Abstract
Natural products offer unmatched chemical and structural diversity compared to other small-molecule libraries, but traditional natural product discovery programs are not sustainable, demanding too much time, effort, and resources. Here we report a strain prioritization method for natural product discovery. Central to the method is the application of real-time PCR, targeting genes characteristic to the biosynthetic machinery of natural products with distinct scaffolds in a high-throughput format. The practicality and effectiveness of the method were showcased by prioritizing 1911 actinomycete strains for diterpenoid discovery. A total of 488 potential diterpenoid producers were identified, among which six were confirmed as platensimycin and platencin dual producers and one as a viguiepinol and oxaloterpin producer. While the method as described is most appropriate to prioritize strains for discovering specific natural products, variations of this method should be applicable to the discovery of other classes of natural products. Applications of genome sequencing and genome mining to the high-priority strains could essentially eliminate the chance elements from traditional discovery programs and fundamentally change how natural products are discovered.
Natural products offer unmatched chemical and structural diversity compared to other small-molecule libraries. Thus, natural products remain the best source of drugs and drug leads and serve as excellent small-molecule probes to investigate biological processes.1,2 The recently discovered diterpenoids platensimycin (PTM) and platencin (PTN) represent two promising natural product drug leads that target fatty acid biosynthesis. PTM and PTN were discovered via the systematic screening of fermentation extracts from 83 000 strains, each of which was fermented in three different media (affording a total of ∼250 000 extracts).3−8 The discovery of PTM and PTN highlighted the exhaustiveness necessary for the successful isolation of novel natural products by the traditional methods.
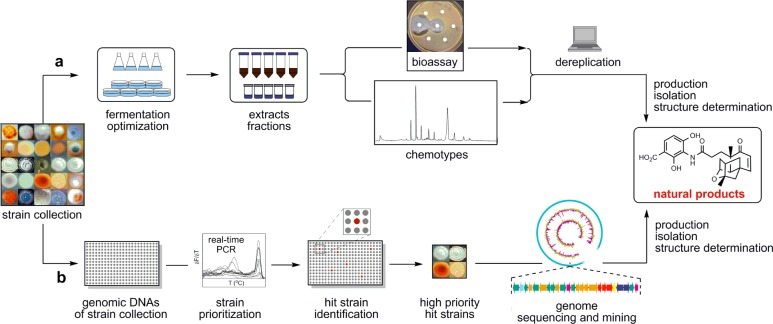
While effective, traditional natural product discovery programs are not sustainable, demanding too much time, effort, and resources (Figure 1a). As the sizes of microbial strain collections continue to grow, innovations in strain prioritization are clearly needed. Rapid strain prioritization could fundamentally change how microbial natural products are discovered. The connection between natural products and the genes encoding their biosynthesis has now been well recognized, and genes, as well as chemistry, are being increasingly exploited to categorize known natural products and discover new ones.9,10 Targeting biosynthetic genes using polymerase chain reaction (PCR) has proven useful for natural product discovery;11−13 however, the lack of high-throughput capabilities has limited their utility in natural product discovery. Real-time PCR, an advancement in PCR technology, has not been fully utilized for natural product discovery efforts. In real-time PCR, each cycle of amplification can be directly monitored based on the detection of fluorescence, hence eliminating gel electrophoresis and staining for visualization as the necessary post-PCR analysis steps.14−16 Real-time PCR also offers improved sensitivity for target detection and melting curve analysis for target specificity.
Figure 1.
Strategies for discovering natural products from microorganisms. (a) Traditional approach to natural product discovery relying on bioassays or chemotypes. (b) Postgenomics approach of natural product discovery featuring a high-throughput real-time PCR method for strain prioritization and genome sequencing and genome mining of the high-priority hit strains.
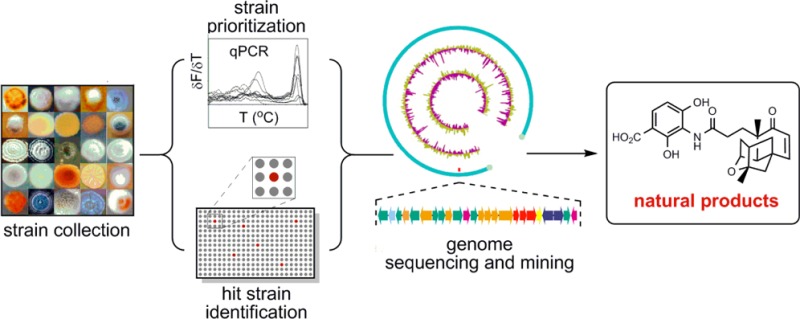
Here we report a high-throughput method for strain prioritization by real-time PCR to identify the most promising strains from a microbial strain collection for natural product discovery (Figure 1b). Considering the tremendous effort spent in discovering the original PTM and PTN producers, Streptomyces platensis MA7327 and MA7339, respectively, as well as the limited genetic amenabilities of the two producers for metabolic pathway engineering, we chose to demonstrate the utility and effectiveness of our method to identify new PTM and PTN producers from a collection of 1911 actinomycete strains. Six new PTM and PTN producers were identified: (i) all six were confirmed to contain the targeted diterpene synthase genes and verified as true PTM and PTN producers, (ii) three of them were demonstrated to overproduce PTM and PTN upon inactivation of the pathway-specific negative regulator, and (iii) two of them were confirmed to harbor the complete PTM–PTN dual biosynthetic gene clusters. The six new PTM and PTN producers, all classified as Streptomyces platensis species on the basis of their housekeeping genes, show distinct morphology, differing from S. platensis MA7327 and MA7339, and three of them showed superior genetic amenability. While the method as described is most appropriate to prioritize strains for discovering specific natural products, variations of this method should be applicable to the discovery of other classes of natural products.
Results and Discussion
Development of a Strain Prioritization Method by Real-Time PCR for Natural Product Discovery
The continued growth of microbial strain collections used for natural product discovery demands a high-throughput method for rapid strain prioritization. A sensitive and reliable survey of microbial genomes in a strain collection for genes characteristic for biosynthesis of the targeted class of natural products would identify the most promising ones for strain prioritization, thereby significantly increasing the likelihood of discovering the targeted class of natural products. Our method exploits the capability of real-time PCR, in a high-throughput manner, to detect DNA regions, encoding biosynthesis of the targeted class of natural products, from the genomic DNA (gDNA) samples of a microbial strain collection for strain prioritization. Subsequent genome sequencing and genome mining of the high-priority hit strains promise to accelerate natural product discovery (Figure 1b).
In real-time PCR applications, fluorescence levels, which correspond to the amount of accumulating products, are generated by fluorophores that intercalate double-stranded (ds) DNA or by fluorophore-conjugated oligonucleotides that prime to DNA targets. We chose to use SYBR Green I as the DNA-intercalating fluorophore, as it enabled us to apply melting curve analysis of the PCR products. Melting curve analysis reveals the apparent melting temperature (Tm) of the PCR products, a value dependent on product length, base composition, and nucleotide mismatch.17 The Tm values therefore can be exploited as an indicator of PCR product specificity without the need for gel electrophoresis.
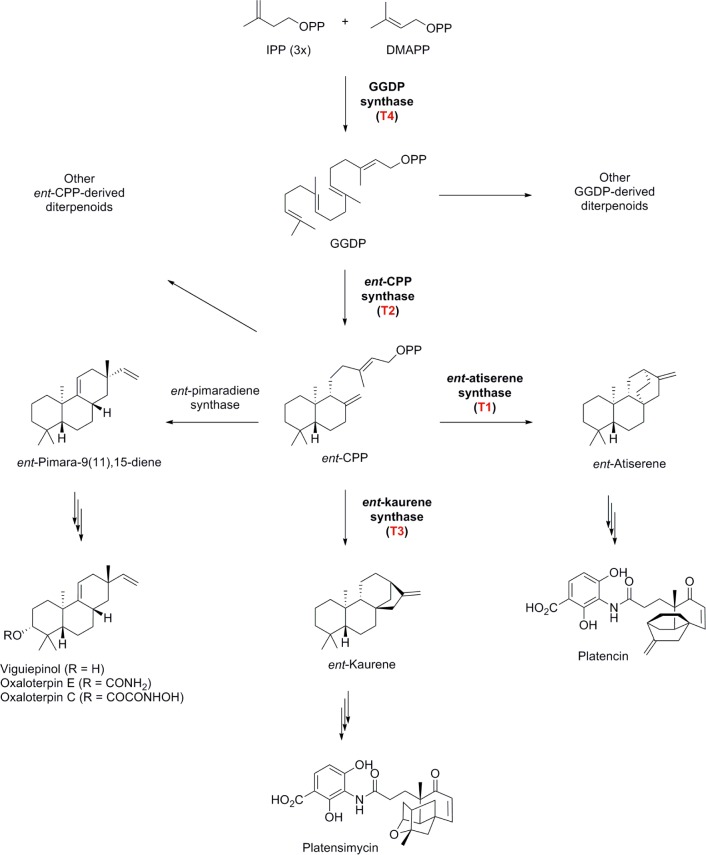
To demonstrate the utility and effectiveness of our strain prioritization method by real-time PCR, we chose to target the diterpene synthase genes to search for diterpenoid producers in general and PTM and PTN producers in particular from the actinomycete strain collection at The Scripps Research Institute. Diterpenoids of bacterial origin are underrepresented among known natural products.18 Diterpene synthases catalyze the critical steps in diterpenoid biosynthesis by morphing geranylgeranyl diphosphate into one of the many diterpenoid scaffolds, further transformations of which by pathway-specific tailoring enzymes afford the vast structural diversity known for diterpenoid natural products (Figure 2a).
Figure 2.
Pathways for the biosynthesis of bacterial diterpenoids, highlighting the four diterpene-related synthases and the tailoring enzymes leading to platensimycin (PTM), platencin (PTN), viguiepinol, oxaloterpin, and other known diterpenoids in bacteria.
We have previously cloned and characterized the biosynthetic gene clusters for PTM and PTN production in S. platensis MA7327 and MA7339 (Figure S1).19 Among the four diterpene-related synthases from the PTM and PTN biosynthetic machineries, geranylgeranyl diphosphate synthase (PtmT4/PtnT4) is common for all bacterial diterpenoids, ent-copalyl diphosphate synthase (PtmT2/PtnT2) is shared by both PTM and PTN, but ent-kaurene synthase (PtmT3) or ent-atiserene synthase (PtmT1/PtnT1) is specific for PTM or PTN, respectively (Figure 2a, S1). We reasoned that targeting ptmT3 or ptmT1/ptnT1 should specifically narrow our search to producers of ent-kaurene-derived or ent-atiserene-derived diterpenoids, such as PTM or PTN, while targeting ptm/ptnT2 or ptm/ptnT4 could broaden our search to include producers that biosynthesize other ent-copalyl diphosphate-derived or all geranylgeranyl diphosphate-derived diterpenoids, respectively (Figure 2).
The primers were designed based on the conserved nucleotide sequences of ptmT1/ptnT1, ptmT2/ptnT2, ptmT3, and ptmT/ptnT4 within the PTM and PTN gene clusters, as well as known homologues of actinomycete origin, and one of each primer for T1 and T3 was selected based on the atypical amino acid motifs (DXXXD) found in bacterial type I diterpene synthases18,19 (Figures S1, S2, and Table S1). The sizes for each of the PCR products were predicted to be 696 bp, 559 bp, 461 bp, or 411 bp, with the calculated Tm of 92.1, 91.6, 91.6, or 91.8 °C for T1, T2, T3, or T4, respectively, according to the ptmT1/ptnT1, ptmT2/ptnT2, ptmT3, and ptmT/ptnT4 sequences (Figure S1). We arbitrarily set the specificity cutoff for each of the targeted PCR products at Tm ±0.8 °C, where Tm is the melting temperature of the positive control, to identify putative hits. The deviation from Tm is inversely proportional to PCR product specificity. Smaller deviations will surely decrease the chance of false positives but might result in the miss of potential hits.
Strain Prioritization by Real-Time PCR for Four Diterpene Synthase Genes, Affording Six PTM and PTN Producers
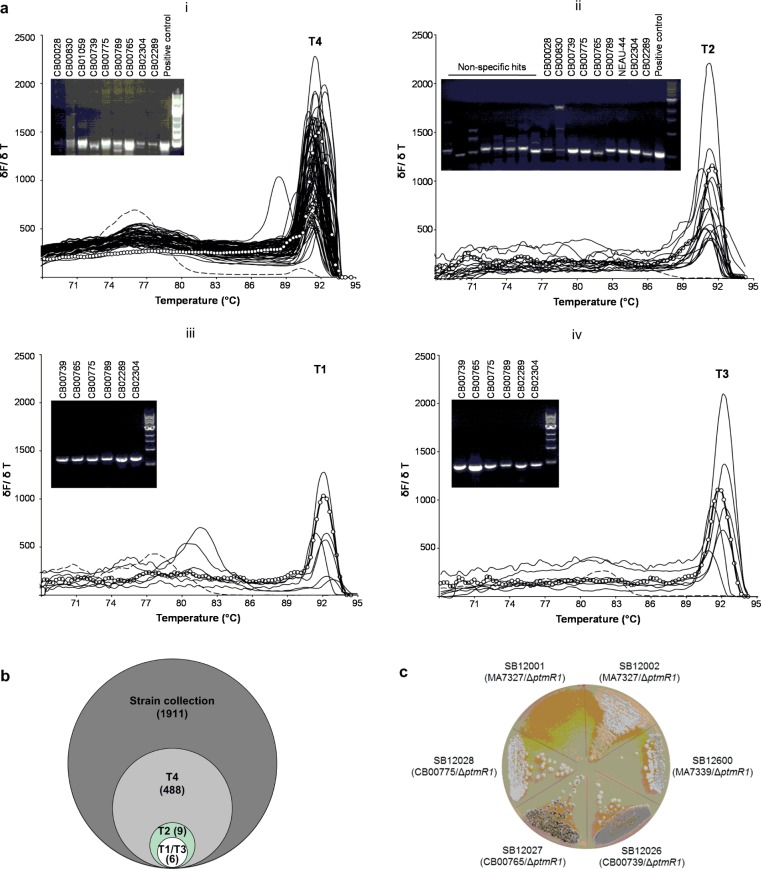
We selected 1911 strains from our actinomycete collection13 and applied the method described above to prioritize the strains for diterpenoid producers in general and PTM and PTN producers in particular. The gDNA of the 1911 strains were individually prepared, normalized, and arrayed in 384-well plates. Each of the plates also contained one blank (i.e., no template DNA) as a negative control and one with the gDNA of S. platensis MA7327 as a positive control. Real-time PCRs were carried out with each of the four sets of primers (Figure S1 and Table S1), and the resultant PCR products were subjected to melting curve analysis to identify putative hits (Figure 3a).
Figure 3.
Strain prioritization for natural product discovery by a high-throughput real-time PCR method, showcasing the identification of platensimycin, platencin, viguiepinol, oxaloterpin, and other diterpenoid producers by targeting selected diterpene-related synthases from the actinomycete collection at The Scripps Research Institute. (a) High-throughput real-time PCR method targeting (i) T4, (ii) T2, (iii) T1, and (iv) T3 and melting curve analysis of the resultant products to identify putative diterpenoid producers from a collection of 1911 strains. Each panel depicts the melting curves with δF/δT (y-axis) representing the rate of change in fluorescence as a function of temperature. Solid lines with open circles represent the positive controls with a normalized melting temperature (Tm). Solid lines represent hits found during the melting step of the real-time PCR experiment with the Tm range at Tm ± 0.8 °C. Dashed lines represent the negative controls with no template DNA. Insets show PCR products of the hits that were analyzed by agarose gel electrophoresis and confirmed by DNA sequencing. Only the melting curves for one of the five 384-well plates were shown for T4, which yielded 71 putative hits. For T1, T2, and T3, the melting curves of the respective hits from all five 384-well plates were combined and depicted together. (b) Euler diagram depicting the 488 putative diterpenoid producers identified from the 1911 strains by targeting T4, among which nine, six, and six were co-identified by targeting T2, T1, and T3, respectively, and confirmed by DNA sequencing. (c) Morphology of the three new platensimycin and platencin overproducers SB12026 (CB00739/ΔptmR1), SB12027 (CB00765/ΔptmR1), and SB12028 (CB00775/ΔptmR1) in comparison with SB12001 (MA7327/ΔptmR1) and SB12002 (MA7327/ΔptmR1), derived from S. platensis MA7327, and SB12600 (MA7339/ΔptmR1) derived from S. platensis MA7339 on an ISP4 agar plate. While SB12001 failed to sporulate and SB12002 sporulated poorly under all conditions examined, SB12026 and SB12027 sporulated well on several media, as exemplified with ISP4.
As summarized in Figure 3b, 488, 17, 6, and 6 out of the 1911 strains were identified as putative hits using the primers, targeting T4, T2, T3, and T1, respectively. Although the 488 hits identified using the T4 primers represented a hit rate that was too high to justify detailed follow-up analysis of each hit, confirmatory PCR on selected hits, using the same T4 primers followed by agarose gel electrophoresis of the resultant products, yielded specific bands with the predicted size (Figure 3a, panel i). However, the resultant PCR products from each of the T4 hits were not all sequenced (Figure S3a), and the possibility of false positives due to nonspecific PCR amplification therefore cannot be excluded. For the 17 hits identified using the T2 primers, follow-up analysis by PCR using the same T2 primers all afforded specific products with the predicted size (Figure 3a, panel ii). DNA sequencing of the resultant products revealed that nine out of the 17 hits were true hits, which contain genes encoding putative ent-copalyl diphosphate synthase (Figure S3b), with the remaining eight hits resulting from nonspecific PCR amplification. Finally, each set of the six hits identified using T1 or T3 primers and, remarkably, both set of six hits afforded specific PCR products with the predicted size (Figure 2b, panels iii and iv, respectively). DNA sequencing of the resultant products confirmed that the six hits were true diterpenoid producers that contain genes encoding the putative ent-kaurene synthase (Figure S3c) and ent-atiserene synthase (Figure S3d).
Cross examination of all hits revealed that six strains, CB00739, CB00765, CB00775, CB00789, CB02289, and CB02304, harbored all four targeted genes. The fact that they possessed both T1 and T3, in addition to T2 and T4, would predict all six hits as PTM–PTN dual producers, a genetic disposition that resembled the PTM–PTN dual producer S. platensis MA7327 but contrasted with the PTN-only producer S. platensis MA7339 (S1).19 Of the remaining three hits identified using T2, they (CB00028, CB00830, and CB01059) all also harbored T4, as would be expected for diterpenoid producers (Figure 2).18 In fact, we previously isolated from CB00830 viguiepinol and oxaloterpins E and C, diterpenoids whose biosynthesis was confirmed to involve T2 and T4.13,20 Therefore, CB00028 and CB01059 are promising producers of new diterpenoid natural products featuring ent-copalyl diphosphate-derived scaffolds (Figure 2).
DNA Sequencing Confirming the Six Hits to Harbor PTM–PTN Dual Biosynthetic Gene Clusters
To verify each of the six hits possessing a functional PTM–PTN dual biosynthetic cluster gene, selected intact genes, encoding the four diterpene synthases T1, T2, T3, and T4, as well as the pathway-specific negative transcriptional regulator PtmR1, were first amplified by PCR using primers that were designed according to the PTM–PTN dual biosynthetic gene cluster from S. platensis MA732719 (Table S1). DNA sequencing of the resultant products confirmed that each of the targeted genes was highly homologous (>95%) to those of the known PTM–PTN producers and each other (Figure S4). The hit strains were next subjected to polyphasic taxonomy studies. Phylogenetic analysis based on four selected housekeeping genes, 16S rRNA, recA, rpoB, and trpB(21,22) (Table S1), assigned all six hits as Streptomyces platensis (Figure S5a). Close examination of the six hit strains, in comparison with S. platensis MA7327 and MA7339, however, revealed that the new strains were distinct, as judged by the morphology and appearance of pigmented spores (Figure S5b). Two of the hit strains, CB00739 and CB00765, were selected and subjected to genome sequencing, confirming that these strains contained complete PTM–PTN gene clusters. The PTM–PTN dual biosynthetic gene clusters from CB00739 and CB00765 have the same number of open reading frames and same genetic organization as those from S. platensis MA7327, and the overall nucleotide sequence identities among the three clusters are >97% (Figure S4).
The Six Positive Strains Produce and Three of Their Engineered Strains Overproduce PTM and PTN
To validate that the identified PTM–PTN dual biosynthetic gene clusters are functional, the six hit strains were first subjected to fermentation optimization for PTM and PTN production and confirmation. Under the same conditions used for S. platensis MA7327 and MA7339,19,23,24 PTM and PTN production was confirmed for all six hit strains (Figure S6), with PTM and PTN titers varying in the ranges 0.66–10 and <0.1–11 mg L–1, respectively (Table 1). These titers correlated well with those reported previously from the S. platensis MA7327 and MA7339 wild-type strains.23,24
Table 1. Titers of Platensimycin and Platencin Produced by Streptomyces platensis Strains.
| titera (mg L–1) for S. platensis strains |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound | MA7327 | SB12001b | SB12002b | CB00739 | SB12026 | CB00765 | SB12027 | CB00775 | SB12028 | CB00789 | CB02289 | CB02304 |
| PTM | 4.1 ± 1.0 | 110 ± 4 | 220 ± 9 | 1.0 ± 0.3 | 310 ± 12 | 5.2 ± 1.0 | 11 ± 0.9 | 4.7 ± 1.3 | 230 ± 20 | 6.5 ± 1.7 | 0.66 ± 0.17 | 10 ± 4 |
| PTN | 0.77 ± 0.21 | 150 ± 14 | 74 ± 3 | 1.6 ± 0.2 | 170 ± 6 | 2.4 ± 0.2 | 12 ± 1 | 0.65 ± 0.05 | 200 ± 25 | 2.2 ± 0.6 | <0.1c | 11 ± 4 |
Unless otherwise noted, values are the averages of at least three independent trials and are reported with standard deviations.
Strains originally reported.24
The PTN peak at λ240 was too small to calculate a reliable titer; PTN was detected by EIC (m/z at 426.20 for the [PTN + H]+ ion) and is shown in Figure S6.
We previously demonstrated that removal of the pathway-specific, negatively acting transcriptional regulator PtmR1 or PtnR1 from S. platensis MA7327 and MA7339 afforded recombinant strains that dramatically overproduced PTM, PTN, or both.23,24 However, the recombinant strains sporulated poorly under all conditions examined (Figure 3c). This has prevented us from further engineering PTM and PTN biosynthesis in these overproducers. We similarly inactivated PtmR1 in three of the six hits, CB00739, CB00765, and CB00775 (Table S2), and the genotypes of the resultant recombinant strains SB12026, SB12027, and SB12028, respectively, were confirmed by PCR (Figure S7). Under the same conditions for PTM and PTN production with the wild-type strains as controls, SB12026, SB12027, and SB12028 indeed overproduced PTM and PTN up to 310-fold, consistent with what has been observed for the S. platensis MA7327 and MA7339 strains (Table 1 and Figure S6). Gratifyingly, SB12026 and SB12027 sporulate well (Figure 3c), opening up the opportunity to further manipulate PTM and PTN biosynthesis in these overproducers for pathway characterization and structural diversity.
High-Throughput Real-Time PCR Method for Natural Product Discovery and Strain Prioritization
In spite of the indisputable track record of natural products as drugs and drug leads and small-molecule probes, the traditional approach to natural product discovery, with its elements of serendipity, demands too much time, effort, and resources.1,2 We now report a high-throughput method using real-time PCR for strain prioritization to identify the most promising strains from a microbial strain collection for natural product discovery. Resources could then be devoted preferentially to the strains that hold the highest promise in producing novel natural products, thereby accelerating detection and isolation of the targeted natural products and cutting the time and cost associated with traditional natural product discovery programs.
Central to our method is the application of real-time PCR, targeting genes characteristic to the biosynthetic machinery of natural products with distinct scaffolds, in a high-throughput format and with superior sensitivity and specificity. The practicality and effectiveness of our method were showcased by targeting four diterpene-related synthase genes, T1, T2, T3, and T4, to prioritize 1911 strains, selected from the actinomycete strain collection at The Scripps Research Institute, for diterpenoid producers in general and PTM and PTN producers in particular (Figure 2). We chose to search for diterpenoid producers because they are underrepresented among microbial natural products18 and for new PTM and PTN producers because of the heroic effort in the discovery of the original S. platensis MA7327 and MA7339 strains,3−8 as well as the need for alternative producers with better genetic amenability.19
We rapidly identified a total of 488 potential diterpenoid producers, among which six were confirmed as PTM–PTN dual producers and one as a viguiepinol and oxaloterpin producer. Although the new PTM–PTN dual producers are all S. platensis strains on the basis of polyphasic taxonomy, they exhibit district morphology, three of which showed superior genetic amenability to the original S. platensis MA7327 and MA7339 strains. Genetic amenability of the producing organisms is of paramount importance in applying microbial genomics to natural product discovery, and new strains, alternatives to genetically recalcitrant original producers for the same natural products or families of natural products with similar scaffolds, represent an innovative solution to this critical challenge. The 25% (i.e., 488 out of 1911) hit rate of diterpenoid producers is higher than expected, given the small number of bacterial diterpenoids known to date,18 but is consistent with an early survey of our strain collection.13 Taken together, these findings support our early proposal that the biosynthetic potential of diterpenoids in bacteria is significantly underestimated.13,18 Further prioritization of the hits identified in this study promises the discovery of novel diterpenoid natural products.
Natural products occupy tremendous chemical structural space that is unmatched by any other small-molecule libraries. While the rich functionality of natural products is, without doubt, one of their great strengths, providing potency and selectivity, the biosynthetic machineries for each of the major molecular scaffolds are highly conserved across the entire family of natural products, as exemplified by the diterpene-related synthases T1, T2, T3, and T4, for diterpenoids. Variations among the myriad of tailoring enzymes associated with each of the biosynthetic machineries further imbue the remarkable structural diversity within each of the natural product families (Figure 2a). Although we have showcased our method by targeting diterpenoids, variation of the method should be readily applicable to the discovery of other classes of natural products by targeting conserved genes within each of the major biosynthetic machineries.9,10 Applications of genome sequencing and genome mining to the high-priority strains could essentially eliminate the chance elements from traditional discovery programs and fundamentally change how natural products are discovered.
Deviations from the targeted Tm value could be correlated to the product specificity. The Tm range used in this study was 0.8 °C, and this range allowed us to identify hits with 97–98%, 63–98%, 96–97%, and 38–98% identity to ptmT1, ptmT2, ptmT3, and ptmT4, respectively (Figure S3). The range of Tm can be exploited to address the needs of each experiment, and a larger range of Tm will surely increase the likelihood of finding diverse hits, albeit with a concomitant increase of nonspecific hits. Finally, we used a DNA intercalating fluorophore in the current method, allowing detection and subsequent melting temperature analysis of the amplified products. One could envision variations of the method with fluorophore-labeled oligonucleotides or molecular beacons in a multiplex approach of real-time PCR to target multiple genes simultaneously within a single reaction, further expanding its utility in strain prioritization, thereby accelerating natural product discovery.
Experimental Section
Preparation of a Genomic DNA (gDNA) Library
The actinomycete collection at The Scripps Research Institute consists of strains isolated from various unexplored and underexplored ecological niches.13 Actinomycete cultivation and gDNA preparation followed standard protocols.25 The concentrations of gDNA samples were estimated using a microplate fluorescence assay with minor modifications.26 Briefly, gDNA samples were diluted in 10 mM Tris-HCl, pH 8.0. SYBR Green I dye (Sigma-Aldrich) was added to each dilution of DNA, mixed, and incubated at room temperature for 10 min in the dark. Fluorescence levels were measured using a SpectraMax M5Multi-Mode microplate reader (Molecular Devices) with excitation and emission wavelengths of 450 and 520 nm, respectively. After the normalization of gDNA concentration, samples were arrayed in 384-well masterblock deep-well plates as working stocks and transferred to microplates using a Biomek FX workstation (Beckman Coulter) for each real-time PCR experiment.
Real-Time PCR
Real-time PCR was performed with an Applied Biosystems 7900HT Fast real-time PCR system. Primer design was based on the ptmT3 sequence and the conserved sequences of ptmT1, ptmT2, and ptmT4 homologues (Figures S1, S2). Genomic DNA (1–100 ng), 0.1 μL of each primer (10 μM stock), 0.5 μL of DMSO, 0.5 μL of 10× SYBR Green I dye, 5 μL of Taq 2X Master Mix, and H2O were mixed to give a reaction volume of 10 μL per well. A “no template” negative control and one positive control with the gDNA of S. platensis MA7327 were included in each 384-well plate. The reaction conditions consisted of a background check at 50 °C for 2 min; initial denaturation at 95 °C for 7 min; and 37–40 cycles of denaturation at 95 °C for 30 s, primer annealing at 64–68 °C for 15 s, extension at 68 °C for 30–60 s, and melting at 95 °C for 15 s with a ramp rate of 2% from 68 to 95 °C. For T4 amplification, three additional cycles of denaturation at 95 °C for 30 s, temperature ramping to 30 °C, and extension at 68 °C for 30 s were included after the initial denaturation step mentioned above. The melting step subjected amplicons to a range of temperatures for determining Tm. Each Tm was normalized to a theoretical Tm calculated using the nearest neighbor model.27−29 For determining each theoretical Tm, the sequence of the target gene was used assuming an initial DNA amount of 100 ng and salt concentration of 50 mM. Samples with Tm ± 0.8 °C compared to the positive control were considered hits. To confirm hits, regular PCR was performed, and the resulting products were analyzed by 1% agarose gel electrophoresis, purified by gel extraction, and sequenced.
DNA Sequencing
The T1, T2, T3, T4, and R1 genes of the six new PTM–PTN dual producers (CB00739, CB00765, CB00775, CB00789, CB02289, and CB02304) were amplified using primers designed according to the PTM–PTN dual biosynthetic gene cluster from S. platensis MA732719 (Table S1). Regions of the four housekeeping genes, 16S rRNA, recA, rpoB, and trpB, were amplified using primers listed in Table S1. The resultant PCR products were cloned and sequenced. The draft genome sequences of S. platensis CB00739 and CB00765 were obtained using the Ion 316 chips and the Ion PGM Sequencing 300 kit (Life Technologies) following the manufacturer’s instructions. Fragment libraries (∼300 bp inserts) and matepair libraries (∼5 kb inserts) were sequenced. For S. platensis CB00739, a final sequence assembly of 5 955 880 reads afforded a 9 441 068 bp draft genome consisting of 23 scaffolds. For S. platensis CB00765, a final sequence assembly of 5 293 288 reads yielded a 9 404 443 bp draft genome consisting of 20 scaffolds. Gaps within the PTM–PTN gene clusters were manually filled by PCR. All DNA sequences were deposited in the NCBI database (see Accession Codes).
Phylogenetic Analysis
Alignment of the concatenated housekeeping genes (partial sequences, 2975-bp total) was generated using MegAlign 10.1.21,22 Phylogenetic analysis was performed using the MEGA 5.2.1 software.30 The phylogenetic tree was constructed using the Tamura-Nei evolutionary distance method and 1000 bootstrap replications.
Gene Disruption
Strains and plasmids/cosmids used in this study are summarized in Table S2. Primers used in the gene disruption experiments are listed in Table S1. Gene disruption of ptmR1 was performed in E. coli BW25113/pIJ790 carrying appropriate cosmids by following the λRED-mediated PCR-targeting mutagenesis method as described previously.19,23,24,31 The ptmR1 genes were replaced with the aac(3)IV + oriT resistance cassette from pIJ773.31 The mutant cosmids were introduced into the S. platensis CB00739, CB00765, and CB00775 by intergeneric conjugation32 after passaging the mutant cosmids through the nonmethylating E. coli ET12567/pUZ8002.25 Apramycin selection and kanamycin sensitivity on ISP4 medium were used to determine double-crossover mutants of the ptmR1 genes. The mutations were confirmed by PCR analysis using primers ptmRidF and ptmRidR (Table S1).
Production of PTM and PTN
PTM and PTN were produced from Streptomyces spp. following previously published procedures.19,23,24 Briefly, Streptomyces spp. spores were inoculated into seed medium and incubated for 48 h, and 2 mL of seed culture was used to inoculate 50 mL of PTM medium19 supplemented with 10 mL L–1 trace elements25 and 1.5 g of Amberlite XAD-16 resin (Sigma-Aldrich). After incubation for 7 days at 28 °C and 250 rpm, the resin was harvested by centrifugation, washed three times with H2O, and extracted with actone (3 × 10 mL). Acetone was removed under reduced pressure, and the resulting oil was resuspended in 1.5 mL of MeOH.
HPLC and LC-MS Analysis
HPLC was carried out on a Varian liquid chromatography system consisting of Varian ProStar 210 pumps and a ProStar 330 photodiode array detector equipped with an Apollo C18 column (250 mm × 4.6 mm, 5 μm, Grace Davison Discovery Sciences, Deerfield, IL, USA). HPLC analysis of PTM and PTN was performed using a 20 min solvent gradient (1 mL min–1) from 15% CH3CN in H2O containing 0.1% formic acid to 90% CH3CN in H2O containing 0.1% formic acid, and PTM and PTN were eluted with retention times of 14.0 and 17.4 min, respectively. The peak area at 240 nm was used to quantify PTM and PTN production on the basis of calibration curves with authentic PTM and PTN standards.
Liquid chromatography–mass spectrometry (LCMS) was carried out on an Agilent 1260 Infinity LC coupled to a 6230 TOF equipped with an Agilent Extend C18 column (50 mm × 2.1 mm, 1.8 μm). Liquid chromatography was performed using a 17 min solvent gradient (0.4 mL min–1) from 10% CH3CN in H2O containing 0.1% formic acid to 100% CH3CN containing 0.1% formic acid, and PTM and PTN were eluted with retention times of 7.4 and 9.7 min, respectively. Extracted ion (m/z at 442.1863 for the [PTM + H]+ ion and m/z at 426.1914 for the [PTN + H]+ ion) chromatograms (m/z at [M + H]+ ± 0.5) verified PTM and PTN production.
Acknowledgments
We thank Dr. S. B. Singh, Merck Research Laboratories, Rahway, NJ, for providing S. platensis MA7327 and MA7339 wild-type strains and the John Innes Center, Norwich, UK, for providing the REDIRECT technology kit. This work is supported in part by the Chinese Ministry of Education 111 Project B08034, National High Technology Joint Research Program of China Grant 2011ZX09401-001, and National High Technology Research and Development Program of China Grant 2012AA02A705 (to Y.D.) and by the Natural Products Library Initiative at TSRI and NIH Grants GM086184 and AI079070 (to B.S.).
Supporting Information Available
Primer designing to target the four diterpene synthase genes, the list of primers used in this study, sequence comparison of T1, T2, T3, and T4 PCR hit fragments, comparison of the PTM–PTN biosynthetic gene clusters, phylogeny and morphology of the new PTM–PTN producers, the list of strains and plasmids used in this work, HPLC analysis of PTM–PTN production, gene inactivation to generate PTM–PTN overproducers, and supplementary references. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The following partial gene sequences were deposited in GenBank for the S. platensis strains MA7327, MA7339, CB00739, CB00765, CB00775, CB00789, CB02289, and CB02304, respectively: 16S rRNA, KJ469279–KJ469286; recA, KJ469287–KJ469294; rpoB, KJ469295–KJ469302; and trpB, KJ469303–KJ469310. The following complete gene sequences were deposited in GenBank for the S. platensis strains CB00739, CB00765, CB00775, CB00789, CB02289, and CB02304, respectively: ptmT1, KJ469311–KJ469316; ptmT2, KJ469317–KJ469322; ptmT3, KJ469323–KJ469328; ptmT4, KJ469329–KJ469334; and ptmR1, KJ469335–KJ469340. The PTM–PTN gene clusters for S. platensis CB00739 and CB00765 were deposited as KJ189771 and KJ189772, respectively.
Author Contributions
‡ Hindra, T. Huang, D. Yang, and J. D. Rudolf contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Newman D. J.; Cragg G. M. J. Nat. Prod. 2012, 75, 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. W.-H.; Vederas J. C. Science 2009, 325, 161–165. [DOI] [PubMed] [Google Scholar]
- Wang J.; Soisson S. M.; Young K.; Shoop W.; Kodali S.; Galgoci A.; Painter R.; Parthasarathy G.; Tang Y. S.; Cummings R.; Ha S.; Dorso K.; Motyl M.; Jayasuriya H.; Ondeyka J.; Herath K.; Zhang C.; Hernandez L.; Allocco J.; Basilio A.; Tormo J. R.; Genilloud O.; Vicente F.; Pelaez F.; Colwell L.; Lee S. H.; Michael B.; Felcetto T.; Gill C.; Silver L. L.; Hermes J. D.; Bartizal K.; Barrett J.; Schmatz D.; Becker J. W.; Cully D.; Singh S. B. Nature 2006, 441, 358–361. [DOI] [PubMed] [Google Scholar]
- Jayasuriya H.; Herath K. B.; Zhang C.; Zink D. L.; Basilio A.; Genilloud O.; Diez M. T.; Vicente F.; Gonzalez I.; Salazar O.; Pelaez F.; Cummings R.; Ha S.; Wang J.; Singh S. B. Angew. Chem., Int. Ed. 2007, 46, 4684–4688. [DOI] [PubMed] [Google Scholar]
- Wang J.; Kodali S.; Lee S. H.; Galgoci A.; Painter R.; Dorso K.; Racine F.; Motyl M.; Hernandez L.; Tinney E.; Colletti S. L.; Herath K.; Cummings R.; Salazar O.; Gonzalez I.; Basilio A.; Vicente F.; Genilloud O.; Pelaez F.; Jayasuriya H.; Young K.; Cully D. F.; Singh S. B. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. B.; Jayasuriya H.; Ondeyka J. G.; Herath K. B.; Zhang C.; Zink D. L.; Tsou N. N.; Ball R. G.; Basilio A.; Genilloud O.; Diez M. T.; Vicente F.; Pelaez F.; Young K.; Wang J. J. Am. Chem. Soc. 2006, 128, 11916–11920. [DOI] [PubMed] [Google Scholar]
- Young K.; Jayasuriya H.; Ondeyka J. G.; Herath K.; Zhang C.; Kodali S.; Galgoci A.; Painter R.; Brown-Driver V.; Yamamoto R.; Silver L. L.; Zheng Y.; Ventura J. I.; Sigmund J.; Ha S.; Basilio A.; Vicente F.; Tormo J. R.; Pelaez F.; Youngman P.; Cully D.; Barrett J. F.; Schmatz D.; Singh S. B.; Wang J. Antimicrob. Agents Chemother. 2006, 50, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genilloud O.; Gonzalez I.; Salazar O.; Martin J.; Tormo J. R.; Vicente F. J. Ind. Microbiol. Biotechnol. 2011, 38, 375–389. [DOI] [PubMed] [Google Scholar]
- Walsh C. T.; Fischbach M. A. J. Am. Chem. Soc. 2010, 132, 2469–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. O.; Van Lanen S. G.; Baltz R. H. J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M.; Baba S.; Takatsu T.; Kizuka M.; Ohata Y.; Tanaka M.; Nonaka K.; Spork A. P.; Ducho C.; Chen W.-C. L.; Van Lanen S. G. Angew. Chem., Int. Ed. 2013, 52, 11607–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen J. G.; Reddy B. V. B.; Ternei M. A.; Charlop-Powers Z.; Calle P. Y.; Kim J. H.; Brady S. F. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 11797–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P.; Ma M.; Rateb M. E.; Shaaban K. A.; Yu Z.; Huang S.-X.; Zhao L.-X.; Zhu X.; Yan Y.; Peterson R. M.; Lohman J. R.; Yang D.; Yin M.; Rudolf J. D.; Jiang Y.; Duan Y.; Shen B. J. Nat. Prod. 2014, 77, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek M. A.; Repa J. J. Adv. Physiol. Educ. 2005, 29, 151–159. [DOI] [PubMed] [Google Scholar]
- VanGuilder H. D.; Vrana K. E.; Freeman W. M. BioTechniques 2008, 44, 619–626. [DOI] [PubMed] [Google Scholar]
- Wilhelm J.; Pingoud A. ChemBioChem 2003, 4, 1120–1128. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 227–259. [DOI] [PubMed] [Google Scholar]
- Smanski M. J.; Peterson R. M.; Huang S.-X.; Shen B. Curr. Opin. Chem. Biol. 2012, 16, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski M. J.; Yu Z.; Casper J.; Lin S.; Peterson R. M.; Chen Y.; Wendt-Pienkowski E.; Rajski S. R.; Shen B. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda C.; Hayashi Y.; Itoh N.; Seto H.; Dairi T. J. Biochem. 2007, 141, 37–45. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Zheng W.; Rong X.; Huang Y. Int. J. Syst. Evol. Microbiol. 2008, 58, 149–159. [DOI] [PubMed] [Google Scholar]
- Labeda D. P. Streptomyces Int. J. Syst. Evol. Microbiol. 2011, 61, 2525–2531. [DOI] [PubMed] [Google Scholar]
- Smanski M. J.; Peterson R. M.; Rajski S. R.; Shen B. Antimicrob. Agents Chemother. 2009, 53, 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Smanski M. J.; Peterson R. M.; Marchillo K.; Andes D.; Rajski S. R.; Shen B. Org. Lett. 2010, 12, 1744–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T.; Bibb M. J.; Buttner M. J.; Chater K. F.; Hopwood D. A.. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Leggate J.; Allain R.; Isaac L.; Blais B. W. Biotechnol. Lett. 2006, 28, 1587–1594. [DOI] [PubMed] [Google Scholar]
- Kibbe W. A. Nucleic Acids Res. 2007, 35Suppl 2W43–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K. J.; Frank R.; Bloecker H.; Marky L. A. Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N.; Nakano S.; Yoneyama M.; Honda K. Nucleic Acids Res. 1996, 24, 4501–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K.; Peterson D.; Peterson N.; Stecher G.; Nei M.; Kumar S. Mol. Biol. Evol. 2011, 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B.; Challis G. L.; Fowler K.; Kieser T.; Chater K. F. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman M.; Logan R.; O’Brien K.; Seno E. T.; Rao R. N.; Schoner B. E. Gene 1992, 116, 43–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.