Abstract
Background
Oral pathology is a commonly reported extraintestinal manifestation of Crohn’s disease (CD). The host–microbe interaction has been implicated in the pathogenesis of inflammatory bowel disease (IBD) in genetically susceptible hosts, yet limited information exists about oral microbes in IBD. We hypothesize that the microbiology of the oral cavity may differ in patients with IBD. Our laboratory has developed a 16S rRNA-based technique known as the Human Oral Microbe Identification Microarray (HOMIM) to study the oral microbiome of children and young adults with IBD.
Methods
Tongue and buccal mucosal brushings from healthy controls, CD, and ulcerative colitis (UC) patients were analyzed using HOMIM. Shannon Diversity Index (SDI) and Principal Component Analysis (PCA) were employed to compare population and phylum-level changes among our study groups.
Results
In all, 114 unique subjects from the Children’s Hospital Boston were enrolled. Tongue samples from patients with CD showed a significant decrease in overall microbial diversity as compared with the same location in healthy controls (P = 0.015) with significant changes seen in Fusobacteria (P < 0.0002) and Firmicutes (P = 0.022). Tongue samples from patients with UC did not show a significant change in overall microbial diversity as compared with healthy controls (P = 0.418).
Conclusions
As detected by HOMIM, we found a significant decrease in overall diversity in the oral microbiome of pediatric CD. Considering the proposed microbe–host interaction in IBD, the ease of visualization and direct oral mucosal sampling of the oral cavity, further study of the oral microbiome in IBD is of potential diagnostic and prognostic value.
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, microbiome, oral microbial biomarkers
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract, likely caused by an aberrant immune response to the microbiota and other environmental factors in genetically susceptible individuals.1,2 Oral mucosal inflammation is commonly described in patients with IBD, particularly Crohn’s disease (CD). Oral pathology has a reported prevalence of 0.5% to 80% in CD.3–8 When studied prospectively in children in collaboration with a dentist, 42% of new diagnoses of CD had oral manifestations.7 Symptoms can span from mild and nonspecific inflammation such as minor aphthous lesions, mucogingivitis, and angular cheilitis to more specific findings, such as mucosal tags, cobblestoning, deep linear ulcerations, and more severe granulomatous swelling isolated to the labia and face known as orofacial granulomatosis (OFG).3,5–7,9 Lesions of the oral mucosa may occur years before the onset of intestinal symptoms, particularly in the pediatric population.6 Specific oral mucosal findings are common in children and in 75%–100% of cases, contain disease-defining, histologic evidence of noncaseating granulomas.3,7,9
Data from animal models suggests that the microbiome is a critical factor in the pathogenesis of IBD in knockout mice that are at risk for the disease. Preliminary studies in humans have found differences in the intestinal microbial populations of IBD patients when compared with healthy controls.2,10–12 However, such studies have largely focused on the lower gastrointestinal tract. The oral cavity provides an easily accessed mucosal surface that may potentially yield valuable information about the microbiome and its interaction with the host. The mouth and its resident flora is a well-characterized microbiome with ≈600 predominant bacterial species, of which about 35% are unable to be grown in culture.13,14 This environment is unlike any other in the body, made up of diverse ecological niches including hard surfaces upon which complex biofilms flourish, anaerobic microclimates, and rapidly shedding mucosal surfaces.
Given the difficulties in studying the oral microbiome using conventional, culture-based techniques, our laboratory has developed a molecular technique using a 16S rRNA-based microarray technology known as the Human Oral Microbe Identification Microarray (HOMIM). Previous investigations in our laboratory and others have implicated distinct changes in the oral microbiome in dental caries and periodontitis.13–15 Oral microbial alterations in systemic diseases have also been identified including atherosclerosis, preterm birth, and pancreatic cancer.13–21
In this case–control study, we demonstrate an overall decrease in the oral bacterial diversity of children with CD when compared with healthy controls. Furthermore, several key phyla were significantly reduced when compared with healthy subjects, as has been identified in studies of the intestinal microbiome.11,12,22–25
PATIENTS AND METHODS
Study Design and Ethical Approval
This exploratory, case–control study was conducted at Children’s Hospital Boston (Boston, MA), and samples analyzed at the Forsyth Institute (Cambridge, MA). The protocol was approved by the Children’s Hospital Committee on Clinical Investigation and Institutional Review Board (IRB #X09100535). Informed consent was obtained from patients (over 18 years of age) or parent (under 18 years of age).
Patient Selection
From October 2009 to December 2010, subjects were recruited from our pediatric gastroenterology practice in a number of venues including the inpatient wards, ambulatory clinic, infusion center, and endoscopy suite. Non-IBD, “healthy” controls were enrolled at oral surgery consultation visits with our collaborating oral surgeon (S.A.), and patients without IBD followed in our gastrointestinal (GI) practice. Subjects were excluded if they had known periodontal disease, received antibiotics in the preceding month, and/or had used antiseptic mouthwash or brushed their tongue within 3 hours of sample collection. A standardized case report form with common oral pathology was utilized for oral examination. Oral pathology was defined as any active swelling, inflammation, or ulceration of the oral mucosa or lips, ulcers, cobblestoning, or other lesions grossly evident. Patients with oral pathology underwent a full oral examination by the oral surgeon (S.A.). Patients in both our control and study arms had oral samples collected once at the time of study enrollment. Metadata collected at time of sampling included demographics, medication history including antibiotic, probiotic, and specific IBD therapies. In addition, family, surgical, and personal history of oral manifestations, Montreal Classification of disease, and measures of disease activity including the Pediatric Ulcer-ative Colitis Activity Index (PUCAI) or Pediatric Crohn’s Disease Activity Index (PCDAI) were collected.26–28
Sample Collection
Oral samples were collected from study subjects using a sterile cytology brush (Medical Packaging, Camarillo, CA) and suspended in 150 µL of TE Buffer (Epicentre Biotechnologies, Madison, WI). Samples were immediately frozen at −80°C until ready for DNA isolation. Samples were collected from dorsum of the tongue. In most subjects, the buccal mucosa was also sampled. If an IBD patient had oral pathology the area of the oral lesion and the contralateral/unaffected mucosa was sampled. Similarly, non-IBD control subjects presenting with nonspecific aphthous lesions or other oral pathology had sampling of the lesion and the contralateral normal mucosa. Clinical patient data was collected and stored on an SPSS (IBM v. 19, Chicago, IL) database.
HOMIM
The HOMIM is a custom-designed, 16S rRNA-based oligonucleotide reverse capture microarray.15 A total of 421 probes, representing roughly 300 of the most predominant oral bacterial species, are arranged phylogenetically and in replicate on each aldehyde-coated glass slide. HOMIM provides information on the nine most common bacterial phyla found in the oral cavity, including: Bacteriodetes, Firmicutes, Proteobacteria, Synergistetes, Fusobacteria, Spirochaetes, Actinobacteria, SR-1, and TM-7. Each array has a total of 24 cluster probes targeting more than two closely related species in addition to multiple positive and negative controls. The lower limit of detection is >104 bacterial cells. More information on HOMIM can be found at http://mim.forsyth.org/homim.html.
DNA Isolation
Samples were incubated at 37°C overnight with 1 µL of Ready-Lyse Lysozyme and subsequently incubated at 65°C for 30 minutes with 150 µL of 2X T and C Lysis Solution and 1 µL of Proteinase K (Epicentre Biotechnologies). The solution is then placed on ice for 7 minutes and later precipitated with 150 µL of MPC Protein Precipitation Reagent. The DNA was precipitated with isopropanol and later washed in 75% ethanol. The DNA-containing pellet was reconstituted in 25 µL TE Buffer.
DNA Preparation
The DNA for hybridization was prepared using two rounds of polymerase chain reaction (PCR). First, a standard PCR using universal bacterial primers for 16S rRNA gene amplification was done. Amplicons of the appropriate size were verified on a 1% agarose gel. Universal 16S rRNA primers were then used for a second “nested” amplification and labeling with a Cy3-dCTP fluorescent dye (Amersham, GE Healthcare, Buckinghamshire, UK).
Blocking and Hybridization
The slides were blocked to remove nonreactive primary alcohols and unreacted aldehyde groups using a solution of sodium borohydride (NaBH4), 1× phosphate-buffered saline (PBS), and 99% ethanol (EtOH). Labeled PCR products were added to hybridization buffer (yeast tRNA, 20× SSC, 10% SDS, and dH2O), heated at 99°C for 5 minutes to denature the DNA, and then added to the slide. Slides were then incubated overnight at 55°C.
Postprocessing
Slides were washed with a solution of SSC and 10% SDS, rinsed with dH2O, spun dry in a centrifuge, and analyzed using a GenePix 4000B scanner and GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA).
Statistical Analysis
Normalization and statistical analysis for HOMIM array raw values were developed initially on a training set including 114 samples (Control = 45, CD = 46, UC = 23). The HOMIM array values were normalized across all arrays by quantile normalization using Bioconductor in R.29,30 To determine oral microbial diversity differences in CD, UC, and control subjects, we implemented Shannon Diversity Index (SDI); as such: , where S is the number of phylum detectable by the HOMIM system, ni is the sum of all probe signal in phylum i, and N is the summation of all probe signal. We calculated SDI for each individual condition. The differences in SDI (ΔSDI) between CD and UC from control subjects for each phylum were determined. The significance of ΔSDI was evaluated by obtaining null distribution of ΔSDI through randomizing observed values for each individual probe across samples from groups of interest. We performed randomization for 1000 permutations, and the significance is evaluated by the following formula: , where i and j are the two groups of subjects being compared. Principal component analysis (PCA) was additionally generated to evaluate clustering of samples.
RESULTS
Patient Demographics and Normalization
A total of 114 unique subjects (162 samples) were included in our final analysis (43 control, 40 CD, 31 UC) (Table 1). Of these samples, there were 111 tongue brushings (43 control, 38 CD, and 30 UC), and 51 buccal mucosa samples (19 control, 18 CD, and 14 UC) (Table 2). When patients with oral pathology had both affected and unaffected samples collected, only the affected samples were used in this analysis to prevent two samples from the same patient potentially confounding our results. Patients overall ranged in age from 4–27 years with age and gender breakdowns similar in the control, CD, and UC groups. Patients did not take antibiotics for a minimum of 1 month prior to study enrollment. Of the IBD patients, 30 (75%) of CD patients and 12 (39%) of UC patients were receiving immunosuppressive agents including: methotrexate, 6-mercaptopurine, azathioprine, tacrolimus, cyclosporine, thalidomide, infliximab, adalimumab, and certolizumab. Our control group was seen for a variety of GI and non-GI issues including: oral surgery consultation (n = 25), abdominal pain (n = 8), recurrent mouth sores (n = 7), irritable bowel syndrome (IBS) (n = 4), cyclic vomiting syndrome (CVS) (n = 2), gastroesophageal reflux disease (GERD) (n = 1), and Behçet disease (BD) (n = 1).
TABLE 1.
Demographics of Study Population
| Cohort | Control (n=43 subjects) |
Crohn’s Disease (n=40 subjects) |
Ulcerative Colitis (n=31 subjects) |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 14 ± 4 | 14 ± 4 | 14 ± 5 |
| Range | 5–24 | 7–23 | 4–24 |
| Male, # (%) | 19 (44%) | 25 (63%) | 18 (58%) |
| Subjects with oral pathology | 7 | 6 | 4 |
| Active disease, # (%) | N/A | 9 (23%) | 14 (45%) |
| Subjects on immunosuppression, # (%) | 0 | 30 (75%) | 12 (39%) |
| Montreal criteria, # (%) | |||
| Ileal disease (L1) | 6 (15%) | ||
| Colonic disease (L2) | 10 (25%) | ||
| Ileocolonic disease (L3) | 24 (60%) | ||
| Isolated upper tract disease | 0 | ||
| Ulcerative colitis type, # (%) | |||
| Pancolitis (E1) | 17 (55%) | ||
| Left sided colitis (E2) | 10 (32%) | ||
| Proctitis < 15cm (E3) | 4 (13%) |
TABLE 2.
Sample Numbers
| Sample Location | Control (n = 43 subjects) |
Crohn’s Disease (n= 40 subjects) |
Ulcerative Colitis (n = 31 subjects) |
|---|---|---|---|
| Tongue samples (n = 111) | 43 | 38 | 30 |
| Buccal mucosal samples (n = 51) | 19 | 18 | 14 |
| Total (n = 162) | 62 | 56 | 44 |
Oral Microbes Differ in Composition and Diversity Based on the Region of the Mouth Sampled
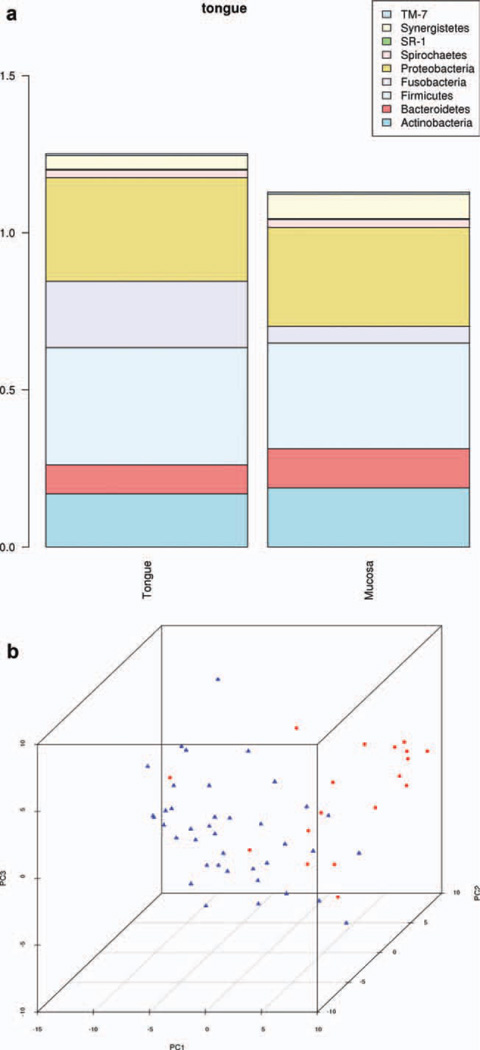
To begin to determine if differences exist between health and disease in the microbes of the oral cavity, we analyzed the oral microbiome at the population level among our study groups. We employed the SDI, as is commonly used in studies of complex microbial environments, to measure overall oral microbial diversity as well as microbial population changes between groups (ΔSDI). To address the variation among regions of the oral cavity, we initially compared the dorsum of the tongue to buccal mucosa. The 43 tongue samples from our control patients were pooled and compared with the 19 buccal samples from unique subjects (as described above using SDI and PCA analysis). Levels of Fusobacteria (ΔSDI = −0.158, P < 0.0001) and Firmicutes (ΔSDI = −0.037, P = 0.037) were significantly reduced in buccal samples as compared with tongue samples of control patients, whereas Bacteroidetes were enriched (ΔSDI = 0.032, P = 0.030). The SDI of buccal samples as a whole (SDI = 1.129) showed an overall trend toward decreased diversity when compared with tongue samples (SDI = 1.252, ΔSDI = −0.122, P = 0.067) (Fig. 1a). Therefore, comparing the locations sampled demonstrated alterations in overall diversity with significant changes among several phyla including Fusobacteria, Firmicutes, and Bacteroidetes. Furthermore, PCA analysis used as a data compression technique to visualize intersample similarity demonstrated clustering based on sample location (Fig. 1b).
FIGURE 1.
(a) SDI of tongue and buccal mucosal samples from control population demonstrating a trend toward overall decreased diversity in control buccal samples as compared with control tongue samples (ΔSDI = −0.122, P = 0.067). Significant differences at the phylum level were seen within this comparison. Fusobacteria (ΔSDI = −0.157, P < 0.0002) and Firmicutes (ΔSDI = −0.038, P = 0.037) were less abundant in buccal samples, whereas Bacteroidetes were more abundant (ΔSDI = 0.033, P = 0.030). (b) PCA of tongue and buccal mucosal samples from control population. Tongue samples are represented by blue triangles, buccal samples by red circles. This type of data compression analysis demonstrates the clustering of samples based on their location and similar microbial profiles.
Overall Oral Microbial Diversity Is Reduced in Patients with CD, However Not UC
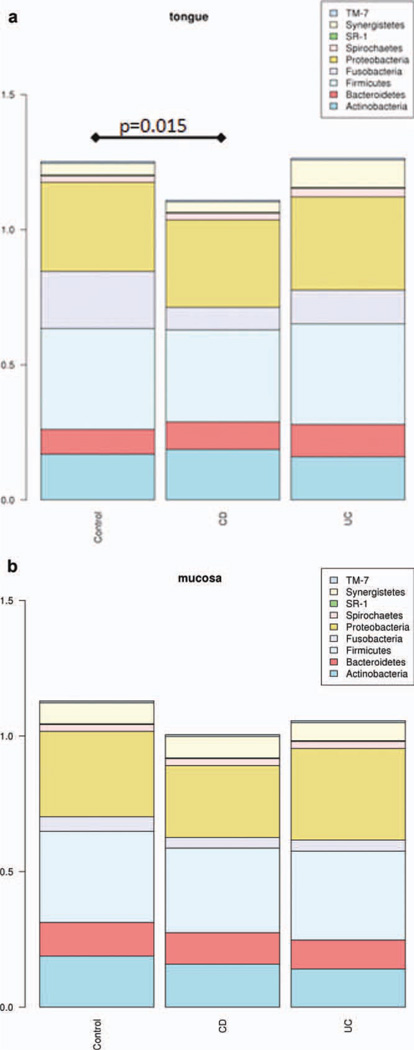
Given the divergence in tongue and buccal mucosal samples seen in healthy individuals, our comparisons of healthy and IBD populations were location-specific (i.e., we compared tongue samples to tongue samples, and buccal samples to buccal samples). Samples were pooled by location across cohorts so that tongue and buccal mucosal brushings could be compared between control, CD, and UC. Using control tongue samples as the reference value (SDI = 1.252), we found a significant decrease in overall diversity of tongue samples in the CD cohort (SDI = 1.108, ΔSDI = −0.143, P = 0.015). In contrast, microbial diversity of tongue samples from UC patients was not significantly different from that of control subjects (SDI = 1.264, ΔSDI = 0.012, P = 0.418) (Fig. 2a).
FIGURE 2.
(a) SDI analysis of tongue samples across cohorts. Overall diversity of CD was significantly reduced as compared with control samples (ΔSDI = −0.143, P = 0.015). In comparison, overall diversity of tongue samples in UC is not significantly different from control samples (ΔSDI = 0.012, P = 0.418). (b) SDI analysis of buccal mucosa samples across cohorts. Overall diversity of CD was reduced as compared with control samples (ΔSDI = −0.125, P = 0.091). Overall diversity of buccal samples in UC is similar to control samples (ΔSDI = 0.073, P = 0.254).
Using the same technique, buccal samples across all groups were compared using SDI. Subjects with CD showed a trend toward decreased overall diversity (ΔSDI = −0.125, P = 0.091). Similar to findings in UC tongue samples, the diversity in buccal samples from subjects with UC was not statistically different from that of control subjects (ΔSDI = −0.073, P = 0.253) (Fig. 2b).
Reduction in Specific Phyla Results in Alterations of Diversity in IBD Oral Samples
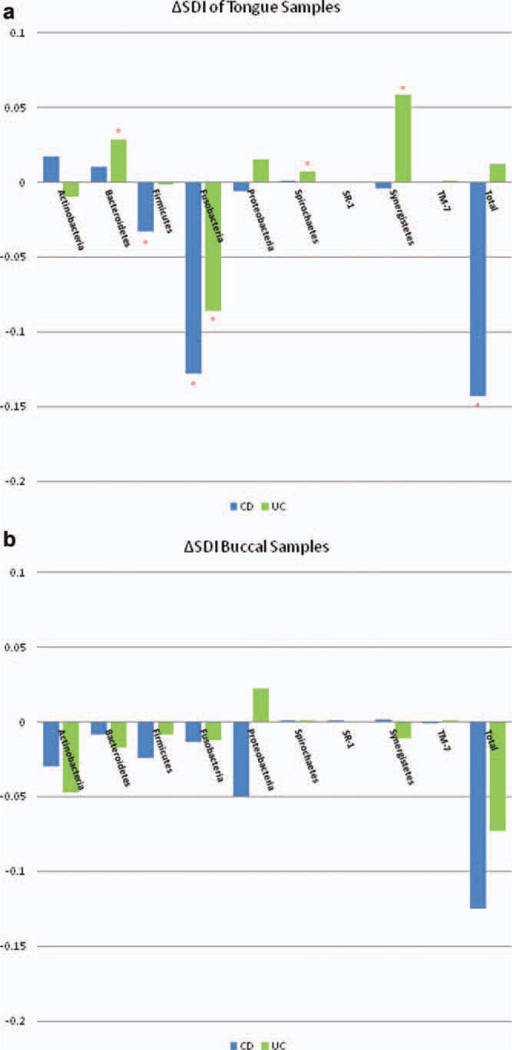
Subsequent analysis included determination of enrichment or loss at the phylum level accounting for the change in overall diversity in our control and study cohorts. Tongue samples in CD showed a significant loss of probe activity from two particular phyla, Fusobacteria (ΔSDI = −0.128, P < 0.0002) and Firmicutes (ΔSDI = −0.033, P = 0.022) (Fig. 3a). Changes seen in tongue samples of subjects with UC were more varied in their loss and enrichment of oral bacterial phyla, yielding a composite lack of significance in overall diversity. However, a statistically significant loss of probe signal was similarly noted in Fusobacteria (ΔSDI = −0.086, P = 0.006), whereas Spirochaetes (ΔSDI = 0.007, P = 0.006), Synergistetes (ΔSDI = 0.058, P = 0.009), and Bacteroidetes (ΔSDI = 0.028, P = 0.030) were all increased as compared with control tongue samples.
FIGURE 3.
(a) ΔSDI of tongue samples from CD and UC patients using control tongue samples as the reference value. Significant phyla are denoted with the asterisk. CD Samples were significantly reduced in Firmicutes (ΔSDI = −0.033, P = 0.022), Fusobacteria (ΔSDI = −0.128, P < 0.0001) and overall diversity (ΔSDI = −0.143, P = 0.015). Tongue samples from UC patients were overall not significantly different from control tongue samples (ΔSDI = 0.012, P = 0.418). Losses, however were noted in Fusobacteria (ΔSDI = −0.086, P = 0.006), whereas Spirochaetes (ΔSDI = 0.007, P = 0.006), Synergistetes (ΔSDI = 0.058, P = 0.009), and Bacteroidetes (ΔSDI = 0.028, P = 0.030) were all enriched. (b) ΔSDI of buccal mucosa samples from CD and UC patients using control buccal samples as the reference value. No individual phyla were significantly different. Overall diversity in buccal samples across cohorts was not statistically significant; however, CD trended toward decreased overall diversity.
Much like tongue samples in the CD cohort, buccal samples from CD subjects showed a trend toward reduced overall diversity (ΔSDI = −0.125, P = 0.091) as compared with healthy controls. However, there were no significant changes seen at the phylum level (Fig. 3b). In contrast, samples from subjects with UC did not show any significant changes or trends in overall diversity and at the phylum level (ΔSDI = −0.073, P = 0.253).
DISCUSSION
To our knowledge, this is the first investigation of the oral microbiome as it relates to IBD. Analogous to studies of the intestinal microbiome in IBD, we demonstrated a marked decrease in both overall microbial diversity and specific phylum levels in CD. While overall diversity was not significantly altered in UC when compared with healthy controls, there were detectable and significant alterations of the oral microbiome within several phyla of this cohort.
Previous studies by our group using HOMIM have shown alterations in the oral microbiome of both local and systemic diseases, suggesting that oral microbial biomarkers may be present in specific disease states. HOMIM represents a rapid, inexpensive, and relatively quantifiable technology in surveying this unique environment in health and disease. As with other microarray technologies, sample investigations are limited to the probes that are present and are therefore subject to detection bias. However, in this preliminary feasibility study using HOMIM the predominant oral species represented allowed for discrimination among oral sampling location and systemic disease from health.
The aberrant interaction between the microbiome and immune system appears to be critical in the pathogenesis of IBD. From experimental models of germfree mice, studies of fecal diversion, and analysis of antibiotic modification of disease, there is an increasing body of evidence that the host–microbe interaction is critical to the development of IBD.1,2,31 Furthermore, numerous studies have shown the increased prevalence of antibody responses to bacterial and yeast-derived proteins in serologic studies of patients with IBD.32–34 Interestingly, patients with CD and oral manifestations have statistically significant higher anti-Saccharomyces cerevisiae antibody (ASCA) titers as compared with individuals without oral findings.35
The oral mucosa is an immunologically active surface with increased cytokine production in children with CD compared with healthy controls, regardless of the presence of oral manifestations.36 The oral cavity serves as a window into the intestinal tract and arguably offers an opportunity to study the complex interaction of the host immune system and microbiome at its epithelial interface. The bacteria of the human microbiome exert enormous metabolic, immunologic, physiologic, and, at times, pathologic influence on our health. We have just begun to establish a “phylogenetic core” of microbiota that is helping to define health in these complex environments including the oral microbiome.37,38 Dysbiosis or deviation from this core has revealed distinct shifts in the enteric microbiota of individuals with CD and UC.12,23,31,39 Indeed, a lack of diversity appears to be a common finding in IBD microbial studies, in which the intestinal microbiome in diseased states appears to lose commensal organisms that typically characterize health.5,24,25,40 It is likely that the myriad organisms that define a healthy microbiome confer protective mechanisms to the host; in the absence of this diversity, pathogens can arise and flourish.
Given the localized inflammation of colonic mucosa in UC in contrast to the transmural, often systemic inflammation found in CD, we theorized that oral microbial alterations would be more likely observed in CD than in UC. We demonstrated that there is a distinct and significant loss of diversity in the oral microbiome of CD as compared with both UC and health. Furthermore, loss of specific phyla such as Fusobacteria and Firmicutes has been demonstrated in studies of the intestinal microbiome in CD and is mirrored in our study of the oral microbiome. 22,41,42 We presented evidence to suggest that the oral microbiome is uniquely altered in patients with IBD, especially in CD. We have begun to assess the diagnostic accuracy of this technology in children in whom IBD is suspected. Future work will evaluate the impact of potential clinical confounders such as disease phenotype, immunosuppression, disease activity, and diet as we expand our statistical and bioinformatic approach. With the prevalence of oral pathology in CD, the ease of visualization, and direct oral mucosal sampling, further study of the oral microbiome in IBD is of potential diagnostic and prognostic value.
ACKNOWLEDGMENT
We thank the Rasmussen, Wolfman and MacInnes families for their generous support of the Children’s Hospital Interdisciplinary Microbiome Project.
Supported in part by a T32 training grant (#5T32DK007477-27) in addition to a Senior Research Award (#1832) from the Crohn’s & Colitis Foundation of America and from a grant from the Harvard Institute of Translational Immunology (HITI)/Leona M. and Harry B. Helmsley Charitable Trust Pilot Grants Program.
REFERENCES
- 1.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 3.Rowland M, Fleming P, Bourke B. Looking in the mouth for Crohn’s disease. Inflamm Bowel Dis. 2010;16:332–337. doi: 10.1002/ibd.20983. [DOI] [PubMed] [Google Scholar]
- 4.Dupuy A, Cosnes J, Revuz J, et al. Oral Crohn disease: clinical characteristics and long-term follow-up of 9 cases. Arch Dermatol. 1999;135:439–142. doi: 10.1001/archderm.135.4.439. [DOI] [PubMed] [Google Scholar]
- 5.Katz J, Shenkman A, Stavropoulos F, et al. Oral signs and symptoms in relation to disease activity and site of involvement in patients with inflammatory bowel disease. Oral Dis. 2003;9:34–40. doi: 10.1034/j.1601-0825.2003.00879.x. [DOI] [PubMed] [Google Scholar]
- 6.Ojha J, Cohen DM, Islam NM, et al. Gingival involvement in Crohn disease. J Am Dent Assoc. 2007;138:1574–1581. doi: 10.14219/jada.archive.2007.0106. quiz 1614–1615. [DOI] [PubMed] [Google Scholar]
- 7.Harty S, Fleming P, Rowland M, et al. A prospective study of the oral manifestations of Crohn’s disease. Clin Gastroenterol Hepatol. 2005;3:886–891. doi: 10.1016/s1542-3565(05)00424-6. [DOI] [PubMed] [Google Scholar]
- 8.Galbraith SS, Drolet BA, Kugathasan S, et al. Asymptomatic inflammatory bowel disease presenting with mucocutaneous findings. Pediatrics. 2005;116:e439–e444. doi: 10.1542/peds.2004-2281. [DOI] [PubMed] [Google Scholar]
- 9.Pittock S, Drumm B, Fleming P, et al. The oral cavity in Crohn’s disease. J Pediatr. 2001;138:767–771. doi: 10.1067/mpd.2001.113008. [DOI] [PubMed] [Google Scholar]
- 10.Schwiertz A, Jacobi M, Frick JS, et al. Microbiota in pediatric inflammatory bowel disease. J Pediatr. 2010;157:240–244. doi: 10.1016/j.jpeds.2010.02.046. e1. [DOI] [PubMed] [Google Scholar]
- 11.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 12.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. e1. [DOI] [PubMed] [Google Scholar]
- 13.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seymour GJ, Ford PJ, Cullinan MP, et al. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect. 2007;13(Suppl 4):3–10. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- 18.Beck JD, Offenbacher S. Systemic effects of periodontitis: epidemiology of periodontal disease and cardiovascular disease. J Periodontol. 2005;76:2089–2100. doi: 10.1902/jop.2005.76.11-S.2089. [DOI] [PubMed] [Google Scholar]
- 19.Joshipura KJ, Rimm EB, Douglass CW, et al. Poor oral health and coronary heart disease. J Dent Res. 1996;75:1631–1636. doi: 10.1177/00220345960750090301. [DOI] [PubMed] [Google Scholar]
- 20.Offenbacher S, Jared HL, O’Reilly PG, et al. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann Periodontol. 1998;3:233–250. doi: 10.1902/annals.1998.3.1.233. [DOI] [PubMed] [Google Scholar]
- 21.Farrell JJZL, Zhou H, Chia D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2011:60. doi: 10.1136/gutjnl-2011-300784. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondot S, Kang S, Furet JP, et al. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm Bowel Dis. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- 24.Kang S, Denman SE, Morrison M, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 25.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prenzel F, Uhlig HH. Frequency of indeterminate colitis in children and adults with IBD — a metaanalysis. J Crohns Colitis. 2009;3:277–281. doi: 10.1016/j.crohns.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Ahn JYL, Ganly I, Morris L, et al. 416 Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. International Association for Dental Research 2011 Annual Meeting. 2011 doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubinsky MC, Lin YC, Dutridge D, et al. Serum immune responses predict rapid disease progression among children with Crohn’s disease: immune responses predict disease progression. Am J Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz J, Kugathasan S, Dubinsky M, et al. Age of diagnosis influences serologic responses in children with Crohn’s disease: a possible clue to etiology? Inflamm Bowel Dis. 2009;15:714–719. doi: 10.1002/ibd.20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papp M, Altorjay I, Lakatos PL. [Relevance of serologic studies in inflammatory bowel diseases.] Orv Hetil. 2007;148:887–896. doi: 10.1556/OH.2007.28064. [DOI] [PubMed] [Google Scholar]
- 35.Russell RK, Ip B, Aldhous MC, et al. Anti-Saccharomyces cerevisiae antibodies status is associated with oral involvement and disease severity in Crohn disease. J Pediatr Gastroenterol Nutr. 2009;48:161–167. doi: 10.1097/MPG.0b013e318183e112. [DOI] [PubMed] [Google Scholar]
- 36.Turkay C, Kasapoglu B. Noninvasive methods in evaluation of inflammatory bowel disease: where do we stand now? An update. Clinics (Sao Paulo) 2010;65:221–231. doi: 10.1590/S1807-59322010000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 38.Zaura E, Keijser BJ, Huse SM, et al. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friswell M, Campbell B, Rhodes J. The role of bacteria in the pathogenesis of inflammatory bowel disease. Gut Liver. 2010;4:295–306. doi: 10.5009/gnl.2010.4.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Femiano F, Lanza A, Buonaiuto C, et al. Pyostomatitis vegetans: a review of the literature. Med Oral Patol Oral Cir Bucal. 2009;14:E114–E117. [PubMed] [Google Scholar]
- 41.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, et al. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 42.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]