Abstract
INTRODUCTION
Little is known about the molecular epidemiology of deafness in sub-Saharan Africa (SSA). Even in Nigeria, the most populous African nation, no genetic studies of deafness have been conducted. This pioneering work aims at investigating the frequencies of gene mutations relatively common in other parts of the world (i.e. those in GJB2, GJB6, and mitochondrial DNA) among subjects from Nigeria with hearing loss (HL) with no evidence of acquired pathology or syndromic findings. In addition, we review the literature on the genetics of deafness in SSA.
METHOD
We evaluated 81 unrelated deaf probands from the Yoruba tribe residing in Ibadan, a suburban city in Nigeria, for the etiology of their deafness. Subjects underwent genetic testing if their history was negative for an environmental cause and physical examination did not find evidence of a syndrome. Both exons of GJB2 and mitochondrial DNA flanking the 1555A>G mutation were PCR-amplified followed by Sanger sequencing. GJB6 deletions were screened via quantitative PCR.
RESULT
We identified 44 probands who had nonsyndromic deafness with no environmental cause. The age at study time ranged between 8 months and 45 years (mean=24 years) and age at onset was congenital or prelingual (<age 2 years) in 37 (84%) probands and postlingual in 7 (16%) probands. Among these, 35 probands were the only affected members of their families (simplex cases), while there were at least two affected family members in 9 cases (multiplex). Molecular analyses did not show a pathogenic variant in any one of the 44 probands studied.
CONCLUSION
GJB2, GJB6 and mitochondrial DNA 1555A>G mutations were not found among this initial cohort of the deaf in Nigeria. This makes imperative the search for other genes in the etiology of HL in this population.
Keywords: Non-syndromic deafness, molecular diagnosis, GJB2, GJB6 deletion, mtDNA mutation, Nigeria
Introduction
Hearing loss (HL) is the most common sensory disorder. One out of every 500 newborns has bilateral permanent sensorineural HL, with the prevalence increasing to 2.7 per 1000 by the age of 5 and 3.5 per 1000 during adolescence [1]. Epidemiological surveys of the deaf have consistently shown that currently about 50% of childhood deafness in developed countries can be attributed to genetic causes [1]. In fact, the causative genomic variants have been documented for most types of deafness in developed countries and efforts are now focused on determining the phenotype-genotype correlation for many of the known genes/variants [1]. In contrast, the literature from sub-Saharan Africa (SSA) on the genetic etiology of deafness is sparse even though Sub-Saharan Africa has a high (1.8%) prevalence for hearing loss affecting communication in children, second only to the south Asia region (2.3%) [2]. Earlier surveys of deafness in Gambia [3] and Nigeria [4] revealed meningitis and chronic middle ear infections as the major diseases causing deafness, while familial factors accounted for less than 10% of the childhood deafness [3]. Consequently, it was recommended that a primarily preventive approach was the most rational way of helping the deaf in these countries [2–5]. However, it is to be noted that genetic services, in particular hearing genetics research, are still at elementary stages in most parts of SSA [6–8]. Hence, the lack of genetic facilities in the investigation of HL in these earlier studies would have resulted in failure to identify genetic factors as a major contributor to the etiology. In addition, these earlier works reported a high proportion (32 and 54.4%) of deafness whose ‘cause is unknown’ leading to the conclusion that these cases of deafness in the unknown category may well be of genetic origin [2–5]. Thus, there is a need to explore the possibility of a genetic etiology for the unknown cause category. In developed countries, with the availability of molecular diagnosis, the cause of childhood deafness is unknown in less than 50% [9, 10]. Furthermore, improvement in health care services, especially the vaccination programme in SSA, has resulted in the control of many infectious diseases, including measles and mumps [11, 12]. By inference, this reduction in the prevalence of these infectious diseases will reduce their contribution as a cause of deafness, thus increasing the relative contribution of genetics. A few molecular analyses in SSA have identified a few genetic mutations as possible etiological factors for deafness in the sub-region [13–23]. It is noted that most of the studies of the molecular genetics of deafness in SSA have been driven by research in the specialist/tertiary hospitals. Notably, there has not been any study on the genetic epidemiology of deafness in Nigeria, the most populous African nation. Hence this pioneering work aims at investigating the frequencies of gene mutations relatively common in other parts of the world (i.e. those in GJB2, GJB6, and mitochondrial DNA) in this population. In addition, this work will review the literature on the genetics of deafness in SSA.
Material and Methodology
Samples
This study has been approved by the Ethics Committee of University of Ibadan (Nigeria) and IRB at the University of Miami (USA). Signed informed consents were collected from all participants or parents. We evaluated 81 unrelated deaf probands from the Yoruba tribe residing in Ibadan, a sub-urban city in Nigeria, for the etiology of their deafness. In order to have deaf subjects from diverse groups, the study participants were selected from various vocational and professional groups, schools and religious groups. The criteria for recruitment was (i) lack of evidence for an environmental cause of HL such as meningitis, measles, mumps or cerebral malaria, as reported by the pupils’ parents or guardians, (ii) lack of evidence for a syndrome obtained by physical examination, and (iii) audiometric findings compatible with a severe to profound sensorineural HL. The study included 44 deaf subjects whose histories were negative for an environmental cause and physical examination did not show syndromic findings. The remaining 37 subjects were excluded because their deafness was either syndromic or secondary to meningitis, viral infection, or associated with other neurological abnormalities such as cerebral palsy. The probands or their representatives were asked for permission to communicate the results of the genetic testing. DNA was extracted using standard procedures with a Qiagen extraction kit at the Institute of Medical Research and Training in University of Ibadan and the DNA samples were subsequently transferred to the Hussman Institute for Human Genomics at the University of Miami for laboratory studies.
Sanger Sequencing and CNV detection
Both exons of GJB2 and mitochondrial DNA flanking the 1555A>G mutation were PCR-amplified followed by Sanger sequencing. [24]. Previously reported 4 large genomic deletions involving GJB6 [25–27] were screened via quantitative PCR. CNV analysis for the genomic region of the GJB6 gene was performed with a TaqMan predesigned probe (Hs03843749, Chr13:20961484 on NCBI build 37) by using a previously described protocol [28]. For the Sanger sequencing, PCR reactions included 25 μg of genomic DNA with Taq DNA polymerase (Roche). Corresponding DNA fragments were amplified using a touchdown protocol. PCR products were visualized on agarose gels cleaned over Sephadex columns or with NucleoFast 96 PCR plates (Clontech) in accordance with the manufacturer’s protocols. Sequence analysis was performed with the ABI PRISM Big Dye Terminator Cycle Sequencing V3.1 Ready Reaction Kit and the ABI PRISM 3730 DNA Analyzer (Applied Biosystems). Sequence traces were analyzed using the Sequencher 4.7 program (Gene Codes Corporation).
Results
Among the 44 probands, there were 32 males and 12 females with age ranging between 8 months and 45 years (mean=24 years). Age at onset was congenital or prelingual (<age 2 years) in 37 (84%) probands and postlingual in 7(16%) probands. Among these, 35 probands were the only affected members of their families (simplex cases), while there were at least two affected family members in 9 cases (multiplex) with likely autosomal recessive and X-linked patterns of inheritance in 8 and 1 families, respectively.
Molecular analyses did not identify a pathogenic or polymorphic variant in GJB2 gene studied in the 44 probands.
Discussion
Our report of the genetics of deafness in Nigeria shows that the DFNB1 locus (containing GJB2 and GJB6 genes) and the mitochondrial 1555A>G mutation are not major genetic causes of deafness among Nigerians. This is in a sharp contrast to the results of many other populations of the world but, not surprisingly, similar to those studies conducted in other sub-Saharan African populations. [29, 30]
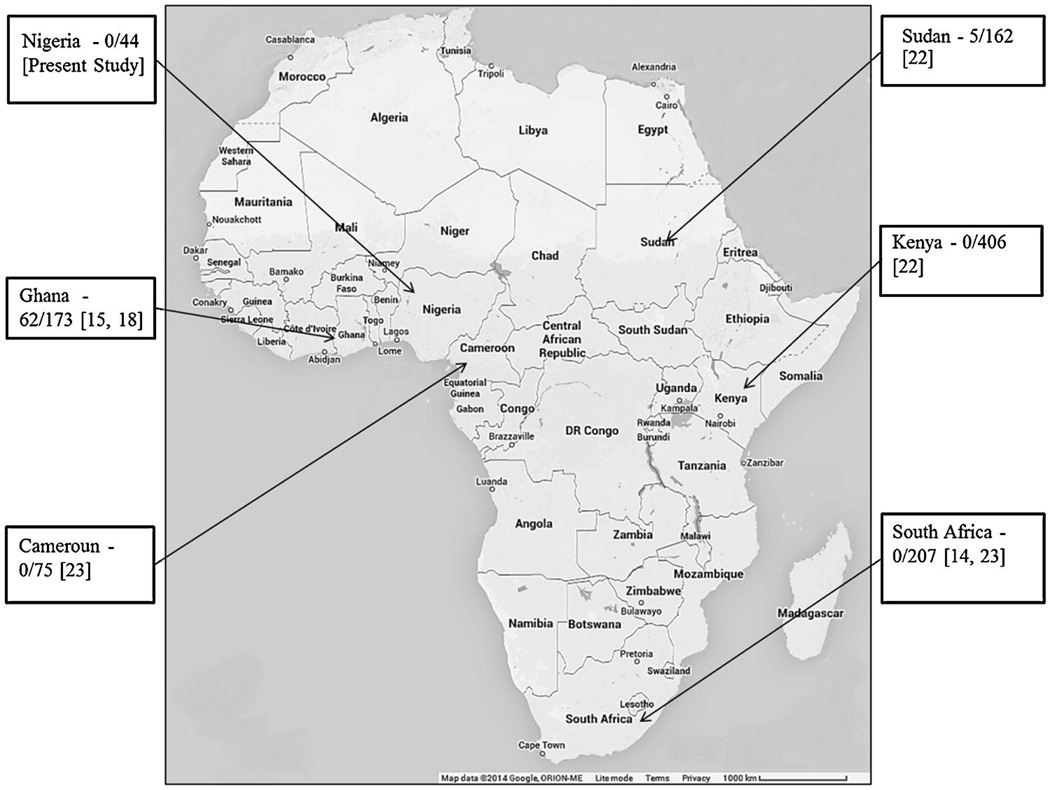
Previous studies on the genetics of deafness in SSA have been mainly from South Africa and Cameroun, with fewer reports from Sudan, Zaire, and Ghana [13–23]. Table 1 shows the detected GJB2 variants reported from SSA and Figure 1 shows the contribution of pathogenic mutations of GJB2 to non-syndromic deafness among the populations in SSA. In contrast to the most of the world, GJB2 mutations are not common in the region [14, 18, 22, 23]. An exception to this has been observed in Ghana where the p.R143W mutation of GJB2 was found as the cause for hearing impairment in all 11 families from a village with an extraordinarily high prevalence of congenital HL, suggesting a founder effect in that population [15].
Table 1.
Pathogenic variants of GJB2 reported among deaf individuals in SSA
| Genotype (cDNA) | Protein change | Number Affecteds |
Sample size |
Ethnic/Country | Reference |
|---|---|---|---|---|---|
| c.427T>C/c.427T>C; | p.R143W/p.R143W | 11 | 11 | Adamarobe/Ghana | Brobby et al [15]* |
| c.427T>C/ c.427T>C | p.R143W/p.R143W | 51 | 365 | Ashanti, Central, Eastern, Greater Accra, Upper East, Upper West, Volta, | Hamelmann et al [18] |
| c.427T>C/c.35insG | p.R143W/p.V13fs | 1 | |||
| c.533T>C/c.533T>C | p.V178A/p.V178A | 2 | |||
| c.236T>C/wt | p.R184Q/wt (Dominant) | 1 | |||
| c.427T>C/p.641T>C | p.R143W/p.L214P | 1 | |||
| c.35delG/c.35delG | p.G12fs | 5 | 162 | Sudanese/Sudan | Gasmelseed et al [22] |
| - | - | 0 | 406 | Kenya | |
| - | - | o | 182 | Pedi, Venda and Tsonga groups in Limpopo, S/A | Kabahuma et al [23] |
[15] total number of families/subjects studied is unknown
Figure 1.
Contribution of GJB2 mutations to non-syndromic deafness among the populations in SSA
Among the very limited data on other genes, Trotta et al [18] documented a single MTRNR1 variant that was suspected to be pathogenic in a molecular screen of 70 deaf children and 67 unaffected controls in Maroua tribe in Cameroon. In addition, linkage analysis revealed a homozypous transition at a splice donor site (c.19+5G>A) of the TMC1 gene among two deaf Sudanese, confirming the role of TMC1 in recessive non-syndromic deafness. [16]
This study did not screen SLC26A4, another common gene for non-syndromic (DFNB4) deafness. In addition, only one probe was designed for detection of GJB6 deletion, hence, GJB6 deletions in regions not covered by this probe could be missed by this study. However, mutations involving these and other genes will be further investigated in future studies.
It is not surprising that the documentation of molecular genetics of deafness in SSA is sparse; indeed the situation is intertwined with the relative low knowledge of the epidemiology of childhood deafness. The prevalence of genetic causes of sensorineural HL reported in Cameroun, Nigeria and South Africa were 14.8%[32], 15%[33] and 36%[34], respectively as shown in Table 2. In those studies, genetic causes were determined based on history of exclusion of putative acquired pathologies and family history. A more qualitative search for the genetics of HL could be facilitated by the inclusion of molecular screening in cases whose HL is diagnosed by newborn hearing screening, which is yet to commence in most parts of SSA [35–38]. However, this becomes imperative for reliable data on genetic epidemiology of deafness in SSA and it is expected to have significant implications for research in and development of protocol for management of genetic deafness in the subregion [39–41].
Table 2.
Genetic epidemiology of deafness reported in sub-Saharan Africa
| Variable | Wonkam et al 2013 (Cameroun) |
Lasisi et al 2005 (Nigeria) |
Sellars et al 1998 (South Africa) |
|---|---|---|---|
| Location | School/Clinic | Clinic | School |
| Proportion of genetic cases | 14.8% | 15% | 36% |
| Proportion of autosomal recessive families | 87.7% | ND | 49% |
| Consanguinity | 15.2% | ND | ND |
| Proportion of severe to profound HL | 100% | 88% | ND |
Footnote: ND- Not documented
In conclusion, we report that GJB2, GJB6 and mitochondrial DNA 1555A>G mutations are not common causes of deafness in Nigeria. This further corroborates earlier findings on the genetic mutations of sensorineural deafness in other parts of SSA. This suggests that this region may harbour unique or infrequent genetic causes (compared to the rest of the world) of hearing loss and provides the impetus for conducting further genetic studies in the SSA.
Acknowledgement
This study was supported by the National Institutes of Health - National Institute on Deafness and Other Communication Disorders grant R01DC009645 to MT and the University of Ibadan Tertiary Education Trust Fund to AOL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Alford RL, Arnos KS, Fox M, et al. American College of Medical Genetics and Genomics Guideline for the clinical evaluation and aetiologic diagnosis of hearing loss. Genet Med. 2014:1–9. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
- 2.WHO global estimates on prevalence of hearing loss, Mortality and Burden of Diseases and Prevention of Blindness and Deafness. WHO; 2012. [Google Scholar]
- 3.Holborow C, Martinson F, Anger N. A study of deafness in West Africa. Int J Pediatr Otorhinolaryngol. 1982;4(2):107–132. doi: 10.1016/0165-5876(82)90087-8. [DOI] [PubMed] [Google Scholar]
- 4.McPherson B, Holborow CA. A study of deafness in West Africa: the Gambian Hearing Health Project. Int J Pediatr Otorhinolaryngol. 1985;10(2):115–135. doi: 10.1016/s0165-5876(85)80024-0. [DOI] [PubMed] [Google Scholar]
- 5.Seely DR1, Gloyd SS, Wright AD, Norton SJ. Hearing loss prevalence and risk factors among Sierra Leonean children. Arch Otolaryngol Head Neck Surg. 1995;121(8):853–858. doi: 10.1001/archotol.1995.01890080023004. [DOI] [PubMed] [Google Scholar]
- 6.Wonkam A, Tekendo CN, Sama DJ, et al. Initiation of a medical genetics service in sub-Saharan Africa: experience of prenatal diagnosis in Cameroon. Eur. J. Med. Genet. 2011;54:e399ee404. doi: 10.1016/j.ejmg.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Oloyede OA. Down syndrome screening in Nigeria. Int J Gynecol Obstet. 2008;100(1):88–92. doi: 10.1016/j.ijgo.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 8.Fraser GR. Profound childhood deafness. J. Med. Genet. 1964;1:118e129. doi: 10.1136/jmg.1.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker MJ, Fortnum H, Young ID, Davis AC. Variations in genetic assessment and recurrence risks quoted for childhood deafness: a survey of clinical geneticists. J. Med. Genet. 1999;36:125–130. [PMC free article] [PubMed] [Google Scholar]
- 10.Hilgert N, Smith RJH, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madhi SA, Bamford L, Ngcobo N. Effectiveness of pneumococcal conjugate vaccine and rotavirus vaccine introduction into the South African public immunisation programme. S Afr Med J. 2014;104(3Suppl1):228–234. doi: 10.7196/samj.7597. [DOI] [PubMed] [Google Scholar]
- 12.CDC. Progress in global measles control and mortality reduction, 2000—2007. MMWR. 2008;57:1303–1306. [PubMed] [Google Scholar]
- 13.Matthijs G, Claes S, Longo-Mbenza B, Cassiman JJ. Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNAgene in a large Zairean pedigree. Eur J Hum Genet. 1996;4(1):46–51. doi: 10.1159/000472169. [DOI] [PubMed] [Google Scholar]
- 14.Bosch J, Lebeko K, Nziale JJ, Dandara C, Makubalo N, Wonkam A. In Search of Genetic Markers for Nonsyndromic Deafness in Africa: A Study in Cameroonians and Black South Africans with the GJB6 and GJA1 Candidate Genes. OMICS. 2014;18(7):481–485. doi: 10.1089/omi.2013.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N Engl J Med. 1998;338:548–550. doi: 10.1056/NEJM199802193380813. [DOI] [PubMed] [Google Scholar]
- 16.Meyer CG, Gasmelseed NM, Mergani A, Magzoub MM, Muntau B, Thye T, Horstmann RD. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25(1):100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- 17.Trotta L, Iacona E, Primignani P, Castorina P, Radaelli C, Del Bo L, Coviello D, Ambrosetti U. GJB2 and MTRNR1 contributions in children with hearing impairment from Northern Cameroon. Int J Audiol. 201I;50(2):133–138. doi: 10.3109/14992027.2010.537377. [DOI] [PubMed] [Google Scholar]
- 18.Hamelmann C, Amedofu GK, Albrecht K, Muntau B, Gelhaus A, Brobby GW, Horstmann RD. Pattern of connexin 26 (GJB2) mutations causing sensorineural hearing impairment in Ghana. Hum. Mutat. 2001;18:84–85. doi: 10.1002/humu.1156. [DOI] [PubMed] [Google Scholar]
- 19.Wonkam A, Jacques J, Noubiap N, Bosch J, Dandara C, Toure CG. Heterozygous p.Asp50Asn mutation in the GJB2 gene in two Cameroonian patients with keratitis-ichthyosis-deafness (KID) syndrome. BMC Med Genet. 2013;14:81. doi: 10.1186/1471-2350-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Human H, Lombard D, de Jong G, Bardien S. A South African family with the mitochondrial A1555G mutation on haplogroup L0d. Biochem Biophys Res Commun. 2009;382(2):390–394. doi: 10.1016/j.bbrc.2009.03.032. [DOI] [PubMed] [Google Scholar]
- 21.Clarke JC, Honey EM, Bekker E, Snyman LC, Raymond RM, Jr, Lord C, Brophy PD. A novel nonsense mutation in the EYA1 gene associated with branchio-otorenal/ branchiootic syndrome in an Afrikaner kindred. Clin Genet. 2006;70(1):63–67. doi: 10.1111/j.1399-0004.2006.00642.x. [DOI] [PubMed] [Google Scholar]
- 22.Gasmelseed NM, Schmidt M, Magzoub MM, Macharia M, Elmustafa OM, Ototo B, Winkler E, Ruge G, Horstmann RD, Meyer CG. Low frequency of deafness associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum Mutat. 2004;23(2):206–207. doi: 10.1002/humu.9216. [DOI] [PubMed] [Google Scholar]
- 23.Kabahuma RI, Ouyang X, Du LL, Yan D, Hutchin T, Ramsay M, Penn C, Liu XZ. Absence of GJB2 gene mutations, the GJB6 deletion (GJB6-D13S1830) and four common mitochondrial mutations in nonsyndromic genetic hearing loss in a South African population. Int J Pediatr Otorhinolaryngol. 2011;75(5):611–617. doi: 10.1016/j.ijporl.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz-Horta O, Duman D, Foster J, II, Sırmacı A, Gonzalez M, et al. Whole-Exome Sequencing Efficiently Detects Rare Mutations in Autosomal Recessive Nonsyndromic Hearing Loss. PLoS ONE. 2012;7(11):e50628. doi: 10.1371/journal.pone.0050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lerer I, Sagi M, Ben-Neriah Z, Wang T, Levi H, Abeliovich D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum Mutat. 2001;18(5):460. doi: 10.1002/humu.1222. [DOI] [PubMed] [Google Scholar]
- 26.Pallares-Ruiz N, Blanchet P, Mondain M, Claustres M, Roux AF. A large deletion including most of GJB6 in recessive non syndromic deafness: a digenic effect? Eur J Hum Genet. 2002;10(1):72–76. doi: 10.1038/sj.ejhg.5200762. [DOI] [PubMed] [Google Scholar]
- 27.del Castillo FJ, Rodríguez-Ballesteros M, Alvarez A, Hutchin T, Leonardi E, de Oliveira CA, Azaiez H, Brownstein Z, Avenarius MR, Marlin S, Pandya A, Shahin H, Siemering KR, Weil D, Wuyts W, Aguirre LA, Martín Y, Moreno-Pelayo MA, Villamar M, Avraham KB, Dahl HH, Kanaan M, Nance WE, Petit C, Smith RJ, Van Camp G, Sartorato EL, Murgia A, Moreno F, del Castillo I. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J Med Genet. 2005;42(7):588–594. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bademci G, Edwards TL, Torres AL, Scott WK, Züchner S, Martin ER, Vance JM, Wang L. A rare novel deletion of the tyrosine hydroxylase gene in Parkinson disease. Hum Mutat. 2010;31(10):E1767–E1771. doi: 10.1002/humu.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci. 2013;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tekin M, Arnos KS, Pandya A. Advances in hereditary deafness. Lancet. 2001;358:1082–1090. doi: 10.1016/S0140-6736(01)06186-4. [DOI] [PubMed] [Google Scholar]
- 31.Hilgert N, Smith RJH, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wonkam A, Noubiap JJ, Djomou F, Fieggen K, Njock R, Toure GB. Aetiology of childhood hearing loss in Cameroon (sub-Saharan Africa) Eur J Med Genet. 2013;56(1):20–25. doi: 10.1016/j.ejmg.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Lasisi OA, Ayodele JK, Ijaduola GTA. Challenges in the management of childhood sensorineural hearing loss in Subsaharan Africa, Nigeria. Int J Paed Otorhinolaryngol. 2005;4:625–629. doi: 10.1016/j.ijporl.2005.08.009. 2005. [DOI] [PubMed] [Google Scholar]
- 34.Sellars S, Groeneveldt L, Beighton P. Aetiology of deafness in white children in the Cape. S Afr Med J. 1976;50(31):1193–1197. [PubMed] [Google Scholar]
- 35.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 36.Denoyelle F, Marlin S, Weil D, Moatti L, Chauvin P, Garabedian EN, et al. Clinical features of the prevalent form of childhood deafness DFNB1, due to a connexin-26 gene defect: Implications for genetic counseling. Lancet. 1999;353:1298–1303. doi: 10.1016/S0140-6736(98)11071-1. [DOI] [PubMed] [Google Scholar]
- 37.Lasisi OA, Onakoya PA, Lasisi TJ, Akinola MD, Tongo O. Neonatal hearing screening in a rural/sub-urban community in nigeria, sub-saharan Africa - A Preliminary Report. Int. J. Paed. Otorhinolaryngol. doi: 10.1016/j.ijporl.2014.06.003. S0165-5876(14)00331-0. [DOI] [PubMed] [Google Scholar]
- 38.Suleiman AO, Suleiman BM, Abdulmajid UF, Suleiman MR, Mustapha AY, Afolabi OA, Yakubu LH, Nathal C, Mohammed GM, Lasisi AO. Paediatric cochlear implantation in north-western Nigeria - Case report and review of our challenges. Int. J. Paed Otorhinolaryngol. 2014;78(2):363–365. doi: 10.1016/j.ijporl.2013.10.061. [DOI] [PubMed] [Google Scholar]
- 39.Brunger JW, Murray GS, O’Riordan M, Matthews AL, Smith RJ, Robin NH. Parental attitudes towards genetic testing for pediatric deafness. Am. J Hum Genet. 2000;67:1621–1625. doi: 10.1086/316901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer CG, Martinez A, Fox M, Zhou J, Shapiro N, Sininger Y, Grody WW, Schimmenti LA. A prospective, longitudinal study of the impact of GJB2/GJB6 Genetic Testing on the beliefs and attitudes of parents of deaf and Hard-of-hearing infants. Am J Med Genet. 2009;149A(6):1169–1182. doi: 10.1002/ajmg.a.32853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardien S1, Human H, Harris T, Hefke G, Veikondis R, Schaaf HS, van der Merwe L, Greinwald JH, Fagan J, de Jong G. A rapid method for detection of five known mutations associated with aminoglycoside-induced deafness. BMC Med Genet. 2009;10:2. doi: 10.1186/1471-2350-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]