Abstract
The exposure of oral antiretroviral (ARV) drugs in the female genital tract (FGT) is variable and almost unpredictable. Identifying an efficient method to find compounds with high tissue penetration would streamline the development of regimens for both HIV preexposure prophylaxis and viral reservoir targeting. Here we describe the cheminformatics investigation of diverse drugs with known FGT penetration using cluster analysis and quantitative structure–activity relationships (QSAR) modeling. A literature search over the 1950–2012 period identified 58 compounds (including 21 ARVs and representing 13 drug classes) associated with their actual concentration data for cervical or vaginal tissue, or cervicovaginal fluid. Cluster analysis revealed significant trends in the penetrative ability for certain chemotypes. QSAR models to predict genital tract concentrations normalized to blood plasma concentrations were developed with two machine learning techniques utilizing drugs' molecular descriptors and pharmacokinetic parameters as inputs. The QSAR model with the highest predictive accuracy had R2test=0.47. High volume of distribution, high MRP1 substrate probability, and low MRP4 substrate probability were associated with FGT concentrations ≥1.5-fold plasma concentrations. However, due to the limited FGT data available, prediction performances of all models were low. Despite this limitation, we were able to support our findings by correctly predicting the penetration class of rilpivirine and dolutegravir. With more data to enrich the models, we believe these methods could potentially enhance the current approach of clinical testing.
The ideal preexposure prophylaxis (PrEP) regimen for protection from HIV remains undefined. Previous studies of FDA-approved antiretrovirals (ARVs), such as tenofovir with or without emtricitabine, using standard treatment doses and dosing frequencies have shown mixed success (0–73%) in preventing HIV acquisition in high-risk populations.1–3 A possible explanation for the failure of some of these trials is the lack of necessary preclinical and early clinical phase data to guide optimal selection and dosing of PrEP regimens.4
The tissues of the female genital tract (FGT) are primary sites of HIV transmission, and ARV concentration in the FGT at the time of HIV exposure is a critical determinant of PrEP success.5,6 Further, because these tissues may act as a latent HIV reservoir, eradication strategies cannot be successful unless adequate ARV concentrations are achieved.7 Unfortunately, ARV penetration into the FGT is highly challenging to prognosticate due to a large amount of interclass and intraclass variability.8 As a result, the extent of FGT penetration can be evaluated only by costly and complex clinical testing. Therefore, alternative mechanisms to identify highly penetrative compounds that can be utilized for PrEP are greatly needed. Identifying structural characteristics of compounds likely to be favorable for FGT penetration can expedite the early phase drug development process, streamline early clinical studies, and possibly lead to faster market approval.
Cheminformatics approaches, such as quantitative structure activity relationship (QSAR) modeling, are extensively utilized in drug discovery and development as effective means to prioritize candidate compounds for experimental testing.9,10 These techniques have been particularly helpful for ADME/Tox predictions, providing a useful and reliable alternative to in vivo assessments. QSAR modeling has also been helpful in predicting drug distribution/penetration into specific biological compartments such as the blood–brain barrier (BBB); for instance, a highly predictive model of penetration (R2=0.80 in an external validation set of 10 compounds) as well as specific contributors to penetration, such as van der Waals surface area and active transport, was reported recently.11 A similarly predictive model for FGT penetration would help inform the development process and assist in the search for ideal PrEP candidates.
To date, no computational technique to predict FGT penetration has been published. Considering recent evaluations of transporter and metabolizing enzyme expression in the FGT showing several possible contributors to penetration,12,13 we hypothesized that the development of a predictive model could benefit from the incorporation of drugs' transporter profiles and other biological parameters as additional molecular descriptors. Zhou et al.12 examined the expression of 19 transporters in the FGT using reverse transcriptase polymerase chain reaction (RT-PCR) and found that several uptake (OCT2, ENT1, OATP-D) and efflux (MDR1, BCRP, and MRPs 1, 4, 5, and 7) transporters were qualitatively expressed at levels equal to or greater than the liver. Moreover, a study by Nicol et al. using real-time PCR (qPCR) provided quantitative evidence of a high expression of MRP4 (120–310% of liver expression) in the epithelial and submucosal cells of vaginal and cervical tissue.13 The findings from these studies thus provide a biologically plausible foundation on which to build a predictive model for FGT penetration using both structural and biological characteristics of drugs.
In this study we attempted both the analysis and modeling of FGT drug penetration based on chemical structures and computed biological properties, specifically drugs' transporter interaction profiles. First we compiled, curated, and integrated from the literature a set of 58 drugs with known FGT penetration. Second, we clustered this set into smaller groups of drugs with similar structures and analyzed the variation (and potential concordance) of FGT penetration within each cluster. Third, we conducted the QSAR modeling of FGT penetration of 58 drugs using both chemical and biological descriptors. The results showed distinctive structure–activity relationships, especially within certain clusters of structurally similar drugs.
A literature search was conducted in the PubMed database focusing on any clinical study reporting pharmacokinetic (PK) data for any compound in the lower FGT using specific search terms (female genital tract or vaginal tissue or cervical tissue or gynecologic tissue and drug penetration or distribution) and filters (human, 1950–2012). EMBASE and Web of Science were also searched. Studies that reported data in vaginal tissue (VT), cervical tissue (CT), or cervicovaginal fluid (CVF) were isolated. The endpoint of interest was the tissue penetration ratio (TPR), defined as a tissue or CVF isolated concentration or area-under-the-concentration-time curve (AUC) divided by a plasma concentration or AUC taken at the same time point or interval. If a publication did not report a specific TPR value, it was calculated when the appropriate data were available. In the event that multiple TPR values generated from different techniques (i.e., steady-state AUC ratio vs. single time point ratio) were reported, the TPR was chosen based on an algorithm created according to pharmacologic rigor as follows: VT or CT steady-state AUC preferred over (>) VT or CT single dose AUC>CVF steady-state AUC>CVF single dose AUC>VT or CT steady-state single time point>VT or CT single dose single time point>CVF steady-state single time point>CVF single dose single time point. If multiple TPR values were reported that were generated from the same technique (i.e., two steady-state AUC ratios), the TPR from the study with the larger sample size was chosen. AUCs were preferentially selected to negate the effect of differential dosing and/or sampling times between studies, though this was not possible for many compounds.
The TPRs from all identified compounds were included in a database with the drug's generic name, therapeutic class, chemical structure and SMILES string (imported and/or checked using Chemspider at www.chemspider.com), and physicochemical properties such as plasma protein binding percentage (PPB%) and volume of distribution (Vd), which were obtained from the drug package inserts. Two-dimensional structures of the 58 drugs were obtained and standardized according to procedures described previously.14 In addition to including the raw TPR data, we grouped compounds into three discrete categories based on their actual TPR value. “Poor” penetrators were defined as compounds with a TPR in the 0.00–0.49 range (compounds that had FGT concentrations or exposures <50% that of plasma exposure), “good” penetrators were defined as compounds with a TPR from 0.50 to 1.49 (compounds with FGT exposures ≥50% and <150% that of plasma exposure), and “excellent” penetrators were defined as compounds with a TPR≥1.50 (compounds that had FGT exposures ≥150% that of plasma exposure).
MOE ver.2009.10 (Chemical Computing Group, Montreal, Canada) was used to calculate the octanol/water partitioning coefficient (log P, a measure of lipophilicity or membrane permeability) and molecular weight (MW). Transporter interaction scores (from 0 to 1 based on the likelihood of being a substrate) were calculated for each drug using a collection of QSAR classification models developed previously.15 Seven transporters were included: efflux transporters MDR1, MRP1, MRP2, MRP3, MRP4, BCRP, and the uptake transporter OCT1. Where available, we used experimental data on the substrate status of our compounds for these transporters instead of predicted values.
To perform hierarchical cluster analysis, the Sequential Agglomerative Hierarchical Non-overlapping (SAHN) method implemented in the ISIDA/Cluster program (http://infochim.u-strasbg.fr) was employed. Each compound was represented by one cluster at the start. Then, m compounds were merged iteratively into clusters using their pairwise Euclidean distances stored in a squared m * m symmetric distance matrix. The two closest objects (molecules or clusters) were iteratively merged to form a new cluster and the distance matrix was updated with the newly formed cluster distances. The process was repeated until one cluster remained. Associations between clusters were analyzed using the Wilcoxon rank-sum test, with alpha set at 0.05. Relationships between TPR and descriptors were determined by linear regression. Any significant relationships identified in linear regression were then incorporated into a multivariate analysis using stepwise regression. Statistical analyses were performed using R 2.15.3 [R Core Team (2013) www.R-project.org/] and SigmaPlot version 11.0 (Systat Software, Inc., San Jose, CA; www.sigmaplot.com). We also conducted an a posteriori evaluation of the effectiveness of our cluster analysis by correctly predicting TPR for compounds not used for model development including the nonnucleoside reverse transcriptase inhibitor (NNRTI) rilpivirine and the integrase strand transfer inhibitor dolutegravir.
Random Forest and k Nearest Neighbors (kNN) machine learning algorithms were used to conduct QSAR modeling of FGT. We followed the predictive QSAR modeling workflow,16 which consists of the following three major steps: (1) data preparation/analysis, (2) model building, and (3) model validation/selection. Here we followed a 5-fold external cross-validation procedure: the full set of 58 compounds with known experimental activity was randomly split into five training (80% of the modeling set) and external validation sets (the remaining 20%). Models were built using the training set compounds only, and the external set compounds were never taken into account to build and/or select the models. Pearson's correlation coefficient (R2) and root mean square error (RMSE) were used to assess the prediction performances of developed models. Then, selected models were applied to the external set compounds to predict their experimental properties. This overall procedure was repeated five times to ensure that every compound from the modeling set was present only once in the external test set.
Our initial literature search identified 60 unique chemical compounds, representing 13 therapeutic classes. Since we used two-dimensional representation of chemical structures, two stereoisomer compounds (levofloxacin and ofloxacin) were excluded from analysis, resulting in 58 compounds with FGT data: 20 poor penetrators, 22 good penetrators, and 16 excellent penetrators. Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/aid) contains a list of the entire set of compound and their associated TPR value used for our computational analysis. ARVs are highlighted in Fig. 1, which illustrates the large variability in reported TPR for these compounds, both between and within therapeutic class. Of the remaining non-ARV compounds, 17 had multiple TPRs reported within the same study, but only two had TPRs reported from different sources. Based on the a priori TPR reporting algorithm, the following were included: VT or CT steady-state AUC (n=1)>VT or CT single dose AUC (n=2)>CVF steady-state AUC (n=19)>CVF single dose AUC (n=0)>VT or CT steady-state single time point (n=5)>VT or CT single dose single time point (n=25)>CVF steady-state single time point (n=3)>CVF single dose single time point (n=3).
FIG. 1.
Distribution of reported tissue penetration ratios (TPRs) for antiretroviral compounds (21 of 58 drugs). Adapted from Thompson et al.4 Black dots represent steady-state AUC ratios in tissue or cervicovaginal fluid (CVF); white dots represent single time point ratios in the CVF. Solid black lines represent the range of TPRs reported in the literature. Dashed black lines correspond to the selected ranges for poor (TPR 0.00–0.49), good (TPR 0.50–1.49), and excellent (TPR≥1.5) penetrators, respectively. Histogram inset shows the distribution of TPRs across all compounds. TPRs for all drugs used in analyses are included in Supplementary Table S1. NRTI, nucleoside reverse transcriptase inhibitor; 3TC, lamivudine; FTC, emtricitabine; TFV, tenofovir; ZDV, zidovudine; ddI, didanosine; d4T, stavudine; ABC, abacavir; PI, protease inhibitor; DRV, darunavir; IDV, indinavir; ATV, atazanavir; APV, amprenavir; LPV, lopinavir; RTV, ritonavir; NNRTI, nonnucleoside reverse transcriptase inhibitor; NVP, nevirapine; EFV, efavirenz; ETV, etravirine; RPV, rilpivirine; DLV, delavirdine; INSTI, integrase strand transfer inhibitor; RAL, raltegravir; DTG, dolutegravir; RA, receptor antagonist; MVC, maraviroc.
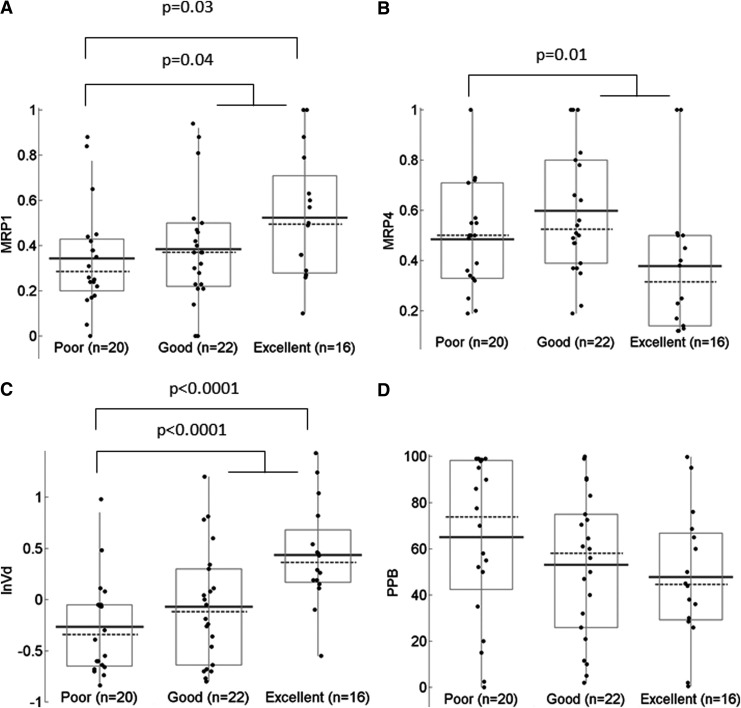
When compounds were stratified according to the penetration class and analyzed for associations with physicochemical and pharmacokinetic parameters, MRP1 and MRP4 substrate scores, and Vd exhibited significant trends (Fig. 2). For MRP1 substrate scores, the difference in median values between “poor” and “excellent” TPR groups, as well as between “poor” and all other compounds, was statistically significant (p=0.03 and 0.04, respectively). For MRP4 substrate scores similar comparisons yielded p-values of 0.06 and 0.01. Among pharmacokinetic parameters, Vd was significant in both comparisons (p<0.001), while %PPB was not significant in either comparison (p=0.12 and 0.22, respectively).
FIG. 2.
Distribution of MRP1 (A) and MRP4 (B) substrate scores (from 0 to 1 with 1 representing a high likelihood of being a substrate), volume of distribution (C), and plasma protein binding values (D) across groups with poor, good, and excellent tissue penetration ratio, shown as boxplots with median (dashed line), mean (solid line), and individual values.
Hierarchical analysis identified seven distinct chemical clusters ranging in size from two to nine compounds. We applied our cluster analysis to the variables identified above in Fig. 3, which shows a detailed distribution for MRP1 and MRP4 scores and volume of distribution values, along with drug classes and structural clusters. Because we previously identified these variables as being significantly associated with TPR, we would expect to see an overall trend among all compounds, with individual clusters in varying degrees of agreement with the overall trend. Figures 3A and B demonstrate a weak but statistically significant relationship between TPR and Vd (R2=0.18; p=0.001) but poor (R2=0.072; p=0.044) correlation between TPR and MRP1 substrate score and no correlation between TPR and MRP4 substrate score (Fig. 3C; R2=0.007; p=0.533). When these variables (in addition to %PPB) were included in our stepwise regression, Vd and %PPB were the only significant variables affecting TPR (R2=0.274; p<0.001), with the MRP1 substrate score dropping below the level of significance (p=0.074). These trends are consistent with those observed in some, but not all individual clusters. Table 1 showcases specific trends within two classes of drugs that differ in their agreement with the results observed in Fig. 3. Among the protease inhibitors (PIs), which cluster together and have trends similar to the overall observation for all variables, indinavir has a high TPR, while the other three PIs penetrate poorly. The volume of distribution and %PPB of indinavir differ from the remaining compounds. Among the NRTIs, which do not agree with the overall trends, abacavir, stavudine, and didanosine have the lowest TPRs while having similar PPB%, Vd, and MRP4 substrate scores but lower MRP1 substrate scores compared to other NRTIs.
FIG. 3.
Scatterplots for the TPR vs. volume of distribution (A) MRP1 substrate scores (B), shown with drug classes (marker shapes) and structural clusters (marker colors); horizontal gray lines denote thresholds for TPR class.
Table 1.
Tissue Penetration Ratio and Property Profile for Two Clusters of Several Protease Inhibitors (n=4) and Nucleoside Reverse Transcriptase Inhibitors (n=2)
| Drug | Class | log TPR | log P | Vd, liter/kg | %PPB | MDR1 | BCRP | MRP1 | MRP4 |
|---|---|---|---|---|---|---|---|---|---|
| Indinavir | PI | 0.66 | 2.8 | 2.7 | 60 | 0.9 | 0.8 | 1.0 | 0.4 |
| Darunavir | PI | 0.18 | 2.4 | 1.8 | 95 | 0.8 | 0.8 | 0.6 | 0.3 |
| Amprenavir | PI | −0.3 | −2.9 | 6.5 | 90 | 1.0 | 0.8 | 0.0 | 0.4 |
| Ritonavir | PI | −0.59 | 5.0 | 0.4 | 99 | 1.0 | 0.8 | 0.7 | 0.5 |
| Atazanavir | PI | −0.74 | 4.7 | 1.3 | 86 | 0.8 | 0.8 | 0.9 | 0.5 |
| Lopinavir | PI | −1.10 | 5.2 | 0.9 | 99 | 0.9 | 0.8 | 0.8 | 0.3 |
| Tenofovir | NRTI | 0.76 | −1.6 | 0.8 | 1 | 0.4 | 0.0 | 0.6 | 1.0 |
| Emtricitabine | NRTI | 0.6 | −0.5 | 1.4 | 2 | 0.3 | 0.1 | 0.3 | 0.1 |
| Lamivudine | NRTI | 0.6 | −0.8 | 1.3 | 36 | 0.3 | 1.0 | 0.3 | 0.1 |
| Zidovudine | NRTI | 0.4 | −1.9 | 1.5 | 38 | 0.3 | 1.0 | 0.4 | 1.0 |
| Didanosine | NRTI | −0.68 | 0.1 | 0.9 | 2 | 0.4 | 0.1 | 0.2 | 0.5 |
| Abacavir | NRTI | −1.1 | 0.4 | 0.8 | 50 | 1.0 | 1.0 | 0.2 | 1.0 |
| Stavudine | NRTI | −1.3 | −1.0 | 0.5 | 0 | 0.2 | 0.1 | 0.2 | 0.2 |
log P was calculated using MOE ver. 2009.10 (Chemical Computing Group, Montreal, Canada); experimental volume of distribution (Vd) and protein binding percentage (%PPB) values were obtained from package inserts. Substrate scores for transporters were calculated based on Sedykh et al.15
PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor.
In our a posteriori analysis, we used known values for each of the significant variables identified in the linear regression model (MRP1 and MRP4 substrate scores and Vd) for rilpivirine and dolutegravir to generate individual TPR estimates based on the regression equations shown in Fig. 3. The TPR for rilpivirine was overpredicted by only 4% using the MRP4 substrate score as the predictive variable (predicted TPR 0.71, actual TPR 0.68), but underpredicted by 25% using the MRP1 substrate score (predicted TPR 0.51) and overpredicted by 50% using Vd (predicted TPR 1.02). The TPR for dolutegravir was systematically overpredicted by 457% using the MRP4 substrate score (predicted TPR 0.32, actual TPR 0.07), by 514% using the MRP1 substrate score (predicted TPR 0.36), and by 985% using Vd (predicted TPR 0.69).
The use of kNN and RF with Dragon17 and SiRMS18 descriptors failed to produce robust QSAR models for both continuous and classification scales. The best continuous model predicted two of the five test sets with R2 values >0.40. When combined, they contained 24 compounds; predictions for these compounds were only modestly successful (R2=0.47; p<0.0001). Performance of classification models did not differ from random models.
ARV FGT penetration is a critical determinant of PrEP success. The lack of reliable approaches to predict the penetration of ARVs into the FGT represents a significant obstacle to develop the next generation of PrEP agents. The time and resources required for conducting clinical studies to determine this characteristic could be drastically reduced if validated predictive models were available. Cheminformatics approaches enable fast, inexpensive, and streamlined analyses of drug characteristics, including tissue distribution, and can help in identifying those compounds capable of penetrating well into the FGT. In this study, we attempted to accurately compute and analyze chemical and biological characteristics that promote FGT penetration using both qualitative and quantitative assessments.
We were able to compile a small dataset of 58 compounds spanning 13 different drug classes. Our attempt to build an externally predictive QSAR model was unsuccessful due to numerous factors, including the small size and high molecular diversity of the dataset. This was unfortunate, as a comprehensive QSAR model that makes predictions based on the influence of many variables in combination most closely represents physiologic activity. To overcome this setback, we conducted a more stepwise approach to identifying important variables, which included chemical cluster analysis and stepwise regression. Our cluster analysis identified the physiologic characteristics of certain drugs that are significantly associated with TPR. Indeed, the data generated from our cluster analysis showed that Vd is predictive of FGT penetration (Fig. 3A). This finding should be self-evident, as Vd depends on the physiologic properties of the body and the physiochemical properties of the drug. For example, a Vd of <0.2 liter/kg typically indicates that a drug primarily resides in blood (e.g., DTG; Vd=0.25 liter/kg, TPR=0.07), whereas higher values indicate distribution in tissues and fat (e.g., IND; Vd=2.6 liters/kg, TPR=4.5).
Among the transporter interactions evaluated in the cluster analysis (MDR1, BCRP, MRP1-4, OCT1), the most significant transporters were found to be MRP1 and MRP4 (Fig. 2A), though neither of these was found to be significant after adjustment for Vd and %PPB. MRP4 and MRP1 are efflux transporters that affect many classes of drugs, including ARVs. Although little is known about the actual activity of these transporters in the FGT, very recent studies have confirmed the presence of MRP1 and MRP4 in these tissues.12,13 The localization of these transporters in tissues may account for the disparate TPRs observed. For example, it may be that substrates of MRP4 would be less likely to accumulate in the FGT if the transporter was located on the luminal membrane. Conversely, MRP1 substrates would tend to accumulate if it was located basolaterally within epithelial tissue. The observation by Nicol that MRP4 and MRP1 are both present in the epithelial layer of vaginal tissue supports this hypothesis.13 Additional evidence is provided by observed trends among NRTIs (Table 1). For example, both tenofovir and abacavir are MRP4 substrates; however, tenofovir has a much higher TPR than abacavir and, unlike abacavir, is a probable substrate of MRP1. It is important to note that our substrate probability descriptor is unrelated to the affinity or avidity of the compound for a particular transporter. For example, tenofovir and abacavir may have different avidities for MRP4, but both have been shown to be MRP4 substrates.
The recognition of Vd and transporter substrate probability as predictive variables for TPR was confirmed in our a posteriori analysis of rilpivirine and dolutegravir. Though dolutegravir's TPR was overpredicted by nearly 10-fold, the predictions for rilpivirine were much closer to the actual value, with one prediction being within 4% of the actual TPR. Importantly, the use of the MRP4 substrate score as the predictive variable resulted in a correct assignment of TPR class (i.e., poor, good, or excellent) for both drugs. Prediction using the MRP1 substrate score also correctly assigned rilpivirine as a good penetrator. Although these cheminformatics approaches did not always identify specific TPR values, the correct estimation of penetrator class still represents an improvement on the current method of clinical testing.
There are several limitations to our analysis, mainly arising from our data set. The large amount of variability in reported TPRs was the primary obstacle to achieving high dataset quality. This was not surprising, as generating pharmacokinetic data for the FGT includes complex sample collection and bioanalytical techniques. Selecting the most accurate TPR for each compound was difficult in cases in which multiple TPR values were reported (which occurred for 40 out of the 58 retrieved compounds). This was particularly problematic for compounds in which disparate TPR values spanned penetrator class assignments (occurring for 10 compounds). In most of these cases, TPR variability was limited to adjacent classes [i.e., poor and good (ABC), good and excellent (TFV)] but larger variation was observed [i.e., poor and excellent (ddI)], increasing the importance of choosing the “correct” value. Although an a priori selection algorithm was used to correct for this, it may be that the TPRs used here did not represent true penetrative ability. For example, dose-dependent changes in transporter activity (e.g., saturation at high doses not used in these studies) could not be accounted for despite our algorithm. Furthermore, it is intuitive to think that a larger data set would result in a more predictive model. Though a larger amount of data may improve the predictive capabilities of our model, successful QSAR models have been developed with data sets of similar size.19
We were further limited in our ability to generate a high-quality predictive model due to the necessity of including TPR values for both CVF and tissue; conducting separate analyses for each compartment would greatly limit our ability to find any significant variables due to small sample size. Some drugs may achieve vastly different concentrations between these two compartments as a result of the physicochemical properties of the drug (e.g., lipophilicity, size) or physiologic factors (e.g., transporter localization). In other words, the factors affecting penetration into the CVF may not be the same as those governing penetration into tissue, and vice versa. Thus, we may have overlooked or misidentified important variables simply because we used a TPR from one compartment versus another. This may explain some of the apparent disparity in our results. For example, it was noted above that transporter localization may explain why TPR associates differentially with MRP4 and MRP1 despite these both being efflux transporters. However, it may be that one of the transporters contributes to CVF penetration and the other contributes to tissue penetration; we would be unable to discern this with our current model. Importantly, data generated in our laboratory have shown that regardless of whether a compound is a good or poor penetrator, it would be classified similarly whether using CVF or tissue measurements.20
Overall this study represents the first attempt at building a predictive model for FGT penetration using cheminformatics approaches applied to 58 compounds. Our best performing QSAR model did not achieve a high predictive ability; however, our cluster analysis identified high MRP1 and low MRP4 substrate probability and high Vd as significant predictors of FGT penetration. Additional compounds are needed to enrich the modeling set and allow for further analysis and an increase in predictability. Once validated, a truly predictive model of FGT penetration could be utilized to screen drug candidates at the early stages of development and isolate those compounds ideally suited for PrEP or active viral reservoir targeting.
Supplementary Material
Acknowledgments
The authors would like to thank K.T. Vaughan, PhD, for her assistance with our literature search, and Kathryn Aldrich for her assistance in identifying additional compounds for our database. C.T., M.N., and A.K. were supported by NIAID U01-A1095031 and the UNC Center for AIDS Research P30-A1050410, and C.T. by the American Foundation for Pharmaceutical Education/Rho Chi 1st Year Graduate Student Fellowship. A.S., E.M., D.F., and A.T. gratefully acknowledge support from NIH Grants R01-GM66940 and R01-GM096967.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Grant RM, Lama JR, Anderson PL, and McMahan V: Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med 2011;363(27):2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karim QA, Karim SSA, Frohlich JA, et al. : Effectiveness and safety of tenofovir gel, and antiretroviral microbicide, for the prevention of HIV infection in women. Science 2011;329(5996):1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Damme L, Corneli A, Ahmed K, et al. : Preexposure prophylaxis for HIV infection among African women. N Engl J Med 2012;367(5):411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson CG, Cohen MS, and Kashuba ADM: Antiretroviral pharmacology in mucosal tissues. J Acquir Immune Defic Syndr 2013;63(Suppl 2):S240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hladik F. and Hope TJ: HIV infection of the genital mucosa in women. Curr HIV/AIDS Rep 2009;6(1):20–28 [DOI] [PubMed] [Google Scholar]
- 6.Tebit DM, Ndembi N, Weinberg A, and Quiñones-Mateu ME: Mucosal transmission of human immunodeficiency virus. Curr HIV Res 2012;10(1):3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saksena NK, Wang B, Zhou L, Soedjono M, Ho YS, and Conceicao V: HIV reservoirs in vivo and new strategies for possible eradication of HIV from the reservoir sites. HIV AIDS (Auckl) 2010;2:103–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicol MR. and Kashuba DM: Pharmacologic opportunities for HIV prevention. Clin Pharmacol Ther 2010;88(5):598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munteanu C, Fernandez-Blanco E, Seoane JA, and Izquierdo-Novo P: Drug discovery and design for complex diseases through QSAR computational methods. Curr Pharm Des 2010;16(24):2640–2655 [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Fourches D, Sedykh A, et al. : Discovery of novel antimalarial compounds enabled by QSAR-based virtual screening. J Chem Inf Model 2013;53(2):475–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Zhu H, Oprea TI, Golbraikh A, and Tropsha A: QSAR modeling of the blood-brain barrier permeability for diverse organic compounds. Pharm Res 2008;25(8):1902–1914 [DOI] [PubMed] [Google Scholar]
- 12.Hu M, Cost M, and Poloyac S: Expression of transporters and metabolizing enzymes in the female lower genital tract: Implications for microbicide research. AIDS Res Hum Retroviruses 2013;29(11):1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicol MR, Fedoriw Y, Mathews M, et al. :. Expression of six drug transporters in vaginal, cervical, and colorectal tissues: Implications for drug disposition in HIV prevention. J Clin Pharmacol 2013. [Epub ahead of print]; DOI: 10.1002/jcph.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fourches D, Muratov E, and Tropsha A: Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 2010;50(7):1189–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sedykh A, Fourches D, Duan J, et al. : Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res 2013;30:996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tropsha A: Best practices for QSAR model development, validation, and exploitation. Mol Inform 2010;29:476–488 [DOI] [PubMed] [Google Scholar]
- 17.Todeschini R. and Consonni V: Handbook of Molecular Descriptors. Wiley-VCH, Weinheim, Germany, 2008 [Google Scholar]
- 18.Kuz'min VE, Artemenko G, and Muratov EN: Hierarchical QSAR technology based on the Simplex representation of molecular structure. J Comput Aided Mol Des 2008;22(6–7):403–421 [DOI] [PubMed] [Google Scholar]
- 19.Muratov EN, Artemenko AG, Varlamova E, and Polishchuk PG: Per aspera ad astra: Application of Simplex QSAR approach in antiviral research. Future Med Chem 2010;2(7):1205–1226 [DOI] [PubMed] [Google Scholar]
- 20.Patterson KB, Prince HA, Kraft E, et al. : Penetration of tenofovir and emtricitabine in mucosal tissues: Implications for prevention of HIV-1 transmission. Sci Transl Med 2012;3:112re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.