Abstract
The objective of this study was to develop a method for categorizing normal individuals (normal, n = 100) as well as patients with osteoarthritis (OA, n = 100), and rheumatoid arthritis (RA, n = 100) based on a panel of inflammatory cytokines expressed in serum samples. Two panels of inflammatory proteins were used as training sets in the construction of two separate artificial neural networks (ANNs). The first training set consisted of all proteins (38 in total) and the second consisted of only the significantly different proteins expressed (12 in total) between at least two patient groups. Both ANNs obtained high levels of sensitivity and specificity, with the first and second ANN each diagnosing 100% of test set patients correctly. These results were then verified by re-investigating the entire dataset using a decision tree algorithm. We show that ANNs can be used for the accurate differentiation between serum samples of patients with OA, a diagnosed RA patient comparator cohort and normal/control cohort. Using neural network and systems biology approaches to manage large datasets derived from high-throughput proteomics should be further explored and considered for diagnosing diseases with complex pathologies.
Keywords: osteoarthritis, cytokines, serum, diagnostic, artificial neural network, machine learning
1. Introduction
While over a 100 variants of arthritis have been described, two clinical phenotypes predominate: rheumatoid arthritis (RA), a chronic inflammatory disease characterized by overt joint inflammation and swelling, joint tenderness and destruction of synovium [1] and osteoarthritis (OA), a heterogeneous group of conditions that develop in distinct, yet overlapping patterns of joint symptoms and signs associated with the loss of articular cartilage integrity [2]. While reliable diagnostics for RA exist, currently only relatively advanced OA can be clinically diagnosed with high confidence using clinical acumen, functional assessment and/or radiographic evidence of joint space narrowing [2]. However, radiography cannot detect the disease in early stages [3], whereas MRI requires some level of cartilage surface degradation to have occurred before a diagnosis can be made [4,5]. Furthermore, MRI is costly and not easily/widely accessible. Because early disease diagnosis and more effective intervention at earlier stages of OA are desired goals, there has been a great emphasis placed on the potential clinical value of biomarker research in OA [6–8], especially those that can be used before the cartilage surface begins to deteriorate.
OA is not just a disease of articular cartilage, it is a disease of the entire joint as an organ [9]. Key joint structures such as the ligaments, menisci, subchondral bone, the joint capsule and synovial tissue of the knee are all affected by, and involved in, the processes that lead to OA. Because the joint acts as an organ, there is compelling evidence that serum biomarkers may be a useful surrogate indicator of joint health. Previously, sera have been examined for cartilage matrix degradation products such as collagen and cartilage oligomeric protein (COMP) [7,10]; however, assays using these analytes have not been widely adopted for clinical use, possibly because of heterogeneity between and within groups (control versus patient), and the variation between baseline levels of a given single biomarker within a group leading to false-positives/negatives. This is particularly an issue in OA biomarker research as no single biomarker has been shown to be completely associated with the disease or disease progression, thereby being not completely present or absent in normal or disease states.
RA is an autoimmune disease characterized by significant inflammation leading to an increased expression of cartilage degradation enzymes and eventual loss of the cartilage surface of the joint (if left untreated). Currently, there are a number of effective interventions in the management of RA [11,12]. In contrast to RA, OA has not been universally viewed as an inflammatory condition; however, recent findings continue to build a case for a central role for chronic inflammation in OA [13]. Cytokines are proteins that are key mediators in the inflammatory response, with numerous cytokines and related signalling pathways implicated in the onset and pathogenesis of OA [14–16]. Recently, our group and others have begun to use intra-articular and systemic inflammatory cytokine profiling to identify and stratify OA patients [17,18]. This is accomplished through quantitative analysis of specific cytokines in small volumes of body fluids (serum, synovial fluid) using multiplexing technologies. These proteomic analyses result in large datasets requiring significant statistical and computational analyses to obtain effective and reproducible outcomes, while maintaining the sensitivity and specificity of the assay.
Artificial neural networks (ANNs) are machine-learning algorithms that are modelled to mimic the way a human brain may process decisions in a simplified manner. Briefly, based on a number of inputs, an ANN is able to create unbiased associations between the inputs and outputs using a number of layers of interconnected nodes (neurons). Each node is designed to receive multiple inputs, which are then multiplied by optimally assigned weights, summed and then passed to a single output through an activation function and on to the next level of operation, either another ‘neuron interface’ or output. In recent years, ANNs have gained acceptance for use in medical research and have been applied to a wide variety of applications in medicine for their predictive capabilities [19]. These examples include predicting the risk of cardiac arrest based on chest pain, optimum renal stone fragmentation by extracorporeal lithotripsy, predicting the efficacy of certain breast cancer treatments as well as determining the likelihood of methicillin-resistant Staphylococcus colonization in patients admitted to intensive care units [20]. ANNs have also been successfully applied in OA research that has investigated the identification of joint locations within proximal hand and wrist radiographs that exhibit arthritic-like changes [21], diagnosis of knee OA using dynamic electrical impedance signals [22] and the diagnosis of knee OA using proteomic analysis of synovial fluid using mass spectrometry [23]. Based on these studies and others, it appears that ANNs are a promising approach to distilling complex datasets down to simple binary outputs (e.g. go/no-go, disease or no-disease). The aim of this study was to develop, train and use an ANN to test the efficacy of a panel of inflammatory cytokines as a potential biomarker that could be used to accurately classify patients as having OA. The output produced by the ANN was then verified by reinvestigating the dataset using a decision tree algorithm.
2. Methods
2.1. Patients
2.1.1. Normal (n = 100; mean age 40.0 ± 9.5 years)
All control normal participants showed no physical signs of OA or RA, and were questioned about both personal and family histories of arthritis or any autoimmune diseases. Patients were excluded from the study if they had a personal or family history of RA, systemic lupus erythematosus, systemic sclerosis, inflammatory myopathy, vasculitis, spondyloarthropathies, inflammatory bowel disease, diabetes mellitus type I and/or thyroid disease.
2.1.2. Osteoarthritis (n = 100; mean age 60.4 ± 10 years)
Inclusion criteria were based on a diagnosis of mild/moderate OA performed by a sports medicine physician at the University of Calgary based on clinical symptoms of three months or greater with radiographic evidence of changes associated with OA. Radiographic evidence of OA of any compartment of the knee with collapsed or near-collapsed joint space of any compartment of the knee.
2.1.3. Rheumatoid arthritis (n = 100, mean age 46.5 ± 14.5)
Inclusion criteria were an age of 40 years or older, and a diagnosis based on the American College of Rheumatology criteria with evidence of X-ray changes consistent with cartilage changes within the knee joint [1]. All RA patients were being managed with various immune suppressive drugs at the time of sample collection.
2.1.4. Sample collection
Serum samples were collected by standard venipuncture with vacuum tubes from normal and RA patients at the University of Manitoba and from OA patients at the University of Calgary. All samples were stored at −80°C until required for analysis.
2.1.5. Multiplexed arrays
Multiplexed array technologies allow for multiple concentrations of specific proteins (cytokines in our study) to be quantified within a single biological fluid sample. This is accomplished in a manner that enables direct comparisons of protein concentration values between samples. In this study, each sample (record) represents each person's (normal, OA and RA) expression panel of 38 select inflammatory cytokines present in their serum samples.
Sample analysis was performed by Eve Technologies (Calgary, AB Canada) using the Milliplex MAP human cytokine/chemokine panel (Millipore) on a Luminex 100 platform (Luminex Corp., Austin, TX, USA), according to the manufacturer's instructions and as a previously published [24]. Serum aliquots were thawed on ice, and 20 µl of fluid was diluted with the Milliplex running buffer (Millipore, Billercia, MD, USA). All samples were assayed at least in duplicate, and prepared standards were included in all runs. Briefly, cytokine-/chemokine-specific antibodies are pre-coated onto colour-coded microparticles and dispensed into microtitre plates by the manufacturer. The standards and test samples were added to the microtitre wells, and after standard incubation, any unbound substances were washed away by vacuum filtration in an apparatus provided by the manufacturer. This was followed by the addition of biotinylated antibodies specific to the protein of interest to each well and after standard incubation protocol any unbound biotinylated antibody was removed by vacuum washes as before. Streptavidin–PE antibodies were then added to each well, followed by a final wash to remove unbound streptavidin–PE. The microparticles were resuspended in Milliplex buffer and read using the Luminex 100 analyser.
2.2. Statistics
ANOVA with Bonferroni post hoc adjustments, Graphpad Prism version 5.0d for Mac OS X (GraphPad software, San Diego, CA, USA), was used to determine the significant differences between patient groups for each cytokine (potential biomarker). Significance was accepted at p ≤ 0.05.
2.3. Artificial neural network
Using Matlab R2011a for PC (Natick, MA, USA) a feedforward ANN was developed. Scaled conjugate gradient back-propagation (trainscg in Matlab) was used as the training function as it has been shown to be robust in classification problems. Two feedforward ANNs were trained using separate datasets as inputs. The first set was composed of all cytokines (38 in total) as inputs and used 10 neurons in a single hidden layer. The second set included only cytokines that were shown to significantly differentiate at least one of the three patient groups from the others (12 in total) and used five neurons in the hidden layer. In both ANNs, there were three output variables, each record was allocated to either the normal, OA or RA patient group. The architectures for the two networks are 38 × 10 × 3 (input × hidden × output) for the full dataset and 12 × 5 × 3 for the reduced dataset. The number of neurons in the hidden layer was chosen to be as small as possible while still maintaining accuracy. Datasets were divided randomly into the training set (70% of data) which was used to train the ANN, the validation set (15%) which was used to test the trained ANN to determine when training has completed and the test set (15%) which is left independent of the training process.
We quantified the performance of our ANNs by calculating the sensitivity and specificity of the output of each training, validation and test dataset used. For this study, sensitivity was defined as the ability of the algorithm to correctly detect the patients of the group in question (normal, OA or RA). This was calculated by the percentage of patients correctly allocated to their physician diagnosed group for each of the three groups. The specificity was defined as the algorithm's accuracy in being selective for only the patients that belong in each of the three physician diagnosed groups. This value was calculated by subtracting the percentage of false-positives (from 100%) for each of the three groups.
2.4. Decision tree verification of artificial neural network
To verify the results produced by ANN, a collection of bagged decision trees were programmed in Matlab R2011a for PC using the TreeBagger algorithm. To train this algorithm, a randomly selected dataset consisting of 85% of each of the three patient groups (normal, OA and RA) was used. The test set consisted of the remaining 15% from each patient group that was not used in the training of the decision tree. The entire dataset was used for this analysis as it provided better results than just the significantly expressed cytokine values used as a secondary training set in the ANN. Sensitivity/specificity was defined as described for the ANN.
3. Results
3.1. Statistics
Of the 38 cytokines investigated in the blood serum samples, 12 were shown to have significantly different expression levels within at least one of three possible patient group comparisons (table 1). Interestingly, in some instances, OA and RA serum samples contained significantly lower levels of some inflammatory cytokines when compared with normal samples. For example, OA samples had significantly lower EGF, growth-regulated oncogene (GRO) and MIP-1beta levels compared with normal samples, whereas RA samples showed significantly lower EGF, fractalkine, GRO, macrophage-derived chemokine (MDC) and CD40 ligand (CD40L) expression levels compared with normal samples.
Table 1.
Summary of cytokine expression and patient group statistics. Mean ± s.d.; x = p ≤ 0.05; dashes = no significance.
| cytokine | mean and standard deviation |
ANOVA with Bonferroni post hoc |
||||
|---|---|---|---|---|---|---|
| normal | OA | RA | norm versus OA | norm versus RA | RA versus OA | |
| EGF | 167 ± 390 | 33 ± 45 | 42 ± 51 | x | x | — |
| Fractalkine | 297 ± 653 | 190 ± 580 | 101 ± 169 | — | x | — |
| GRO | 1611 ± 1555 | 583 ± 300 | 533 ± 209 | x | x | — |
| IFNgamma | 26 ± 70 | 68 ± 153 | 11 ± 24 | x | — | x |
| IL-7 | 6 ± 11 | 6 ± 8 | 10 ± 9 | — | x | x |
| IL-8 | 13 ± 35 | 24 ± 33 | 15 ± 21 | x | — | — |
| IL-17 | 13 ± 47 | 34 ± 70 | 6 ± 12 | x | — | x |
| MCP-1 | 221 ± 91 | 475 ± 249 | 408 ± 158 | x | x | x |
| MDC | 1935 ± 1634 | 1909 ± 1366 | 729 ± 399 | — | x | x |
| MIP-1beta | 141 ± 193 | 65 ± 107 | 109 ± 154 | x | — | — |
| CD40L | 2280 ± 948 | 5656 ± 4801 | 771 ± 1022 | x | x | x |
| VEGF | 40 ± 69 | 102 ± 138 | 37 ± 41 | x | — | x |
| eotaxin | 64 ± 83 | 112 ± 73 | 132 ± 420 | |||
| FGF-2(basic) | 56 ± 58 | 65 ± 51 | 61 ± 76 | |||
| Flt-3 ligand | 28 ± 43 | 32 ± 47 | 34 ± 36 | |||
| G-CSF | 20 ± 42 | 12 ± 19 | 17 ± 64 | |||
| GM-CSF | 61 ± 251 | 31 ± 94 | 39 ± 128 | |||
| IFNalpha2 | 63 ± 244 | 50 ± 136 | 28 ± 48 | |||
| IL-1alpha | 27 ± 93 | 13 ± 34 | 10 ± 15 | |||
| IL-1beta | 24 ± 114 | 10 ± 30 | 25 ± 99 | |||
| IL-1ra | 63 ± 292 | 53 ± 204 | 45 ± 258 | |||
| IL-2 | 20 ± 103 | 15 ± 52 | 30 ± 82 | |||
| IL-3 | 11 ± 77 | 0 ± 1 | 0 ± 0 | |||
| IL-4 | 22 ± 88 | 7 ± 20 | 16 ± 59 | |||
| IL-5 | 1 ± 9 | 1 ± 5 | 1 ± 1 | |||
| IL-6 | 5 ± 15 | 12 ± 22 | 29 ± 225 | |||
| IL-9 | 3 ± 24 | 2 ± 10 | 2 ± 9 | |||
| IL-10 | 7 ± 36 | 7 ± 10 | 11 ± 60 | |||
| IL-12(p40) | 112 ± 589 | 61 ± 206 | 98 ± 356 | |||
| IL-12(p70) | 32 ± 215 | 32 ± 89 | 13 ± 44 | |||
| IL-13 | 8 ± 38 | 10 ± 26 | 4 ± 13 | |||
| IL-15 | 33 ± 165 | 17 ± 51 | 39 ± 134 | |||
| IP-10 | 109 ± 102 | 149 ± 112 | 204 ± 206 | |||
| MCP-3 | 33 ± 44 | 22 ± 31 | 31 ± 44 | |||
| MIP-1alpha | 47 ± 89 | 58 ± 97 | 38 ± 34 | |||
| TGFalpha | 10 ± 45 | 13 ± 25 | 16 ± 20 | |||
| TNFalpha | 5 ± 20 | 8 ± 14 | 9 ± 14 | |||
| TNFbeta | 17 ± 93 | 15 ± 41 | 14 ± 56 | |||
3.2. Artificial neural network
Based on the findings produced using classical statistics, two ANNs were trained using two separate training sets. The first ANN was trained using all available cytokines (38 in total), and the second trained using only significantly expressed cytokines (12 in total). The training, validation and test set summaries for each ANN are reported in table 2. Numbers within table 2 represent actual patent numbers, not percentages. For example, in table 2—top (for all cytokines), the training set consisted of 75 normal individuals, 67 OA patients and 68 RA patients. All of the normal individuals were correctly categorized, whereas one OA patient and one RA patient were categorized as normal with the remainder correctly diagnosed. These numbers are used to calculate the sensitivity/specificity reported.
Table 2.
Summary of artificial neural network results.
| diagnosis |
||||
|---|---|---|---|---|
| normal | OA | RA | ||
| ANN: all cytokines (n = 38) | ||||
| training set | ||||
| patient type | normal | 75 | 0 | 0 |
| OA | 1 | 66 | 0 | |
| RA | 1 | 0 | 67 | |
| validation set | ||||
| patient type | normal | 11 | 0 | 0 |
| OA | 0 | 21 | 0 | |
| RA | 0 | 0 | 13 | |
| test set | ||||
| patient type | normal | 12 | 0 | 0 |
| OA | 0 | 13 | 0 | |
| RA | 0 | 0 | 20 | |
| ANN: significant cytokines (n = 12) | ||||
| training set | ||||
| patient type | normal | 66 | 2 | 1 |
| OA | 0 | 68 | 2 | |
| RA | 2 | 0 | 69 | |
| validation set | ||||
| patient type | normal | 17 | 0 | 1 |
| OA | 0 | 15 | 0 | |
| RA | 0 | 0 | 12 | |
| test set | ||||
| patient type | normal | 15 | 0 | 0 |
| OA | 0 | 15 | 0 | |
| RA | 0 | 0 | 15 | |
High levels of sensitivity and specificity were obtained within the test set of the first ANN using all 38 cytokines (sensitivity/specificity—norm: 100/100, OA: 100/100, RA: 100/100; table 2—top). The second training set consisted of only significant levels of cytokines and produced the same result (sensitivity/specificity—norm: 100/100, OA: 100/100, RA: 100/100; table 2—bottom). The decision to test a second, smaller dataset (analysis of fewer molecular markers of inflammation) was made to potentially minimize the inclusion of superfluous data to better understand the ‘key’ factors in cohort identification.
3.3. Decision tree
The training set, test set and all data summaries for each decision tree are reported in table 3. As with the representation of the results from the ANNs, numbers within table 3 represent actual patient numbers, not percentages. These numbers are used to calculate the sensitivity and specificity.
Table 3.
Summary of decision tree results.
| diagnosis |
||||
|---|---|---|---|---|
| normal | OA | RA | ||
| TreeBagger: all cytokines (n = 38) | ||||
| training set | ||||
| patient type | normal | 76 | 0 | 0 |
| OA | 0 | 71 | 0 | |
| RA | 0 | 0 | 63 | |
| test set | ||||
| patient type | normal | 24 | 0 | 0 |
| OA | 0 | 29 | 0 | |
| RA | 1 | 1 | 35 | |
| all data | ||||
| patient type | normal | 100 | 0 | 0 |
| OA | 0 | 100 | 0 | |
| RA | 1 | 1 | 98 | |
| single tree: all cytokines (n = 38) | ||||
| training set | ||||
| patient type | normal | 84 | 1 | 0 |
| OA | 0 | 85 | 0 | |
| RA | 2 | 4 | 79 | |
| test set | ||||
| patient type | normal | 14 | 0 | 1 |
| OA | 1 | 14 | 0 | |
| RA | 0 | 1 | 14 | |
| all data | ||||
| patient type | normal | 98 | 1 | 1 |
| OA | 1 | 99 | 0 | |
| RA | 2 | 5 | 93 | |
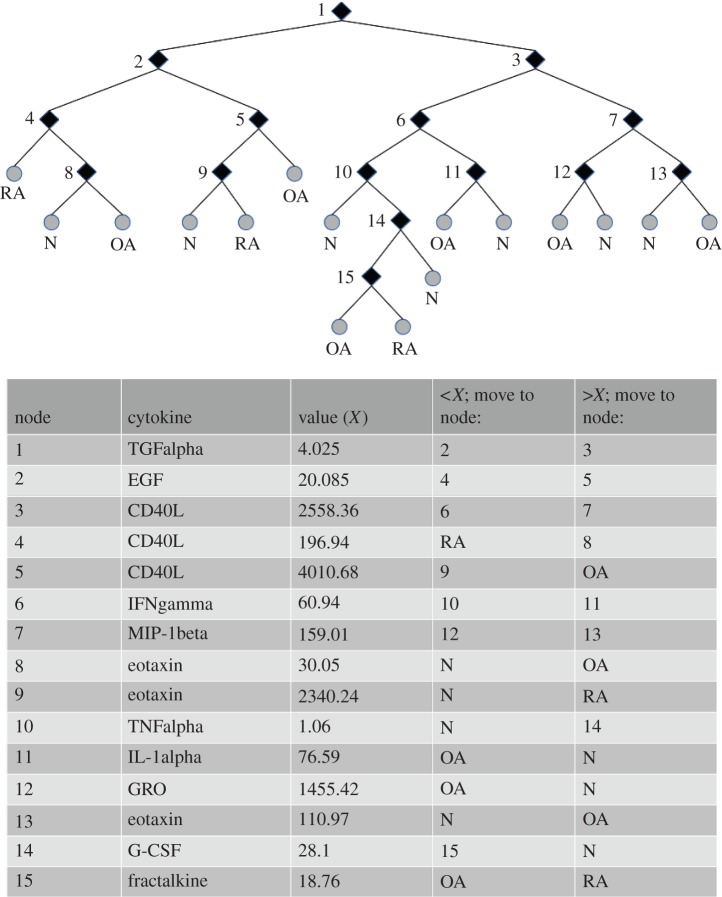
The decision tree algorithm, TreeBagger (Matlab R2011a), executed using all proteins (38 cytokines) as inputs, also returned a high level of sensitivity/specificity (norm: 100/96, OA: 100/97, RA: 95/100; table 3—top). The TreeBagger algorithm yielded results in a manner similar to ANN, meaning that its internal complexity does not allow the user to observe what variables (in this case, cytokines) lead the program to the returned output, thus creating a ‘black box’ scenario. For future research, it would be extremely useful to know which biomarkers (or concentration of biomarkers) are associated with each of the three possible groups (OA, RA and normal). For this reason, a single-decision tree, summarized in table 3 bottom, was programmed. The sensitivity/specificity of the single-decision tree was not as high as that returned by the multi-tree approach (norm: 93/93, OA: 93/93, RA: 93/93); however, information on the specific cytokines involved in the decision process was obtained. As depicted in figure 1, TGFalpha, EGF, CD40L, IFNgamma, MIP-1beta, eotaxin, TNFalpha, IL-1-alpha, GRO, G-CSF and fractalkine were all used to allocate patients into one of the three possible patient groups. Interestingly, there were seven subtypes of normal individuals, six subtypes of OA patients and three subtypes of RA patients based on these inflammatory biomarkers.
Figure 1.
Depiction of single-decision tree with descriptive table.
Decision trees work by finding the optimal cut-off values and combinations of input variables to best allocate samples (or patients) into one of the bins (three treatment groups in this study) requested by the programmer. In this study, analyte levels will be referred to as ‘high’ or ‘low’ relative to cut-off expression level that has been determined by this decision tree algorithm. As described in figure 1, individuals within this study can be described by belonging to one of two broad groups, those with high expression of TGFalpha or those with low expression of TGFalpha. Of the group with low expression of TGFalpha, there were two ways to receive the diagnosis of normal, OA and RA. Normal patients were diagnosed by expressing low EFG, high CD40L and low eotaxin or by expressing high EGF, low CD40L and low eotaxin. OA patients were diagnosed by expressing low EGF, high CD40L and high eotaxin or by expressing high EGF and high CD40L. RA patients were diagnosed by expressing low EGF and low CD40L or expressing high EGF, low CD40L and high eotaxin.
Individuals with high levels of TGFalpha (figure 1, node 1) are further divided into two subgroups: those with high or low CD40L (figure 1, node 3) levels. Of the group with high TGFalpha and low CD40L, there are three ways to be categorized as normal diagnosis, two ways to be categorized as OA and one way to be categorized as RA. Normals were categorized based on low levels of IFNgamma and low TNFalpha, expressing low IFNgamma, high TNFalpha and high G-CSF or expressing high IFNgamma and high IL-1alpha. OA patients were categorized on the basis of low levels of IFNgamma, high TNFalpha, low G-CSF and low fractalkine or by high levels of IFNgamma and low IL-1alpha. RA patients were categorized on the basis of low levels of IFNgamma, high TNFalpha, low G-CSF and high fractalkine.
Within the group of high expression of TGFalpha (figure 1, node 1) and high expression of CD40L (figure 1, node 3), there are two ways to be categorized as normal and two ways to be categorized as OA, it was not possible to be categorized as RA in this group. Normal individuals were categorized on the basis of low MIP-1beta and high GRO or by high MIP-1beta and low eotaxin. OA patients were categorized by low MIP-1beta and low GRO or by high MIP-1beta and high eotaxin.
Interestingly, CD40L and eotaxin were each used in three separate combinations to categorize cohorts. Combinations with CD40L included individuals with high levels of TGF-beta, and with low levels of TGF-beta in combination with high or low EGF. Combinations with eotaxin-included individuals with low levels of TGF-beta and high EGF with low CD40L or low EGF with high CD40L, as well as high TGF-beta with high CD40L and high MIP-1beta. This suggests that in a complex disease it is important to evaluate the levels of biomarkers in the context of other biomarkers.
Single-decision trees were further investigated to see how lowering the number of inputs would affect the sensitivity and specificity of the algorithm (table 4). In the above single tree analysis, 11 markers were identified as important for decision-making, yet 12 were important for programming the tree. The protein VEGF was important for programming the tree, but not included in any of the decision nodes that were quantified. As we were able to obtain a high sensitivity and specificity with 12 markers, we decided to use this as the starting point and programmed the algorithm to produce the best possible outcome with six, four and two inputs (markers). In descending order of importance for decision-making, CD40L, TGFalpha, EGF, INFgamma, eotaxin, MIP-1beta, TNFalpha, IL-1alpha, G-CSF, fractalkine, GRO and VEGF were the most important markers. As summarized in table 4, overall sensitivity and specificity declined as fewer markers were used (as the algorithm omitted the least important markers). Of note, the use of classical statistics did not identify all of the 12 markers that were deemed essential for the highest sensitivity and specificity that the algorithm achieved. Eotaxin, G-CSF, IL-1alpha, TGFalpha and TNFalpha did not show statistical significance in treatment group comparisons (p > 0.05), yet were essential in achieving the high level of accuracy. This result appears to reaffirm the idea that what may be important in the creation of a diagnostic for a complex disease such as OA and RA could be the interpretation of combinations of markers, and not just the statistical significance of a few.
Table 4.
Summary of pruned single-decision tree.
| single tree no. inputs | inputs | normal |
OA |
RA |
|||
|---|---|---|---|---|---|---|---|
| sensitivity (%) | specificity (%) | sensitivity (%) | specificity (%) | sensitivity (%) | specificity (%) | ||
| 12 | CD40L, TGFalpha, EGF, INFgamma, eotaxin, MIP-1beta, TNFalpha, IL-1alpha, G-CSF, fractalkine, GRO, VEGF | 93 | 93 | 93 | 93 | 93 | 93 |
| 6 | CD40L, TGFalpha, EGF, INFgamma, eotaxin, MIP-1beta | 93 | 82 | 93 | 93 | 80 | 92 |
| 4 | CD40L, TGFalpha, EGF, INFgamma | 100 | 75 | 80 | 100 | 87 | 100 |
| 2 | CD40L, TGFalpha | 73 | 55 | 53 | 67 | 80 | 92 |
4. Discussion
When we applied ANN processing to a dataset collected from multiplexing of blood serum samples collected from OA patients and compared with samples from normal and RA cohorts, we were able to improve the potential diagnostic efficacy of our panel of inflammatory cytokines over statistical approaches such as PCA and clustering analysis [17]. Using the entire panel of 38 cytokines relevant to inflammation, we were able to train an ANN to distinguish between our three patient groups with a high degree of sensitivity and specificity. These results were verified using the multi-decision tree algorithm. Furthermore, a second single-decision tree was programmed to provide insights into which cytokines were used to allocate individuals into specific cohort groups, and at what cut-off value each unique decision was made.
For the diagnosis of OA, innovative joint imaging techniques are emerging, but the need still exists for a reliable (non-invasive) diagnostic tool to at least support an early and accurate diagnosis. Using a panel of well-documented inflammatory cytokines, we have developed a non-invasive test that can be used to identify patients with OA of the knee. Although there is currently no disease modifying treatment for early OA, the panel of cytokines that we have developed may prove useful for the development and evaluation of future pharmacological, behavioural and/or mechanical (including surgical) interventions.
Using a similar approach, the efficacy of analysing synovial fluid proteins as diagnostic biomarkers of arthritic conditions has previously been reported [23]. Using an ANN on proteins quantified within synovial fluid, Han et al. differentiated between samples from OA and RA patients with a high degree of sensitivity and specificity. Additionally, another study by Swan et al., used an ANN to identify potential biomarkers in OA articular cartilage [25]. This same group has also discussed the benefits of using bioinformatic lessons learned from the genomics era for application to large proteomic datasets, and specifically in chronic diseases such as OA [26]. Our study was accomplished using serum, a less invasive sampling method. Furthermore, owing to the less invasive nature of our sampling method, we were able to effectively compare the results with samples from a group of normal non-OA and non-RA controls. Historically, in contrast to the more clinically obvious signs and symptoms of inflammation in RA, for the most part, OA has been regarded as a non-inflammatory condition. With the addition of samples from 100 normal individuals, we were able to detect an inflammatory phenotype within our OA patients that led to their discrimination from normal and RA cohorts.
Biomarkers of OA have been previously described, yet none has been widely clinically adopted or Food and Drug Administration/Health Canada approved for use in OA diagnosis. We have investigated multiplexed inflammatory data with ANNs, and decision trees and it appears that a combination of investigated cytokines within a given sample is a more important consideration for accurate disease classification than the levels of unique individual cytokines to achieve validation of the clinical assessment. It is possible that the combination of certain cytokines describes a more global picture, a latent variable or relationship that is lost when each cytokine is investigated separately for significance within the patient population. Therefore, in the future, it may be prudent to not exclude markers that are individually found not to discriminate cohorts based on standard statistical techniques. Retrospectively, it could be quite interesting and rewarding to apply this ANN methodology to historical biomarkers of OA and RA. Specifically in OA, biomarkers such as collagen II fragments, COMP, HA, matrix metalloproteinases and single inflammatory proteins (IL-1B, IL-6, TNFalpha, among others) have not garnered significant clinical adoption. This may be due to heterogeneity and variability between individuals and more than likely the disease itself; however, applying this type of ANN methodology to these historical biomarkers may shed light on clouded and contradictory results published within this field of research.
This work, specifically the interpretation of the single-decision tree, has allowed us to begin to capture and quantify, the phenotypic complexity of arthritic disease and the normal state. This methodology suggests that in order to achieve an accurate diagnosis (between normal, OA or RA states), each biological marker (in this case, cytokines) needs to be considered in relation to all other biological markers being examined, and not as independent measures. The outputs from this algorithm also suggest that the variation seen within individual biological markers of a patient population can be extremely useful when investigated in the context of variation present in additional markers. Thus, classical statistics comparing group means and standard deviations alone may not be appropriate in this instance. Arthritis is a complex disease with a complex phenotype, it is not unrealistic to consider that the effective diagnoses will likely require a relatively complex interpretation of data that can account for heterogeneity of a patient population in addition to heterogeneity of a disease state (e.g. onset, pathogenesis and progression).
While ANNs have gathered significant attention in the basic sciences for analysing large and complex datasets, there have been relatively few clinical diagnostic applications of this methodology, and even fewer cases in which this methodology has been applied to the diagnosis of OA. In this study, we have demonstrated a proof of principle for this algorithm when combined with a high-throughput proteomic inflammatory assay in the serum of patients with OA. The synergy of these methodologies may represent an effective starting point to develop a reliable non-invasive diagnostic for early OA.
In conclusion, through the multiplexed analysis of a panel of inflammatory cytokines quantified in the sera of OA and RA patients, and normal individuals, we were able to accurately classify OA patients as discriminated against normal individuals and RA patients. This methodology could be further developed into a diagnostic tool to aid and complement physician diagnosis of OA.
Acknowledgements
We thank Jenelle McAllister for collecting OA serum samples, as well as Mark Fritzler and Riley Sullivan (Eve Technologies).
This study protocol was approved by the University of Calgary Human Research Ethics Board and the Research Ethics Board of the University of Manitoba. All participants provided written consent to participate.
References
- 1.Aletaha D, et al. 2010. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 62, 2569–2581. ( 10.1002/art.27584) [DOI] [PubMed] [Google Scholar]
- 2.Altman R, et al. 1986. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 29, 1039–1049. [DOI] [PubMed] [Google Scholar]
- 3.Kraus VB, Nevitt M, Sandell LJ. 2010. Summary of the OA biomarkers workshop 2009—biochemical biomarkers: biology, validation, and clinical studies. Osteoarthritis Cartilage 18, 742–745. ( 10.1016/j.joca.2010.02.014) [DOI] [PubMed] [Google Scholar]
- 4.Bowyer J, Heapy CG, Flannelly JK, Waterton JC, Maciewicz RA. 2009. Evaluation of a magnetic resonance biomarker of osteoarthritis disease progression: doxycycline slows tibial cartilage loss in the Dunkin Hartley guinea pig. Int. J. Exp. Pathol. 90, 174–181. ( 10.1111/j.1365-2613.2008.00634.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruyere O, et al. 2007. Longitudinal study of magnetic resonance imaging and standard X-rays to assess disease progression in osteoarthritis. Osteoarthritis Cartilage 15, 98–103. ( 10.1016/j.joca.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 6.Kokebie R, Aggarwal R, Lidder S, Hakimiyan AA, Rueger DC, Block JA, Chubinskaya S. 2011. The role of synovial fluid markers of catabolism and anabolism in osteoarthritis, rheumatoid arthritis and asymptomatic organ donors. Arthritis Res. Ther. 13, R50 ( 10.1186/ar3293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golightly YM, Marshall SW, Kraus VB, Renner JB, Villaveces A, Casteel C, Jordan JM. 2011. Biomarkers of incident radiographic knee osteoarthritis: do they vary by chronic knee symptoms? Arthritis Rheum. 63, 2276–2283. ( 10.1002/art.30412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catterall JB, Stabler TV, Flannery CR, Kraus VB. 2010. Changes in serum and synovial fluid biomarkers after acute injury (NCT00332254). Arthritis Res. Ther. 12, R229 ( 10.1186/ar3216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. 2012. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 64, 1697–1707. ( 10.1002/art.34453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsaid KA, Chichester CO. 2006. Review: collagen markers in early arthritic diseases. Clin. Chim. Acta. 365, 68–77. ( 10.1016/j.cca.2005.09.020) [DOI] [PubMed] [Google Scholar]
- 11.Haraoui B, Pope J. 2011. Treatment of early rheumatoid arthritis: concepts in management. Semin. Arthritis Rheum. 40, 371–388. ( 10.1016/j.semarthrit.2010.10.004) [DOI] [PubMed] [Google Scholar]
- 12.Blumberg SN, Fox DA. 2001. Rheumatoid arthritis: guidelines for emerging therapies. Am. J. Manag. Care 7, 617–626. [PubMed] [Google Scholar]
- 13.Wang Q, et al. 2011. Identification of a central role for complement in osteoarthritis. Nat. Med. 17, 1674–1679. ( 10.1038/nm.2543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb GR, Westacott CI, Elson CJ. 1998. Osteoarthritic synovial fluid and synovium supernatants up-regulate tumor necrosis factor receptors on human articular chondrocytes. Osteoarthritis Cartilage 6, 167–176. ( 10.1053/joca.1998.0109) [DOI] [PubMed] [Google Scholar]
- 15.Jacques C, Gosset M, Berenbaum F, Gabay C. 2006. The role of IL-1 and IL-1Ra in joint inflammation and cartilage degradation. Vitam. Horm. 74, 371–403. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. 2011. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33–42. ( 10.1038/nrrheum.2010.196) [DOI] [PubMed] [Google Scholar]
- 17.Heard BJ, Fritzler MJ, Wiley P, McAllister J, Martin L, El-Gabalawy H, Hart DA, Frank CB, Krawetz R. 2013. Intra-articular and systemic inflammatory profiles identify patients with osteoarthritis. J. Rheumatol. 40, 1379–1387. ( 10.3899/jrheum.121204) [DOI] [PubMed] [Google Scholar]
- 18.Kraus VB. 2011. Osteoarthritis year 2010 in review: biochemical markers. Osteoarthritis Cartilage 19, 346–353. ( 10.1016/j.joca.2011.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Begg R, Kamruzzaman J, Sarker R (eds). 2006 Neural networks in healthcare: potential and challenges . Hershey, PA: Ideal Group Publishing. ( ) [DOI]
- 20.Hsu CC, Lin YE, Chen YS, Liu YC, Muder RR. 2008. Validation study of artificial neural network models for prediction of methicillin-resistant Staphylococcus aureus carriage. Infect. Control Hosp. Epidemiol. 29, 607–614. ( 10.1086/588588) [DOI] [PubMed] [Google Scholar]
- 21.Duryea J, Zaim S, Wolfe F. 2002. Neural network based automated algorithm to identify joint locations on hand/wrist radiographs for arthritis assessment. Med. Phys. 29, 403–411. ( 10.1118/1.1446099) [DOI] [PubMed] [Google Scholar]
- 22.Gajre SS, Anand S, Singh U, Saxena RK. 2006. Novel method of using dynamic electrical impedance signals for noninvasive diagnosis of knee osteoarthritis. Conf. Proc. IEEE Eng. Med. Biol. Soc. 1, 2207–2210. [DOI] [PubMed] [Google Scholar]
- 23.Han MY, Dai JJ, Zhang Y, Lin Q, Jiang M, Xu XY, Liu Q. 2012. Identification of osteoarthritis biomarkers by proteomic analysis of synovial fluid. J. Int. Med. Res. 40, 2243–2250. ( 10.1177/030006051204000622) [DOI] [PubMed] [Google Scholar]
- 24.Djoba Siawaya JF, et al. 2008. An evaluation of commercial fluorescent bead-based luminex cytokine assays. PLoS ONE 3, e2535 ( 10.1371/journal.pone.0002535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swan AL, Hillier KL, Smith JR, Allaway D, Liddell S, Bacardit J, Mobasheri A. 2013. Analysis of mass spectrometry data from the secretome of an explant model of articular cartilage exposed to pro-inflammatory and anti-inflammatory stimuli using machine learning. BMC Musculoskelet. Disord. 14, 349 ( 10.1186/1471-2474-14-349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J. 2013. Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. OMICS 17, 595–610. ( 10.1089/omi.2013.0017) [DOI] [PMC free article] [PubMed] [Google Scholar]