiple myeloma. Blood. 2011;118(14):3901–3910. doi: 10.1182/blood-2010-11-317438. [DOI] [PubMed] [Google Scholar]
- 60.Whitecross KF, Alsop AE, Cluse LA, et al. Defining the target specificity of ABT-737 and synergistic antitumor activities in combination with histone deacetylase inhibitors. Blood. 2009;113(9):1982–1991. doi: 10.1182/blood-2008-05-156851. [DOI] [PubMed] [Google Scholar]
- 61.Matthews GM, Lefebure M, Doyle MA, et al. Preclinical screening of histone deacetylase inhibitors combined with ABT-737, rhTRAIL/MD5-1 or 5-azacytidine using syngeneic Vk*MYC multiple myeloma. Cell Death Dis. 2013;4:e798. doi: 10.1038/cddis.2013.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson RC, Vardinogiannis I, Gilmore TD. The sensitivity of diffuse large B-cell lymphoma cell lines to histone deacetylase inhibitor-induced apoptosis is modulated by BCL-2 family protein activity. PLoS ONE. 2013;8(5):e62822. doi: 10.1371/journal.pone.0062822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4(5):600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boya P, González-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao DJ, Wang ZV, Battiprolu PK, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. 2011;108(10):4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delgado ME, Dyck L, Laussmann MA, Rehm M. Modulation of apoptosis sensitivity through the interplay with autophagic and proteasomal degradation pathways. Cell Death Dis. 2014;5:e1011. doi: 10.1038/cddis.2013.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2(12):a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]