Abstract
Analysis of 4D medical images presenting pathology (i.e., lesions) is significantly challenging due to the presence of complex changes over time. Image analysis methods for 4D images with lesions need to account for changes in brain structures due to deformation, as well as the formation and deletion of new structures (e.g., edema, bleeding) due to the physiological processes associated with damage, intervention, and recovery. We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. We demonstrate the effectiveness of our novel approach with the analysis of 4D images of traumatic brain injury (TBI), using a synthetic tumor database as the source domain.
1 Introduction
Traumatic brain injury (TBI) is a critical problem in healthcare that impacts approximately 1.7 million people in the United States every year [3]. The varying cause and degree of injury (falls, car accidents, etc.) presents significant challenges in the interpretation of image data but also in quantitative assessment of brain pathology via image analysis. Determining effective therapy and intervention strategies requires the ability to track the image changes over time, which motivates the development of segmentation and registration methods for longitudinal 4D Magnetic Resonance (MR) images. Such methods need to account for changes in brain structures due to deformation, as well as the formation and deletion of new structures (e.g., edema, bleeding) due to physiological processes associated with damage, therapeutical intervention, and recovery.
In 4D image analysis, researchers have proposed methods [9, 2, 5, 6] to register images with lesions over time accounting for appearance of new structures. However, these methods have not been evaluated for mapping healthy subjects to a patient with lesions. Niethammer et al. proposed a registration method for TBI images using geometric metamorphosis that maps TBI over time using known, pre-segmented lesion boundaries defined manually [5]. Wang et al. [14] proposed a registration-segmentation method for 4D TBI images using personalized atlas construction that combines information from multiple time points, accounting for diffeomorphic changes (smooth deformation) and non-diffeomorphic changes (creation/deletion of lesions) over time. However, their method requires manual initialization in the form of user-defined spheres covering the lesions and only provides modeling of intra-patient changes without providing explicit mapping to normative healthy brain anatomy.
We propose a novel framework that models changes in 4D pathological anatomy across time and provides explicit mapping from a healthy template to TBI subject images. This aids analysis of TBI patients by enabling the mapping of parcellation labels describing anatomical regions of interest and quantitative comparison against a common reference space defined by the normative template. Moreover, our framework uses transfer learning [7] to leverage rich information from a “known source” domain, where we have a large collection of fully segmented images, to yield effective models for the “input target” domain (TBI images). This is essential as such a database does not exist for TBI imaging, and thus we explore and demonstrate the use of an existing database of multi-modal tumor imaging that serves as a well-studied source domain. The information in the learned tumor model are transferred to the domain of TBI images using importance weighting based domain adaptation [12], a well known transfer learning technique, resulting in a fully automatic method that does not require user input. In this paper, we propose importance weighting based domain adaptation for generative kernel density models, thus extending its applications beyond standard discriminative models available in machine learning literature [1].
2 Method
We propose a framework that constructs 4D models of pathological anatomy starting from a healthy template, to describe changes at different time points accounting for the complete 4D information. Our framework also leverages known domains, such as brain tumors, where we have a rich collection of information in the form of segmented tumor images with varying size, shape, deformations, and appearance. The database of tumor images is obtained by using the brain tumor simulator1 developed by Prastawa et al.[11]. It is capable of generating synthetic images for a large variety of tumor cases with complete 3D segmentations. Fig. 1 shows a conceptual overview of our mathematical framework.
Fig. 1.
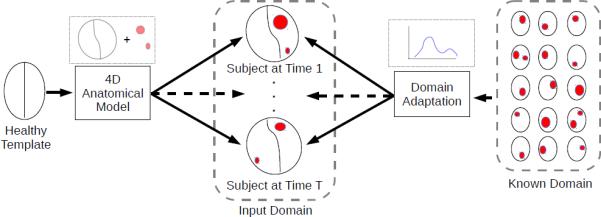
Conceptual overview of the proposed framework. Our framework maps a healthy template to input TBI images at different time points using a 4D anatomical model which provides spatial context. The model leverages information from a different known domain, in this case tumor images that are fully segmented. Data from the known domain with lesions (indicated in red) at different locations with varying size, shape, and deformations are used to estimate an appearance model for the input TBI images.
2.1 4D Modeling of Pathological Anatomy
We model the anatomical changes over time as a combination of diffeomorphic image deformation and non-diffeomorphic changes of probabilities for lesion categories, accounting for temporally smooth deformations and abrupt changes, e.g., due to lesions appearing and disappearing over time. Specifically, the spatial prior for each class c at time point t is modeled as
| (1) |
where A is the tissue class probability that is initially associated with the healthy template, ϕt is the diffeomorphic deformation from time t to the atlas, and Qt is the non-diffeomorphic probabilistic change for time t. This approach follows the metamorphosis framework of Trouvé and Younes [13]. Our method estimates a common subject-specific atlas A for all time points.
Given the model and 4D multimodal images It at timepoints t, we estimate model parameters that minimize the following functional:
| (2) |
where represents the data functional (the negative total log-likelihood)
| (3) |
and represents the regularity terms:
| (4) |
T denotes the number of observed time points, C denotes the number of tissue classes, is the image likelihood function for class c with parameter , A(0) is the initial atlas A obtained from the healthy template, and d(id, ) is the distance to the identity transform. enforces the sparsity of Q, prevents extreme deviations in A from the initial model, and enforces the smoothness of the deformations ϕt. These regularization functionals are weighted by user-defined parameters α, β, γ respectively.
2.2 Image Appearance Model using Domain Adaptation
We compute our image appearance model using the well-known domain adaptation technique, where we adapt an appearance model from a known domain (tumor images) to the input domain (TBI images). We use a simulator [11] to generate a large collection of synthetic tumor images that resemble TBI images, and we use the rich information in this database to automatically compute the likelihood density model and then transfer this model to the TBI domain. Fig. 2 shows examples of fully segmented synthetic tumor images from the known domain and unsegmented TBI images in the input domain.
Fig. 2.
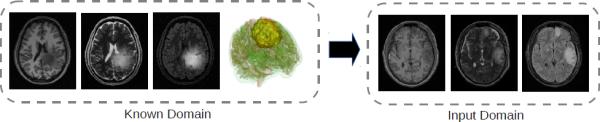
Example data in different domains, containing the T1, T2, and FLAIR (Fluid Attenuated Inversion Recovery) modalities. Appearance information from the known tumor domain, which contains 3D anatomical labels, are transferred to the input TBI domain using domain adaptation.
We select a tumor image from the database that has the smallest earth mover's distance [8] compared to the input TBI images. We then obtain training samples in the known or “source” domain as a subset of the completely segmented tumor data , with Î representing the tumor intensities, representing the discrete segmentations, representing the probabilistic segmentations, and x̂ representing the coordinates in the tumor image domain. The transfer of learned appearance models is accomplished via domain adaptation that incorporates importance weighting. We weight intensity observations I using the weights with p̂ being the density in the source domain. In practice, w is estimated using KLIEP (Kullback-Leibler Importance Estimation Procedure) [12] which minimizes the Kullback-Leibler divergence between the density of the input domain and the weighted density of the source domain KL(p(I) ∥ w(I) p̂(I)).
Using the estimated weights w, we compute the density parameter ^ that maximize the data likelihood in the tumor domain:
| (5) |
We use the kernel density model for the image appearance, parametrized by the kernel bandwidths for each class . The image likelihood in the TBI domain is modeled in the same fashion, where we initialize TBI parameter θ using the “domain adapted” tumor parameter from Eq. (5).
2.3 Model Parameter Estimation
We perform model parameter estimation by minimizing the overall objective function (Eq. 2) with respect to each parameter. Fig. 3 provides a conceptual view of the parameter estimation process, which incorporates gradient descent updates that are effectively image registration and segmentation operations. In particular, we use these gradient equations to optimize the data functional :
| (6) |
| (7) |
| (8) |
where |Dϕ| denotes the determinant of the Jacobian of ϕ. The updates show that Qt moves to the data likelihood specific to time t, A moves to the average data likelihood over time, and ϕt deforms A to match the boundaries between data and atlas. Constraints are enforced using projected gradient descent [10]. The image likelihood model obtained from domain adaptation is fitted to the input image data using gradient descent update , which finds the set of widths that best matches data to the current estimate ofF the atlas at timepoint t, .
Fig. 3.
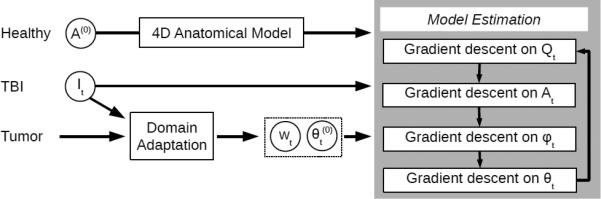
Model parameter estimation process. The healthy template provides the initial personalized atlas A in the 4D anatomical model. Input images together with the tumor database are used to generate densities represented by importance weights wt and kernel width parameter t. All parameters are updated using alternating gradient descent, where initially Qt is zero and ϕt is the identity transform.
3 Results
We evaluate the performance of our new approach on 4D TBI image data containing two time points: acute and chronic (≈ 3 days and 6 months post-surgery). The performance of our proposed method is show ≈ in Tab. 1, where we compare our method against those that do not use 4D modeling, with and without domain adaptation. Dice overlap values comparing automatic lesion segmentations against a human expert rater are relatively low, which is a well known fact when dealing with small objects with complex and fuzzily defined boundaries. However, our method not only provides improved lesion segmentation but also better overall segmentation, as shown qualitatively in Fig. 4.
Table 1.
Dice overlap values for segmentations of acute-stage lesions comparing our proposed method to a direct application of tumor appearance model and an application of domain adaption, both without using a 4D model. The new integrated method yields improved results by combining 4D anatomical information and adapting tumor appearance information.
| Method | 4D Model | Appearance Model | Subject 1 | Subject 2 | Subject 3 |
|---|---|---|---|---|---|
| I | None | Not adapted | 0.2536 | 0.1211 | 0.5238 |
| II | None | Domain adapted | 0.3053 | 0.1131 | 0.5238 |
| III | Proposed | Domain adapted | 0.3792 | 0.1367 | 0.6035 |
Fig. 4.
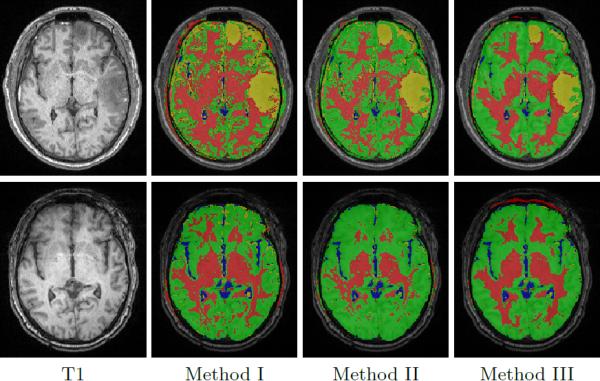
Segmentation results for subject 1 at acute (top) and chronic (bottom) stages using different methods. Our proposed method (III) has the best segmentation quality overall. Red: white matter, green: gray matter, blue: cerebrospinal fluid, and yellow: lesion.
The estimated 4D spatial priors for TBI subject 3 are illustrated in Fig. 5, incorporating template deformation to match image boundaries and non-diffeomorphic changes due to lesions. Subject 3 provides an interesting and revealing example of longitudinal pathology progression. The acute scan reveals gross pathology in the left frontal region, which results in considerable atrophy in this region at the chronic stage. However, the subject's chronic scan features an additional large lesion in the mid-frontal region due to the occurrence of a large abscess between acute and chronic scans. This is an excellent example of the dynamic and complex longitudinal changes that can occur in TBI patients.
Fig. 5.
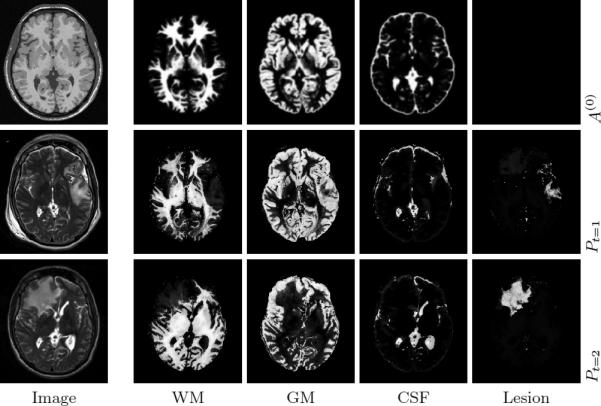
Estimated 4D anatomical priors for TBI subject 3. First row shows the initial atlas A(0) in the template space, with the healthy T1 image as a reference. Second and third row show the personalized atlas Pt = A ○ ϕt + Qt for acute and chronic stages, with input T2 images shown. Our method is able to account for changes in the left-frontal and mid-frontal regions across time.
The proposed method brings the advantage of providing a mapping from a normative template to a TBI subject. In Fig. 6, we show a parcellation label image, provided by the International Consortium for Brain Mapping (ICBM), that has been mapped to a TBI subject. The mapping of a normal anatomy to pathological anatomy will be potentially important to compare type, locality and spatial extent of brain damages in the context of anatomically relevant regions with associated brain function information.
Fig. 6.
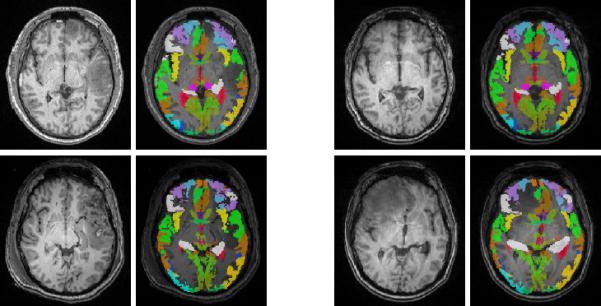
Example brain parcellation labels mapped to the acute (left) and chronic (right) time points of subject 1 (top) and 3 (bottom). For each time point, we show the input T1 image and the overlaid parcellation labels. Our method generates parcellation maps that match tissue boundaries and account for lesions.
4 Conclusions
We demonstrate work in progress towards a framework that estimates 4D anatomical models from longitudinal TBI images. Our framework is fully automatic and leverages information from a different domain (brain tumor) to generate appearance models via domain adaptation. In addition to the new 4D anatomical modeling, we also presented a new domain adaptation method for generative kernel density models, integrated with our anatomical model in a single objective function (Eq. 2). Results on 3 TBI subjects show that our automatic method yields segmentations that match ground truth of manual segmentations. Furthermore, our method generates diffeomorphic deformation models as well as non-diffeomorphic probabilistic changes that have potential for analyzing and characterizing changes of normal appearing tissue and lesions. In the future, we will quantify temporal brain changes across a large set of TBI patients which were exposed to different treatment strategies. Our approach has potential to significantly improve regional and connectivity analysis of individuals relative to a population [4], by making use of the mapping of a normative template with associated parcellation labels to TBI subjects, without tedious manual input.
Acknowledgments
This work has been supported by National Alliance for Medical Image Computing (NA-MIC) U54 EB005149 (Guido Gerig) and the Utah Science Technology and Research (USTAR) initiative at the University of Utah.
Footnotes
References
- 1.Beijbom O. Domain adaptations for computer vision applications. Tech. rep. University of California; San Diego: Apr, 2012. arXiv:1211.4860. [Google Scholar]
- 2.Chitphakdithai N, Duncan JS. Non-rigid registration with missing correspondences in preoperative and postresection brain images. MICCAI. 2010:367–374. doi: 10.1007/978-3-642-15705-9_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faul M, Xu L, Wald M, Coronado V. Traumatic brain injury in the United States: Emergency department visits, hospitalizations and deaths 2002-2006. CDC, National Center for Injury Prevention and Control; Atlanta (GA): 2010. [Google Scholar]
- 4.Irimia A, Wang B, Aylward S, Prastawa M, Pace D, Gerig G, Hovda D, Kikinis R, Vespa P, Van Horn J. Neuroimaging of structural pathology and connectomics in traumatic brain injury: Toward personalized outcome prediction. NeuroImage: Clinical. 2012;1(1):1–17. doi: 10.1016/j.nicl.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niethammer M, Hart GL, Pace DF, Vespa PM, Irimia A, Horn JDV, Aylward SR. Geometric metamorphosis. MICCAI. 2011;(2):639–646. doi: 10.1007/978-3-642-23629-7_78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Medical Image Analysis. 2011;15(4):622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan SJ, Yang Q. A survey on transfer learning. Knowledge and Data Engineering, IEEE Transactions on. 2010 Oct;22(10):1345–1359. [Google Scholar]
- 8.Pele O, Werman M. Computer vision, 2009 IEEE 12th international conference on. IEEE; 2009. Fast and robust earth mover's distances. pp. 460–467. [Google Scholar]
- 9.Periaswamy S, Farid H. Medical image registration with partial data. Medical Image Analysis. 2006;10(3):452–464. doi: 10.1016/j.media.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Prastawa M, Awate S, Gerig G. Building spatiotemporal anatomical models through joint segmentation, registration, and 4D-atlas estimation. MMBIA. 2012:49–56. doi: 10.1109/mmbia.2012.6164740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prastawa M, Bullitt E, Gerig G. Simulation of brain tumors in MR images for evaluation of segmentation efficacy. Medical image analysis. 2009;13(2):297–311. doi: 10.1016/j.media.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiyama M, Nakajima S, Kashima H, Von Buenau P, Kawanabe M. Direct importance estimation with model selection and its application to covariate shift adaptation. Advances in NIPS. 2008;20:1433–1440. [Google Scholar]
- 13.Trouvé A, Younes L. Metamorphoses through lie group action. Foundations of Computational Mathematics. 2005;5(2):173–198. [Google Scholar]
- 14.Wang B, Prastawa M, Awate S, Irimia A, Chambers M, Vespa P, van Horn J, Gerig G. Segmentation of serial MRI of TBI patients using personalized atlas construction and topological change estimation. ISBI. 2012:1152–1155. doi: 10.1109/isbi.2012.6235764. [DOI] [PMC free article] [PubMed] [Google Scholar]


