Abstract
Plasmodium protozoa, the source of malarial infections, catabolize large quantities of hemoglobin during an intra-erythrocytic phase. During this process, free heme is detoxified through biomineralization into an insoluble heme aggregate, hemozoin (Hz). In its native state, Hz is associated with a variety of lipid peroxidation products including 4-hydroxy-2-nonenal (HNE). In the present study, gene expression profiles were used to compare responses to two of the individual components of Hz in a model macrophage cell line. LPS-stimulated RAW 264.7 cells were exposed to HNE and the synthetic form of Hz, β-hematin (BH), for 6 or 24 h. Microarray analysis identified alterations in gene expression induced by exposure to HNE and opsonized BH (fold change ≥1.8, p-value ≤0.01). Patterns of gene expression were compared to changes induced by an opsonized control latex bead challenge in LPS-stimulated cells and revealed that the BH response was predominantly phagocytic. Ingenuity Pathway Analysis demonstrated that HNE mediated a short term oxidative stress response and had a prolonged effect on the expression of genes associated with categories of ‘Cell Cycle’, ‘Cellular Assembly and Organization’, ‘DNA Replication, Recombination, and Repair’, and ‘Cellular Development’. Comparisons of expression changes caused by BH and HNE with those observed during malarial infection suggest that BH and HNE are involved in inflammatory response modulation, altered NF-κB signal transduction, extracellular matrix (ECM) degradation, and dyserythropoiesis. HNE exposure led to several significant steady-state expression changes including repressed chemokine (C-C motif) ligand 5 (Ccl5) indicative of dyserythropoiesis, and a severe matrix metalloproteinase 9 (Mmp9)/tissue inhibitor of metalloproteinase 1 (Timp1) imbalance in favor of ECM proteolysis.
Introduction
Parasitic resistance to drugs, vector resistance to insecticides, and climatic and environmental changes, have caused a global resurgence of the Plasmodium infection, malaria (1). Progress towards the treatment and prevention of infection is hindered by the complex parasite lifecycle and host-pathogen interactions. Many of the hallmarks of malaria are associated with the rupture of parasitized erythrocytes, release of cellular and parasite debris into the vasculature, and subsequent immune response. Although immune activity is essential for protective immunity, a modulated response likely contributes to malaria pathogenesis (2). Accumulating evidence suggests that many adverse effects are caused by endogenous toxins generated during interactions of parasite-derived species with host tissues (3). Consequently, the interplay between various parasite-derived species, including hemozoin (Hz), and the host immune system is of extreme interest.
Hz is a heme detoxification biomineral formed during the intraerythrocytic parasite stage as a result of the high concentration of free heme released during hemoglobin catabolism. Structurally, the biomineral is an aggregate of hydrogen bonded five-coordinate ferric protoporphyrin IX [Fe(III)PPIX] dimers, joined by reciprocating monodentate carboxylate linkages between the central iron of one monomer and a propionic acid side chain of the other (4). In its native state, Hz is coated by an array of proteins, nucleic acids, host- and parasite-derived lipids (5), and racemic lipid peroxidation products (6). Notably, the level of the secondary oxidation product 4-hydroxy-2-nonenal (HNE) measured in Hz-laden monocytes (7) is the highest intracellular HNE concentration in any biological system observed to date (8).
At schizogony, it is estimated that cellular debris, including 200 µmol of particulate Hz, is released into the circulation of a P. falciparum-infected patient (9) initiating an innate immune response. Hz has been shown to perturb the expression of cytokines (9–11) and impair both re-phagocytosis (12) and phorbol ester mediated oxidative burst (13). Given the activity of native Hz against an activated immune response, it is of interest to understand the effects of individual Hz components on macrophage function. Recently, the immunomodulatory activity of native Hz was modeled using constitutive components in a cell culture system. Membrane lipids from erythrocyte ghosts were incubated with BH, and RAW 264.7 macrophage-like cells subsequently exposed. These cells demonstrated impaired PMA-activated NADPH oxidase and LPS-stimulated inducible nitric oxide synthase (iNOS) activities. Upon treatment with either BH- or ghost-supernatant alone, no inhibition was observed indicating that lipid peroxidation products generated during reactions between BH and ghost membranes were responsible for the effects.
The known cellular responses to several lipid peroxidation components of Hz suggests possible involvement in malaria pathophysiology (6, 14). The current study aims to dissect the components of Hz in a model system and explore potential roles of key players involved in disrupting the programmed function of the triggered immune response during infection. Since the effects of the individual components of Hz remain largely unexplored in the context of malaria, a microarray approach was used to explore gene expression changes in activated cells. Given the unquestionable reactivity of HNE (8, 15, 16), and ability of the heme moiety to mediate lipid peroxidation (17–19), HNE and BH were targeted as the native Hz components. Gene expression patterns were analyzed in the context of biological processes to examine the downstream effects of both specific and non-specific damage, and comparisons were made with cellular alterations that are observed during malarial infection.
Experimental Procedures
β-hematin Synthesis and Characterization
BH was synthesized as previously described (20, 21) using purified hemin chloride (Fluka). Exhaustive washings in MeOH, 0.1 M NaHCO3 (pH 9.0), and DMSO removed excess free heme and small aggregates. Formation was confirmed by powder X-ray diffraction (Scintag X1 h/h automated powder difractometer with a copper target, a Peltier-cooled solid-state detector and a zero-background silica (510) sample support) and FTIR spectroscopy (ATI Mattson Genesis Series). BH was suspended in ethanol, sonicated, applied to a polished aluminum specimen mount, and dried at 25 °C overnight. The sample was sputter-coated with gold for 20 s and imaged using a Hitachi S4200 scanning electron microscope at 1.0 kV accelerating voltage. Particles were determined to have an average length of 0.9 ± 0.3 µM (Supporting Information Figure 1).
Cell Culture and Treatment
Murine macrophage-like RAW 264.7 cells (American Type Culture Collection) were cultured under standard incubation conditions (37 °C, 5% CO2) and grown in RPMI supplemented with 5% FBS (Atlanta Biologicals) and 1µg/mL P/S (Cellgro MediaTech). Cells were untreated or treated with 35 µM HNE (EMD Biosciences), 0.1 mg/mL serum-opsonized BH, or serum-opsonized latex bead (0.1µm, 10 µL of 0.05% per 1 × 106 cells), and immediately stimulated with LPS (1 µg/mL). Opsonization was performed as previously described (22).
RNA Isolation and Expression Analysis
Microarray and qRT-PCR analyses were performed by the Vanderbilt Microarray Shared Resource. Total RNA was isolated using the Versagene RNA purification and DNase treatment kits, following manufacturer’s recommendations. Three biological replicates per sample were analyzed for quality (Agilent 2100 Bioanalyzer, Agilent Technologies). One (1) µg of Total RNA (30 ng mRNA) was used to generate First Strand cDNA using the NanoAmp RT-IVT labeling kit according to manufacturer’s protocol. Following first strand synthesis, second strand synthesis was completed. The resulting cDNA was then purified using an ABI kit provided column and the entire reaction was used in an IVT reaction to generate cRNA or DIG labeled cRNA. cRNA was purified using a kit provided column, assessed for quality on an Agilent Bioanalyzer, and reverse transcribed to make ss cDNA. Samples prepared from 6 h incubations were fragmented, labeled with terminal deoxy transferase with biotin, hybridized to an Affymetrix mouse gene 1.0ST arrays per manufacturer’s protocol, and detected with Streptavidin-Phycoerythrin. Samples obtained from 24 h incubations were fragmented, hybridized to an ABI mouse genome survey microarray per manufacturer’s protocol, and detected with the addition of the chemiluminescence reaction substrate. Expression values were quantile normalized and filtered (S/N >3 and flag value <5000, ABI arrays). Partek 6.4 and GeneSpring GX 7.3.1 software were used to determine statistically significant differentially expressed genes from probes altered by ≤ or ≥ 1.8-fold (0.01 p-value cutoff, Benjamini-Hochberg multiple testing correction) in treated stimulated cells (experimental) relative to stimulated cells (control). In accordance with MIAME procedure, microarray data have been submitted to the NCBI Gene Expression Omnibus and can be found under series number GSE13281. Ingenuity Pathways Analysis was used for gene expression analysis (Ingenuity Systems®, www.ingenuity.com).
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was used to validate the expression levels of several genes identified as differentially expressed (quadruplicate measurements of three biological replicates per sample). cDNA was reverse-transcribed from 0.5 µg of total RNA using random hexamer primers and Superscript II Reverse Transcriptase (Invitrogen). Reactions were purified using Qiagen’s PCR Purification Kit following the manufacturer’s protocol. Following RT, all assays were performed with Applied Biosystems TaqMan FAM labeled 20× probes (Table 1). Ywhaz was chosen as the endogenous control based on results obtained from an Applied Biosystems mouse endogenous control array. cDNA amplification was performed using TaqMan 2× Universal PCR Master Mix (Applied Biosystems) and standard Taqman cycling conditions were used as specified by the manufacturer. Cycling and data collection were performed using the Applied Biosystems 7900 HT instrument and analysis performed using SDS software to calculate Ct values for each detector. Ct values were processed based on the comparative Ct method.
Table 1.
Taqman Gene Expression Assays Used for Quantitative Real-Time RT-PCRa
| Treatment | Gene | Assay ID | Amplicon length |
|---|---|---|---|
| 6 h BH, 6 h HNE | Csf2 | Mm00438328_m1 | 71 |
| 6 h HNE | Ccl2 | Mm00441242_m1 | 74 |
| 6 h HNE | Il1b | Mm00434228_m1 | 90 |
| 6 h HNE, 24 h BH | Il1a | Mm00439620_m1 | 68 |
| 6 h, 24 h HNE | Mmp9 | Mm00442991_m1 | 76 |
| 6 h, 24 h HNE | Timp1 | Mm00441818_m1 | 90 |
| 6 h, 24 h HNE | Csf3 | Mm00438334_m1 | 106 |
| 24 h HNE | Ccl5 | Mm01302428_m1 | 71 |
| 24 h HNE | Tnf | Mm00443258_m1 | 81 |
| 24 h HNE | Nfkbie | Mm00500796_m1 | 78 |
| 24 h HNE | Ikbke | Mm00444862_m1 | 66 |
| 24 h BH | Fos | Mm00487425_m1 | 59 |
Each assay consists of two unlabeled PCR primers and a FAM™ dye-labeled TaqMan MGB (minor groove binder) probe.
ELISA
Cells (4 × 106 cells/well in 6 well plates) were plated and incubated for 24 h. The cells were washed once with DPBS and treated in triplicate with LPS (1 µg/mL) or HNE (35 µM) + LPS (1 µg/mL). Cell culture medium was collected at 0 or 24 h, and GCSF and MMP9 protein levels were determined using commercial ELISA reagents (R&D Systems) according to manufacturer’s protocol.
Flow Cytometric Analysis
Phagocytosis and cell death assays were performed on a BD LSRII flow cytometer. At least 10,000 events per sample were collected for the determination of cell populations using FACSDiva v6.1.1. After the indicated time post serum opsonized-BH or -latex bead treatment, cells were washed with DPBS and incubated at 37 °C for 15 minutes in CellStripper non-enzymatic cell dissociation buffer (Cellgro MediaTech). Potential HNE-mediated cell death was determined by the Vybrant Apoptosis Assay Kit II (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were harvested, resuspended in assay buffer, and stained with Alexa Fluor 488 conjugated Annexin V and propidium iodide (PI). Sample analysis was performed using FloJo v8.8.2 (Treestar).
Results
Analysis of gene expression changes in BH- or HNE-treated, LPS-stimulated RAW 264.7 cells
LPS stimulated macrophage-like RAW 264.7 cells were exposed to 0.1 mg/mL BH or 35 µM HNE for 6 or 24 h. The concentrations of BH and HNE were chosen based on reported estimates of Hz (100 µM) in brain capillaries of malaria victims (23) and HNE (40 µM) levels in Hz-fed monocytes (7). Phagocytosis of opsonized-latex beads and -BH was examined by flow cytometry. Latex bead fluorescence was detected in 86 % and 99 % of the gated parent population at 6 and 24 h, respectively, demonstrating the level of phagocytosis (Supporting Information Figure 2). Accumulation of Hz within monocytes has been shown to, apart from an increase in depolarized side scatter, considerably increase conventional side scatter (24). In BH treated cells, phagocytosis of the total population was indicated by a marked increase in conventional side scatter mean fluorescence versus control cells (Supporting Information Figure 3). It was previously shown by confocal microscopy that opsonized BH was ingested by RAW 264.7 cells and localized within the phagolysosome (21). Given the possibility of HNE to induce cell death, the effect of 35 µM HNE on RAW 264.7 cell death was investigated. Changes in viable, apoptotic, and dead cell populations were measured by flow cytometry 24 h after treatment of LPS stimulated cells (Supporting Information Figure 4). Evaluation of cells with apoptosis- and necrosis-specific stains demonstrated that HNE only altered the percentage of apoptotic cells by 3.4 % relative to stimulated cells. The population of necrotic cells was unchanged regardless of HNE treatment. These values demonstrate that 24 h incubation of 35 µM HNE is well tolerated by RAW 264.7 cells.
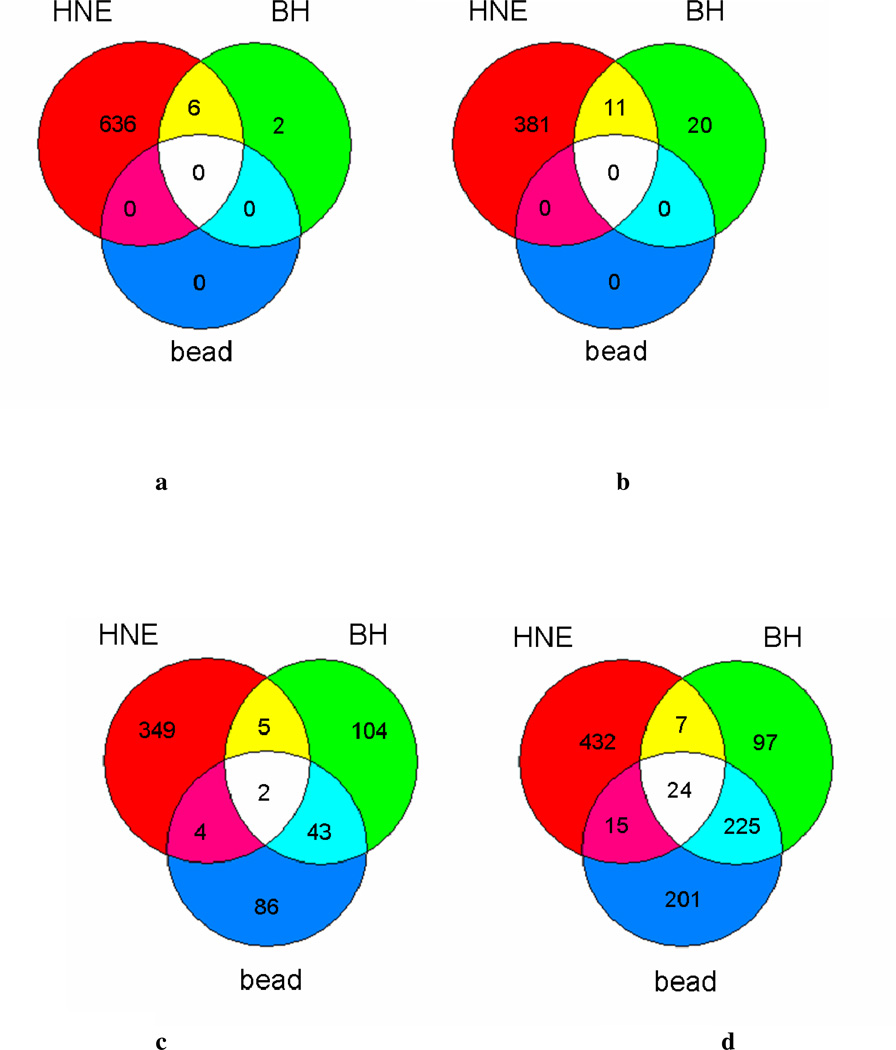
Statistically significant (p ≤ 0.01) gene expression changes (fold change ≥1.8 relative to control), where expression is considered a measurement of the RNA abundance at the time of isolation, were identified by microarray analysis. Within each treatment category, differentially expressed genes were sorted into lists based on the direction of regulation and compared to identify common changes relative to stimulated cells (Figure 1a–d). In order to identify expression changes dependent on interactions of BH rather than those due to phagocytosis, differentially expressed genes were controlled by a corresponding particulate latex bead challenge. Six hours post-challenge, there were no significant expression changes mediated by latex bead phagocytosis, and only a small group (i.e., 39 genes) altered by BH phagocytosis. Steady-state mRNA levels (24 h) demonstrate that nearly 70% of the genes differentially expressed by BH are in common with the ‘inert’ latex bead control, indicating that the response to BH is predominantly phagocytic. Consequently, phagocytosis-related genes were identified and disregarded for remaining analyses. The number of genes differentially expressed by HNE or BH treatment indicates the degree of perturbation by each of the native Hz-associated components. HNE treatment altered a significantly larger group of genes than BH treatment, suggesting a more serious impact on cellular function.
Figure 1.
Overlapping genes with significant differential expression mediated by BH and HNE. Venn diagrams show the intersection of genes that were altered by 0.1 mg/mL BH with those altered by either latex bead or 35 µM HNE treatment. Numbers represent statistically significant (p ≤ 0.01) genes up- or down-regulated ≥1.8-fold relative to LPS stimulated cells at 24 h. (a) down-regulated genes identified at 6 h, (b) up-regulated genes identified at 6 h, (c) down-regulated genes identified at 24 h, (d) up-regulated genes identified at 24 h.
Ingenuity Pathway Analysis (IPA) was used to perform a functional analysis of each dataset. IPA functional analysis ranks molecular and cellular functions according to Fischer’s Exact Test p-value. Those categories exhibiting p < 0.001 are shown in Table 2 for both BH and HNE treatment datasets at 6 and 24 h timepoints. The magnitude of response to either BH or HNE treatment is evident from the number of significantly affected biological processes identified by IPA. At 6 h, both BH and HNE affect a diverse group of functions including ‘Cell Signaling’ and ‘Small Molecule Biochemistry’ among others. By 24 h, the cellular response to BH is minimal. Given that BH does not impair microbicidal functions, is sensitive to microbicidal agents, and is degraded upon phagocytosis in RAW 264.7 cells (21), it was not surprising to find that the steady-state response to BH was modest. The degree of perturbation by HNE at 24 h complements literature observations regarding its extensive reactivity with cellular nucleophiles (15, 16, 25–29). Expression changes mediated by BH and HNE indicate differential cellular responses to both treatments as a function of time. There is, however, some overlap in the early and late response to HNE, primarily associated with ‘Stress Response’, ‘Immune Response’, and ‘Gene Expression’ (listed within Table 3).
Table 2.
Functional analysis of BH and HNE datasetsa
| Biological Function | p-valuea | |||
|---|---|---|---|---|
| BH | HNE | |||
| 6 h | 24 h | 6 h | 24 h | |
| Amino Acid Metabolism | < 10−04 | 2.03 × 10−04 | ||
| Carbohydrate Metabolism | 5.54 × 10−04 | |||
| Cell Cycle | 5.54 × 10−04 | 7.24 ×10−04 | < 10−04 | < 10−04 |
| Cell Death | 2.41 × 10−04 | < 10−04 | < 10−04 | |
| Cell Morphology | 7.10 × 10−04 | < 10−04 | < 10−04 | |
| Cell Signaling | 5.61 × 10−04 | < 10−04 | ||
| Cell-To-Cell Signaling and Interaction | < 10−04 | < 10−04 | < 10−04 | |
| Cellular Assembly and Organization | 1.61 × 10−04 | < 10−04 | < 10−04 | |
| Cellular Compromise | < 10−04 | |||
| Cellular Development | 5.54 × 10−04 | < 10−04 | < 10−04 | |
| Cellular Function and Maintenance | 2.84 × 10−04 | < 10−04 | < 10−04 | |
| Cellular Growth and Proliferation | < 10−04 | < 10−04 | ||
| Cellular Movement | < 10−04 | < 10−04 | ||
| DNA Replication, Recombination, and Repair | < 10−04 | < 10−04 | ||
| Free Radical Scavenging | 5.54 × 10−04 | |||
| Gene Expression | < 10−04 | < 10−04 | ||
| Lipid Metabolism | 5.54 × 10−04 | |||
| Molecular Transport | 5.54 × 10−04 | < 10−04 | ||
| Post-Translational Modification | 6.25 × 10−04 | < 10−04 | < 10−04 | |
| Small Molecule Biochemistry | 5.54 × 10−04 | < 10−04 | 2.03 × 10−04 | |
| Vitamin and Mineral Metabolism | < 10−04 | |||
Ingenuity Pathway Analysis uses a right-tailed Fisher Exact Test to calculate p-values. Significance values for each dataset indicate the probability that the association between the genes and the given molecular and cellular functions are due to random chance.
Table 3.
Select Gene Expression Changes Mediated by HNEa
| Gene Symbol |
Description | Fold Change | MGI Gene ID |
||
|---|---|---|---|---|---|
| 6 h | 24 h | ||||
| Cell Cycle | Atm | ataxia telangiectasia mutated homolog (human) | 1.9 | 107202 | |
| Atr | Ataxia telangiectasia and rad3 related | 3.8 | 108028 | ||
| Bub1 | budding uninhibited by benzimidazoles 1 homolog (S. cere |
−2.7 | 1100510 | ||
| Bub1b | budding uninhibited by benzimidazoles 1 homolog, beta (S. |
−2.8 | 1333889 | ||
| Ccna2 | cyclin A2 | −5.1 | 108069 | ||
| Ccnb1 | cyclin B1 | −5.2 | 88302 | ||
| Ccnd1 | cyclin D1 | 1.9 | 88313 | ||
| Ccnf | cyclin F | −4.1 | 102551 | ||
| Ccng2 | cyclin G2 | −2.9 | 1095734 | ||
| Ccr1 | chemokine (C-C motif) receptor 1 | −3.1 | 104618 | ||
| Cdc20 | cell division cycle 20 homolog (S. cerevisiae) | −2.3 | 1859866 | ||
| Cdc25a | cell division cycle 25 homolog A (S. pombe) | −1.9 | 103198 | ||
| Cdc25c | cell division cycle 25 homolog C (S. pombe) | −3.5 | 88350 | ||
| Cdk6 | cyclin-dependent kinase 6 | 1.9 | 1277162 | ||
| Chek1 | checkpoint kinase 1 homolog (S. pombe) | −2.3 | 1202065 | ||
| Dbf4 | DBF4 homolog (S. cerevisiae) | −2.0 | 1351328 | ||
| E2f2 | E2F transcription factor 2 | 2.0 | 1096341 | ||
| Fen1 | flap structure specific endonuclease 1 | 2.5 | 102779 | ||
| Gadd45a | growth arrest and DNA-damage-inducible 45 alpha | 2.5 | 107799 | ||
| Mki67 | antigen identified by monoclonal antibody Ki 67 | −4.0 | 106035 | ||
| Msh5 | mutS homolog 5 (E. coli) | 6.2 | 1329021 | ||
| Mutyh | mutY homolog (E. coli) | 3.1 | 1917853 | ||
| Mxd1 | MAX dimerization protein 1 | −4.7 | 96908 | ||
| Ndc80 | NDC80 homolog, kinetochore complex component (S. cerevisia |
−2.4 | 1914302 | ||
| Pa2g4 | proliferation-associated 2G4 | 2.3 | 894684 | ||
| Pcna | proliferating cell nuclear antigen | 2.3 | 97503 | ||
| Plk1 | polo-like kinase 1 (Drosophila) | −8.6 | 97621 | ||
| Rad23a | RAD23a homolog (S. cerevisiae) | 2.2 | 105126 | ||
| Rad51 | RAD51 homolog (S. cerevisiae) | 16.3 | 97890 | ||
| Rbl1 | retinoblastoma-like 1 (p107) | 2.7 | 103300 | ||
| Riok3 | RIO kinase 3 | 1.9 | 1914128 | ||
| Sass6 | spindle assembly 6 homolog (C. elegans) | −2.4 | −4.6 | 1920026 | |
| Suv39h1 | suppressor of variegation 3–9 homolog 1 (Drosophila) |
2.4 | 1099440 | ||
| Tgfb | transforming growth factor, beta 1 | 2.0 | 98725 | ||
| Xaf1 | XIAP associated factor 1 | −2.1 | −87.4 | 3772572 | |
| Zwilch | Zwilch, kinetochore associated, homolog (Drosophila) |
−2.3 | 1915264 | ||
| Cell Signaling | Fcgr1a | Fc fragment of IgG, high affinity Ia, receptor (CD64) | −4.4 | −13.6 | 95498 |
| Fcgr2b | Fc fragment of IgG, low affinity IIb, receptor (CD32) | −7.7 | −2.9 | 95499 | |
| Cellular Development | Cd83 | CD83 molecule | −2.4 | −2.4 | 1328316 |
| Ifi16 | interferon, gamma-inducible protein 16 | −4.1 | −11.6 | 96429 | |
| Irf7 | interferon regulatory factor 7 | −3.4 | −51.3 | 1859212 | |
| Mafb | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) |
−4.4 | −2.2 | 104555 | |
| Dyserythropoiesis | Ccl5 | chemokine (C-C motif) ligand 5 | −22.5 | 98262 | |
| Traf3 | Tnf receptor-associated factor 3 | −2.3 | 108041 | ||
| Tsc22d3 | TSC22 domain family, member 3 | −3.6 | 1196284 | ||
| ECM degradation | Mmp9 | matrix metalloproteinase 9 | −4.0 | 5.3 | 97011 |
| Timp1 | tissue inhibitor of metalloproteinase 1 | −8.5 | - | 98752 | |
| Gene Expression | Axud1 | AXIN1 up-regulated 1 | −3.0 | −1.8 | 2387989 |
| Batf2 | basic leucine zipper transcription factor, ATF-like 2 | −1.8 | −3.1 | 1921731 | |
| Ddx58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −2.4 | −14.2 | 2442858 | |
| Mx1 | myxovirus (influenza virus) resistance 1, interferon- inducible protein p78 (mouse) |
−7.9 | −43.4 | 97243 | |
| Pcgf5 | polycomb group ring finger 5 | −1.8 | −2.1 | 1923505 | |
| Phf11 | PHD finger protein 11 | −2.9 | −13.7 | 1918441 | |
| Sp100 | SP100 nuclear antigen | −2.8 | −4.4 | 109561 | |
| Glutathione Metabolism | G6pd2 | glucose-6-phosphate dehydrogenase 2 | 1.8 | 2.0 | 105977 |
| G6pdx | glucose-6-phosphate dehydrogenase X-linked | 2.1 | 105979 | ||
| Gclc | glutamate-cysteine ligase, catalytic subunit | 8.6 | 104990 | ||
| Gclm | glutamate-cysteine ligase , modifier subunit | 7.6 | 104995 | ||
| Gss | glutathione synthetase | 2.4 | 95852 | ||
| Gsta1 | glutathione S-transferase, alpha 1 (Ya) | 7.2 | 30.7 | 1095417 | |
| Gstp1 | glutathione S-transferase, pi 1 | 1.8 | 95865 | ||
| Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | 3.3 | 96413 | ||
| Pgd | phosphogluconate dehydrogenase | 2.4 | 97553 | ||
| Immune Response | Adrb2 | adrenergic, beta-2-, receptor, surface | 4.1 | 2.2 | 87938 |
| C5r1 | complement component 5, receptor 1 | 3.5 | 88232 | ||
| Casp4 | caspase 4, apoptosis-related cysteine peptidase | −2.2 | −2.4 | 107700 | |
| Ccl17 | chemokine (C-C motif) ligand 17 | 3.1 | 1329039 | ||
| Ccl2 | chemokine (C-C motif) ligand 2 | −44.8 | 98259 | ||
| Ccl22 | chemokine (C-C motif) ligand 22 | −12.3 | 1306779 | ||
| Ccl4 | chemokine (C-C motif) ligand 4 | −2.1 | 98261 | ||
| Ccl6 | chemokine (C-C motif) ligand 6 | −3.1 | 98263 | ||
| Ccl7 | chemokine (C-C motif) ligand 7 | −22.6 | −2.3 | 99512 | |
| Ccr1 | chemokine (C-C motif) receptor 1 | −3.1 | 104618 | ||
| Cd14 | CD14 antigen | 2.2 | 88318 | ||
| Cd300lf | CD300 antigen like family member F | −3.9 | 2442359 | ||
| Cd40 | CD40 antigen | −4.3 | 88336 | ||
| Cd44 | CD44 antigen | −2.6 | 88338 | ||
| Cd86 | CD86 antigen | −1.8 | 101773 | ||
| Cenpa | centromere protein A | −2.3 | 88375 | ||
| Cfb | complement factor B | −7.4 | 105975 | ||
| Clec12a | C-type lectin domain family 12, member a | −1.9 | 3040968 | ||
| Clec2d | C-type lectin domain family 2, member d | −4.1 | 2135589 | ||
| Clec4n | C-type lectin domain family 4, member n | −2.4 | 1861231 | ||
| Clec5a | C-type lectin domain family 5, member a | −1.8 | 1345151 | ||
| Csf2 | colony stimulating factor 2 (granulocyte- macrophage) |
−12.1 | 1339752 | ||
| Csf3 | colony stimulating factor 3 (granulocyte) | −18.2 | 18.0 | 1339751 | |
| Cxcl1 | chemokine (C-X-C motif) ligand 1 | 2.7 | 108068 | ||
| Cxcl14 | chemokine (C-X-C motif) ligand 14 | −2.7 | 1888514 | ||
| Cxcr4 | chemokine (C-X-C motif) receptor 4 | 2.5 | 109563 | ||
| Dcn | decorin | −4.0 | 94872 | ||
| Ercc1 | excision repair cross-complementing rodent repair deficient |
−1.8 | 95412 | ||
| F10 | coagulation factor X | 4.1 | 103107 | ||
| Fbxo5 | F-box protein 5 | −4.0 | 1914391 | ||
| Gbp1 | guanylate binding protein 1 | −3.1 | −16.8 | 95666 | |
| Gbp3 | guanylate nucleotide binding protein 3 | −3.7 | 1926263 | ||
| Gbp5 | guanylate nucleotide binding protein 5 | −5.9 | 2429943 | ||
| H28 | histocompatibility 28 | −6.3 | 95975 | ||
| Hdc | histidine decarboxylase | −8.8 | −3.1 | 96062 | |
| Icam1 | intercellular adhesion molecule 1 | 1.8 | 7.5 | 96392 | |
| Igf1 | insulin-like growth factor 1 | 2.1 | 96432 | ||
| Il10 | interleukin 10 | −4.7 | 96537 | ||
| Il10ra | interleukin 10 receptor, alpha | 3.7 | 96538 | ||
| Il13ra1 | interleukin 13 receptor, alpha 1 | −3.4 | 105052 | ||
| Il18 | interleukin 18 | −2.0 | 107936 | ||
| Il18rap | interleukin 18 receptor accessory protein | −2.8 | 1338888 | ||
| Il1a | interleukin 1 alpha | −66.6 | 96542 | ||
| Il1b | interleukin 1 beta | −32.7 | 96543 | ||
| Il1f6 | interleukin 1 family, member 6 | −12.6 | 1859324 | ||
| Il1rl1 | interleukin 1 receptor-like 1 | −3.6 | 98427 | ||
| Il1rn | interleukin 1 receptor antagonist | −6.8 | 96547 | ||
| Il27 | interleukin 27 | −6.4 | 2384409 | ||
| Il4ra | interleukin 4 receptor, alpha | −2.9 | 105367 | ||
| Il6 | interleukin 6 | −43.3 | −11.2 | 96559 | |
| Isg20 | interferon stimulated exonuclease gene 20kDa | −3.1 | −7.6 | 1928895 | |
| Ltb | lymphotoxin B | −1.9 | 104796 | ||
| Nlrc4 | NLR family, CARD domain containing 4 | 2.3 | 2.7 | 3036243 | |
| Oasl2 | 2'–5' oligoadenylate synthetase-like 2 | −5.1 | 1344390 | ||
| Pgdfb | platelet derived growth factor, B polypeptide | 3.2 | 97528 | ||
| Pla2g7 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
3.1 | 10.7 | 1351327 | |
| Pou2f2 | POU domain, class 2, transcription factor 2 | −1.9 | 101897 | ||
| Rsad2 | radical S-adenosyl methionine domain containing 2 | −4.4 | 1929628 | ||
| Tnf | tumor necrosis factor | 8.7 | 104798 | ||
| Traf3ip2 | Traf3 interacting protein 2 | −1.8 | 2143599 | ||
| Ube2L6 | ubiquitin-conjugating enzyme E2L 6 | −1.8 | −2.3 | 1914500 | |
| Vegfa | vascular endothelial growth factor A | −2.4 | 103178 | ||
| Interferon-associated Signaling or Regulation |
H2-Bf | histocompatibility 2, complement component factor B |
−51.0 | 105975 | |
| H2-DMb2 | histocompatibility 2, class II, locus Mb2 | −5.1 | 95923 | ||
| H2-Q1 | histocompatibility 2, Q region locus 1 | −3.2 | 95928 | ||
| H2-Q5 | histocompatibility 2, Q region locus 5 | −2.1 | 95934 | ||
| H2-T23 | histocompatibility 2, T region locus 23 | −9.4 | 95957 | ||
| H2- T9/H2- T22 |
histocompatibility 2, T region locus 9;histocompatibility 2, T region locus 22 |
−5.2 | 95965 | ||
| Ifi202b | interferon activated gene 202B | −3.8 | 1347083 | ||
| Ifi203 | interferon activated gene 203 | −3.7 | 96428 | ||
| Ifi204 | interferon activated gene 204 | −3.7 | 96429 | ||
| Ifi205 | interferon activated gene 205 | −4.3 | 101847 | ||
| Ifi47 | interferon gamma inducible protein 47 | −3.0 | 99448 | ||
| Ifih1 | interferon induced with helicase C domain 1 | −2.0 | 1918836 | ||
| Ifit1 | interferon-induced protein with tetratricopeptide repeats 1 |
−3.6 | 99450 | ||
| Ifit2 | interferon-induced protein with tetratricopeptide repeats 2 |
−6.3 | 99449 | ||
| Ifit3 | interferon-induced protein with tetratricopeptide repeats 3 |
−3.3 | 1101055 | ||
| Irf8 | interferon regulatory factor 8 | −2.6 | 96395 | ||
| Jak2 | Janus kinase 2 | −1.7 | 96629 | ||
| Mx2 | myxovirus (influenza virus) resistance 2 | −7.3 | 97244 | ||
| Oas1a | 2’–5’ oligoadenylate synthetase 1A | −2.0 | 2180860 | ||
| Ptges | prostaglandin E synthase | 4.7 | 1927593 | ||
| Stat1 | signal transducer and activator of transcription 1 | −2.2 | −10.8 | 103063 | |
| Stat3 | signal transducer and activator of transcription 3 | −2.1 | 103038 | ||
| Stat5a | signal transducer and activator of transcription 5A | −2.1 | 103036 | ||
| Metabolic Process | Adh7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
8.0 | 2.3 | 87926 |
| Hbp1 | high mobility group box transcription factor 1 | 2.9 | 894659 | ||
| Mov10 | Mov10, Moloney leukemia virus 10, homolog (mouse) |
−2.2 | −2.5 | 97054 | |
| Oas3 | 2'–5'-oligoadenylate synthetase 3, 100kDa | −2.4 | −8.3 | 2180850 | |
| Parp12 | poly (ADP-ribose) polymerase family, member 12 | −2.1 | −4.9 | 2143990 | |
| Serpinb1b | serine (or cysteine) peptidase inhibitor, clade B, member 1b |
2.8 | 4.3 | 2445361 | |
| Tiparp | TCDD-inducible poly(ADP-ribose) polymerase | −1.8 | −2.9 | 2159210 | |
| NF-kB signaling | Ikbke | inhibitor of kappaB kinase epsilon | 3.2 | 1929612 | |
| Nfkbia | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha |
1.8 | 104741 | ||
| Nfkbie | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
3.6 | 1194908 | ||
| Oxidative Stress Response |
Abcc1 | ATP-binding cassette, sub-family C (CFTR | 2.1 | 102676 | |
| Akr1a4 | aldo-keto reductase family 1, member A4 (aldehyde reducta |
1.8 | 1929955 | ||
| Aox1 | aldehyde oxidase 1 | 2.2 | 88035 | ||
| Cat | catalase | 3.1 | 88271 | ||
| Dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | 5.3 | 1914285 | ||
| Ephx1 | epoxide hydrolase 1, microsomal | 2.0 | 3.2 | 95405 | |
| Hmox1 | heme oxygenase (decycling) 1 | 6.0 | 96163 | ||
| Mapk14 | mitogen-activated protein kinase 14 | 2.0 | 1346865 | ||
| Pik3cb | phosphatidylinositol 3-kinase, catalytic, beta polypeptid |
2.3 | 1922019 | ||
| Prdx1 | peroxiredoxin 1 | 2.9 | 3.1 | 99523 | |
| Raf1 | v-raf-leukemia viral oncogene 1 | 2.2 | 97847 | ||
| Sod2 | superoxide dismutase 2, mitochondrial | 1.7 | 98352 | ||
| Sqstm1 | sequestosome 1 | 3.1 | 107931 | ||
| Txnrd1 | thioredoxin reductase 1 | 2.1 | 1354175 | ||
| Xdh | xanthine dehydrogenase | 2.0 | 98973 | ||
| Signal Transduction | Fcrl1 | Fc receptor-like 1 | 3.5 | 3.9 | 2442862 |
| Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | −1.9 | 96892 | ||
| Rasgrp3 | RAS guanyl releasing protein 3 (calcium and DAG- regulated) |
3.9 | 4.4 | 3028579 | |
| Rit1 | Ras-like without CAAX 1 | 2.1 | 108053 | ||
| Small Molecule Biochemistry |
Cp | ceruloplasmin (ferroxidase) | −7.7 | −6.0 | 88476 |
| Slc7A2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
−5.2 | −5.2 | 99828 | |
| Cell Structure | Gsn | gelsolin | 2.5 | 95851 | |
| Stmn1 | stathmin 1 | 10.8 | 96739 | ||
| Tuba4 | tubulin, alpha 4 | 2.5 | 1095410 | ||
| Ubiquitin-Proteasome Pathway |
Fbxl17 | F-box and leucine-rich repeat protein 17 | 1.8 | 1354704 | |
| Fbxl20 | F-box and leucine-rich repeat protein 20 | 2.1 | 1919444 | ||
| Fbxo22 | F-box only protein 22 | 3.0 | 1926014 | ||
| Fbxo30 | F-box protein 30 | 2.0 | 1919115 | ||
| Fbxo31 | F-box protein 31 | 1.8 | 1354708 | ||
| Herc3 | hect domain and RLD 3 | 2.3 | 1921248 | ||
| Map1lc3b | microtubule-associated protein 1 light chain 3 beta | 2.0 | 1914693 | ||
| Psmc3ip | proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
6.4 | 1098610 | ||
| Psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
2.0 | 1914247 | ||
| Rnf128 | ring finger protein 128 | 4.2 | 1914139 | ||
| Rnf167 | ring finger protein 167 | 1.9 | 1917760 | ||
| Ube2d3 | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 | 2.4 | 1913355 | ||
| Ube2i | ubiquitin-conjugating enzyme E2I | 3.0 | 107365 | ||
| Ube2t | ubiquitin-conjugating | 7.1 | 1914446 | ||
| Ube4b | ubiquitination factor E4B, UFD2 homolog (S. | 1.8 | 1927086 | ||
| Uchl1 | ubiquitin carboxy-terminal hydrolase L1 | 4.4 | 103149 | ||
| Usp18 | ubiquitin specific protease 18 | −224.6 | 1344364 | ||
| Other | Arrdc3 | arrestin domain containing 3 | 7.0 | 2145242 | |
| Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | 2.0 | 1197519 | ||
| Epsti1 | epithelial stromal interaction 1 (breast) | −2.1 | −10.6 | 1915168 | |
| Gabarapl1 | gamma-aminobutyric acid (GABA(A)) receptor- | 2.8 | 1914980 | ||
| Id1 | inhibitor of DNA binding 1 | −2.0 | 96396 | ||
| Ier3 | immediate early response 3 | −2.0 | 104814 | ||
| Ifi203 | interferon activated gene 203 | −3.7 | −5.2 | 96428 | |
| Klhl6 | kelch-like 6 (Drosophila) | −1.9 | −3.8 | 2686922 | |
| Map1d | methionine aminopeptidase 1D | 1.8 | 1.9 | 1913809 | |
| Ms4A6D | membrane-spanning 4-domains, subfamily A, | −2.3 | −3.9 | 1916024 | |
| Top2a | topoisomerase (DNA) II alpha | 13.2 | 98790 | ||
| Trim30 | tripartite motif-containing 30 | −7.2 | −31.8 | 98178 | |
| Uvrag | UV radiation resistance associated gene | −1.9 | −1.8 | 1925860 | |
| Zak | sterile alpha motif and leucine zipper containing | −1.9 | −4.0 | 2443258 | |
| Zbp1 | Z-DNA binding protein 1 | −5.7 | −69.4 | 1927449 | |
| Zbtb20 | zinc finger and BTB domain containing 20 | 3.6 | 1929213 | ||
| Zfp36 | zinc finger protein 36 | −1.9 | 99180 | ||
Genes altered ≥1.8-fold (p ≤ 0.01) up or down in LPS stimulated HNE-treated cells relative to LPS stimulated cells. Fold changes (FC) represent the average of three independent biological experiments.
Differential gene expression in the context of malaria pathogenesis
Microarray data from this study were compared to two groups of genes. The first group consists of specific genes or gene products that are associated with human (30, 31) or murine (32, 33) models of malarial infection, and/or BH or Hz (9, 10) exposure. The second group includes genes that are classified with specific biological processes that are over-expressed in a murine P. yoelii model (33) and/or naturally-acquired P. falciparum infections (34) (e.g., cell-cell signaling, defense response, immune response, inflammatory response, and signal transduction). Complete lists of differentially expressed genes altered by either HNE or BH treatment that fall within these categories are listed in Supporting Information Tables 1–9.
Validation of microarray results
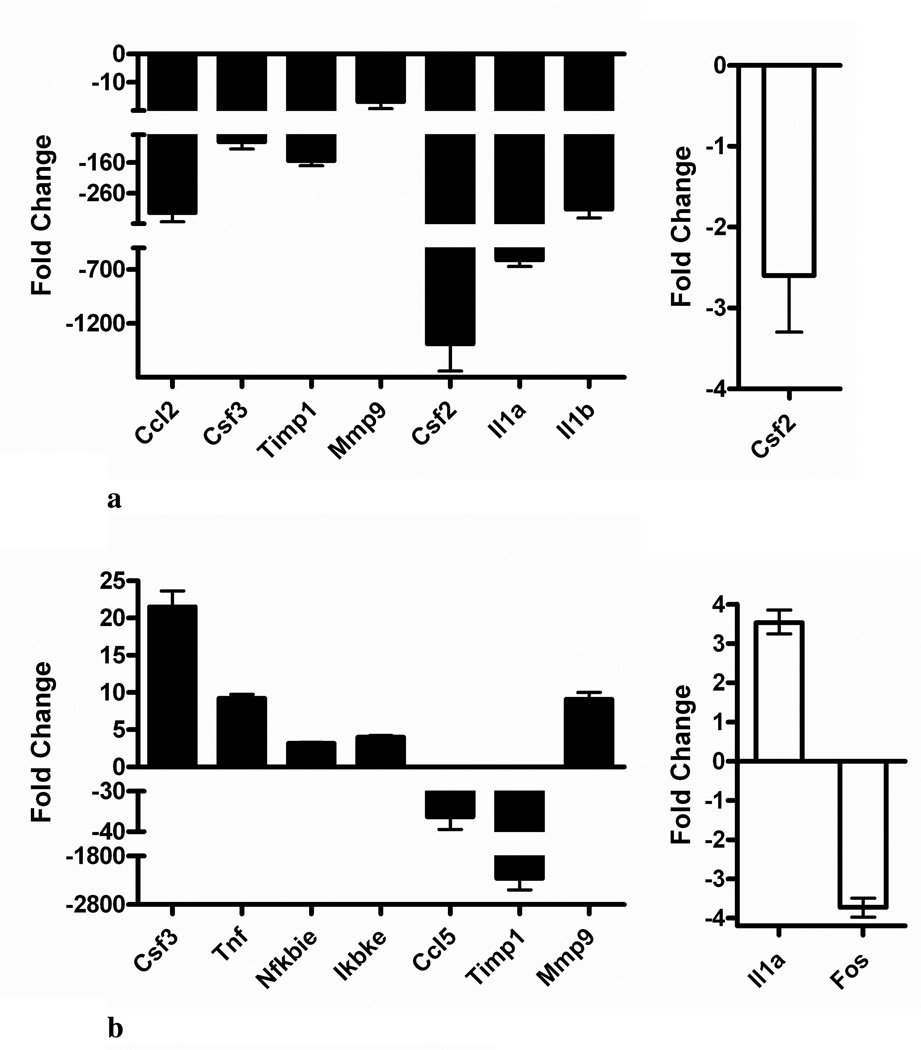
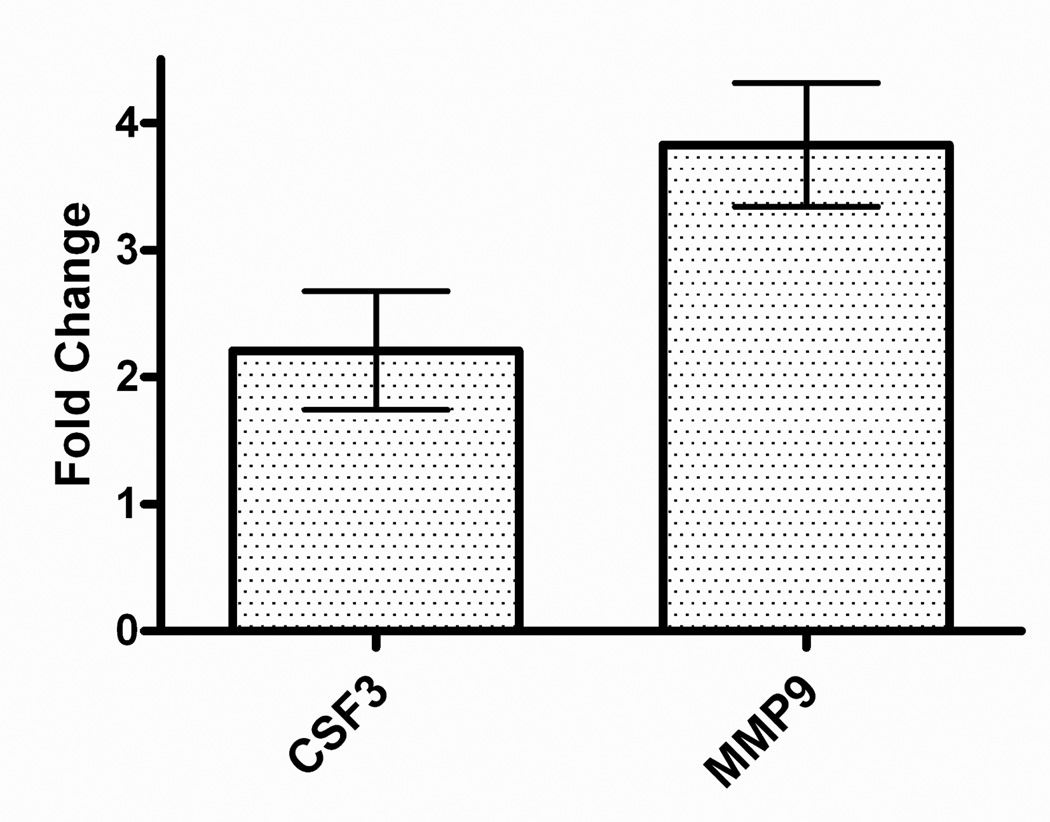
qRT-PCR was used to confirm several genes susceptible to differential regulation by HNE and BH at 6 h and 24 h (Table 1). Analysis was focused on selected genes implicated in the host response to malaria. The results shown in Figure 2 are expressed as fold change relative to stimulated cells (control). At 6 h, RT-PCR confirmed that HNE repressed the expression of chemokine (C-C motif) ligand 2 (Ccl2), colony stimulating factor 3 (granulocyte) (Csf3), tissue inhibitor of metalloproteinase 1 (Timp1), matrix metalloproteinase 9 (Mmp9), Csf2, interleukin (Il) 1 alpha (Il1a) and 1 beta (Il1b), and BH down-regulated colony stimulating factor 2 (granulocyte-macrophage) (Csf2) relative to stimulated cells. At 24 h, HNE enhanced expression of Mmp9, nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon (Nfkbie), inhibitor of kappaB kinase epsilon (Ikbke), tumor necrosis factor (Tnf), and Csf3. Concurrently, Timp1 and chemokine (C-C motif) ligand 5 (Ccl5) genes were repressed. RT-PCR analyses of BH-treated cells at 24 h confirmed Il1a stimulation and FBJ osteosarcoma oncogene (Fos) suppression. These results are consistent with the microarray data. ELISA studies showed that HNE treatment induced MMP9 and CSF3 translation and release relative to stimulated cells (Figure 3).
Figure 2.
Quantitative real-time RT-PCR validation of microarray results. RAW 264.7 cells were stimulated with 0.1 µg/mL LPS and untreated or treated (A) for 6 h or (B) for 24 h with either 35 µM HNE (black bars) or 0.1 mg/mL BH (white bars). Fold-changes (treated, stimulated cells relative to stimulated controls) are shown (X̄ ± 99% confidence interval for quadruplicate measurements of n = 3 biological replicates). Abbreviations: chemokine (C-C motif) ligand (Ccl); colony stimulating factor (Csf); tissue inhibitor of metalloproteinase 1 (Timp1); matrix metalloproteinase 9 (Mmp9); interleukin 1 (Il1); tumor necrosis factor (Tnf); nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon (Nfkbie); inhibitor of kappaB kinase, epsilon (Ikbke); and FBJ osteosarcoma oncogene (Fos).
Figure 3.
ELISA validation of microarray results. Equivalent numbers of cells (4 × 106/well) were stimulated with LPS (1 µg/mL) in the absence or presence of 35 µM HNE for 24 h. CSF3 (colony stimulating factor 3 (granulocyte)) and MMP-9 (matrix metalloproteinase 9) released into the culture medium were analyzed by ELISA. Fold changes (HNE-treated stimulated cells relative to stimulated controls) are shown (X̄ ± SD for triplicate measurements, representative of three independent experiments).
Discussion
It has been suggested that the immunological activity resulting from native Hz may not be due to the biomineral itself, but to a toxin that is presented on its surface (35). Lipid peroxidation products have been implicated as potential non-specific Hz toxins. Heme compounds are effective mediators of non-enzymatic lipid oxidation, supporting the premise that these products arise from the oxidation of lipids by Hz. Further, purified Hz and BH have been shown to drive the oxidation of arachidonic acid to racemic mixtures of HETEs, HODEs, (17, 19) and HNE (18). In a native state, these molecules are adsorbed to the surface of the biomineral and introduced into the cell during phagocytosis of Hz.
Several groups have reported conflicting results concerning the effects of the biomineral in both in vitro and in vivo systems (36–38). As Hz is complex in nature, the state of the material must be well-defined in order to identify which components are responsible for a given immune response. Native Hz has been isolated from parasitized erythrocytes and gently washed to remove cellular debris but left with a lipid coat adsorbed onto the hydrophobic porphyrin plane (12). Purified Hz refers to native Hz whose lipid coat has been removed (39, 40). Finally, BH has been prepared and purified from hemin, and is consequently devoid of all biologically-derived components (20, 37, 41, 42). Many of the reported differences in cellular response are likely attributable to the source of the biomineral (native vs. synthetic), purification procedure, and the method of macrophage activation (e.g., LPS, interferon-γ (IFN-γ), LPS+IFN-γ, or PMA). In order to facilitate consistent interpretation of the response to individual Hz components in the current study, the impact of BH and HNE was investigated on LPS-activated macrophage-like cells.
Given the relationship between high levels of Hz observed in victims, severity of infection, and disruption of macrophage function, microarray technology was used to profile expression changes mediated by two defined Hz-derived components, BH and HNE, in LPS-stimulated macrophage-like RAW 264.7 cells. Expression changes are calculated relative to LPS-stimulated cells to measure the response to each hemozoin component in an activated cell.
Stress response
Regulation of antioxidant response element (ARE) gene expression is controlled by the transcription factor NRF2, whose activity is suppressed through the formation of a complex with its inhibitor Keap1. Disruption of NRF2/Keap1 complex liberates NRF2, allowing its nuclear translocation and subsequent activation of ARE-dependent gene expression. Consistent with previous findings in a variety of cell types treated with HNE (8, 15), a potent “oxidative stress response mediated by Nrf2” was observed at both 6 h and 24 h following HNE treatment, suggesting an effort to reduce cellular damage (Table 3). Expression of genes encoding phase I and II metabolizing enzymes and antioxidant response proteins was significantly enhanced by HNE. Activation of a stress response is consistent with an increase of Prdx1 in the spleens of P. berghei infected mice (32) and Sod2 and Hmox1 in both the blood of acute pediatric malaria victims (30) and Hz-loaded placental tissue (31). A common stress response is heme oxygenase (decycling) 1 (Hmox1) induction. In the current study Hmox1 expression is induced by both HNE and BH at 6 h. This observation is in agreement with up-regulated Hmox1 expression in mouse peritoneal macrophages (PM) treated with BH (43).
Cell cycle checkpoint signaling
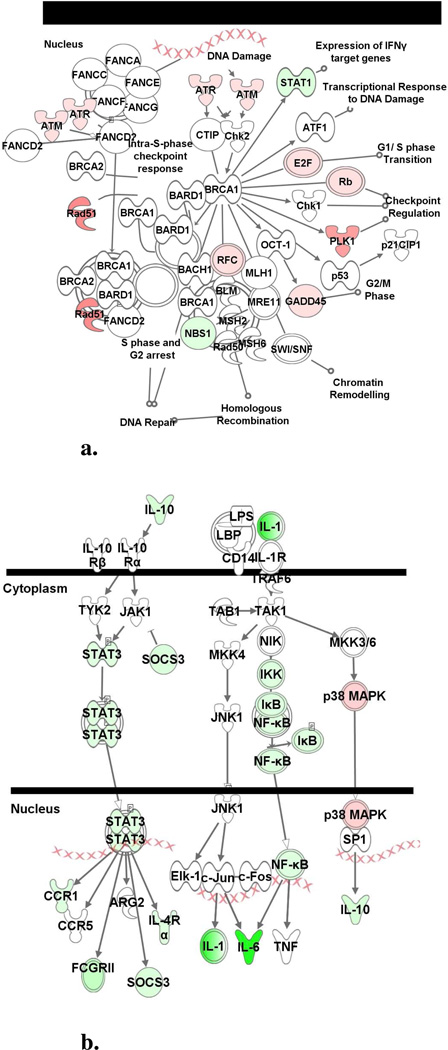
The present data indicate a broad ‘DNA replication, Recombination, and Repair’ response to HNE at 6 and 24 h. Early expression of several genes associated with checkpoint control is repressed. By 24 h, however, a dramatic DNA damage response including the induction of a number of genes associated with G1/S cell cycle checkpoint regulation was observed (Figure 4 and Table 3). Since HNE can form adducts with deoxyguanosine residues (44), activation of cell cycle checkpoint signaling genes suggests an effort to ameliorate DNA injury. The presence of damaged DNA is consistent with enhanced expression of excision repair and mismatch repair genes at 24 h. Notably, increased ATM and RAD23A expression has also been identified in whole blood of P. falciparum infected children (30).
Figure 4.
Ingenuity canonical pathway analysis. Differentially expressed genes were mapped to the Ingenuity Canonical Pathway library to identify significantly altered canonical signaling pathways. (a) ‘IL-10 Signaling’ and (b) ‘Role of BRCA1 in DNA Damage Response’ were influenced by 35 µM HNE at 24 h and 6 h, respectively. Genes or gene products are represented as nodes and the intensity of the node color indicates the degree of up- (red) or down- (green) regulation.
Macrophage activation
An exacerbated inflammatory response is assumed to play a significant role in malaria pathogenesis. In an effort to uncover the basis of the inflammatory activity, Hz has been examined as a contributing agent (9, 10). This microarray analysis demonstrates that BH had a modest effect on the induction of immune response genes at 6 h and 24 (Table 4). HNE treatment, however, drastically repressed expression of immune/inflammatory response genes (Figure 4b) and interferon-signaling and -regulated genes at 6h. By 24 h, the response was reversed and the induction of a large group of immune/inflammatory response genes was identified (Table 3). Notably, this change in response over time may explain the often contradictory interpretation of Hz and immune cell interactions reported in the literature (9, 10, 12, 36–38, 45).
Table 4.
Select Gene Expression Changes Mediated by BHa
| Gene Symbol |
Description | Fold Change | MGI Gene ID |
||
|---|---|---|---|---|---|
| 6 h | 24 h | ||||
| Interferon- associated Signaling or Regulation |
H2-Ab1 | histocompatibility 2, class II antigen A, beta 1 |
−1.8 | 103070 | |
| H2-Q1 | histocompatibility 2, Q region locus | −4.8 | 95928 | ||
| Immune Response |
Ccl2 | chemokine (C-C motif) ligand 2 | 2.2 | 98259 | |
| Ccl6 | chemokine (C-C motif) ligand 6 | 5.4 | 98263 | ||
| Cd2 | CD2 antigen | 102.4 | 88320 | ||
| Csf2 | colony stimulating factor 2 (granulocyte-macrophage) |
−12.1 | 1339752 | ||
| Cxcl2 | chemokine (C-X-C motif) ligand 2 | 1.9 | 1340094 | ||
| Ereg | epiregulin | 13.2 | 107508 | ||
| Fos | FBJ osteosarcoma oncogene | −3.6 | 95574 | ||
| Il1a | interleukin 1 alpha | 6.0 | 96542 | ||
| Il20 | interleukin 20 | 2.4 | 1890473 | ||
| Oxidative Stress Response |
Hmox1 | heme oxygenase (decycling) 1 | 6.0 | 96163 | |
| Cell Structure | Pstpip2 | proline-serine-threonine phosphatase interacting protein 2 |
6.1 | 1335088 | |
| Tuba4 | tubulin, alpha 4 | 1.9 | 1095410 | ||
Genes altered ≥1.8-fold (p ≤ 0.01) up or down in LPS stimulated BH-treated cells relative to LPS stimulated cells. Fold changes (FC) represent the average of three independent biological experiments.
A number of specific studies probing the inflammatory response to malaria are consistent with the observed HNE-mediated gene expression changes: murine kidneys infected with P. berghei display increased expression of TNF-α (46), and P. falciparum exposed CHO cells exhibit increased levels of TNF-α, G-CSF, and TGF-β (47). Several individual expression changes also correspond with responses observed in microarray analyses of genuine or experimental malaria. For example, messages for LY96, CD14 and C5R1 in whole blood of pediatric victims (30) CD2, C5R1 and IL10RA in placental malaria (31), and Cd2 in a P. yoelii infected mice (33) are consistent with enhanced expression by HNE in the current analysis.
Although HNE and BH augment the expression of genes involved in mounting an immune response, they repress the expression of a number of genes central to a cell’s antigen presenting ability (Tables 3 and 4). The present study identified significant (p < 0.05) steady-state repression of antigen major histocompatibility complex (MHC) class II-associated genes in both HNE- and BH-treated cells. Notably, Hz loading has been implicated as a factor contributing to both impaired monocyte MHC class II antigen presentation (22) and defective dendritic cell function (14, 48). The ability of HNE, and to a lesser extent BH, to suppress MHC II expression implies two Hz components involved in the defective responses. Furthermore, elevated levels of TGF-β and PGE2 are associated with T-cell inhibition (49). HNE enhanced the expression of genes encoding Tgfb and Ptges and may contribute to the impaired T-cell activity observed upon Hz phagocytosis.
NF-κB signaling
The LPS mediated pathway to produce nitric oxide (NO) is well characterized and entails NF-κB signal transduction. In quiescent cells, NF-κB is sequestered by inhibitory proteins (IκB) in the cytoplasm. Upon activation, IκB kinase (IKK) phosphorylates IκB, triggering its polyubiquitination and subsequent degradation. Once NF-κB is liberated from IκB, NF-κB translocates to the nucleus and regulates gene expression, including iNOS.
It has been shown that serum withdrawal in RAW 264.7 cells results in the activation of NF-κB, expression of iNOS, and synthesis of NO. However, serum withdrawal-mediated IκB phosphorylation and downstream signaling was abolished in HNE treated cells (50). In accord with this data, HNE prevented NO production in LPS-stimulated RAW 264.7 cells (21). HNE has been shown to covalently adduct to IKK, inhibiting kinase activity thus preventing the phosphorylation of IκB (28). As a result, IκB degradation and NF-κB translocation are impaired.
Unlike HNE, there are a wide range of observations regarding the effects of Hz and BH on iNOS activity and NO synthesis. For example, both BH and purified Hz do not inhibit IFN-γ mediated NO in B10R murine macrophage cells (40). Similarly, RAW 264.7 cells stimulated by LPS and loaded with BH exhibit normal NO levels (21). In contrast, LPS-mediated NO production is reduced in BH treated murine PM cells (43). Skorokhod et al. found that levels of NO are not impaired in several murine phagocytic cell lines after crude Hz or BH loading, but determined that human monocytes are unable to produce NO when stimulated with either LPS or IFN-γ (51). Further, native Hz decreases NO in LPS or IFN-γ stimulated murine PM suggesting that a non-heme moiety component is responsible for the dysfunction (52). This varied group of results demonstrates the need for careful extrapolation of NO production data based on cell type, stimulatory molecule, and the state of the Hz preparation.
In the present study, HNE, and not BH, had an impact on NF-κB related gene expression (Table 3). Early changes in expression indicate repression of the NF-κB pathway (down-regulated Cd40, Nfkb1, and Nfkbiz levels) by HNE. However, at 24 h, IKK (i.e., Ikbke) and IκB (Nfkbia, Nfkbie) expression is enhanced. Notably, transcript abundance does not necessarily correlate with protein level or kinase activity. In accord with the HNE studies mentioned above, IKK expression may be increased because the available enzyme is inactivated by HNE. Due to the numerous gene expression modulations mediated by HNE, it seems probable that most are a result of downstream effects of HNE interactions. The increase of IKK expression, however, may be a direct response to dysregulated kinase activity.
ECM degradation
A current hypothesis is that expression changes of ECM genes may have direct involvement in malaria pathogenesis, particularly in cases of cerebral malaria (CM). CM is a severe complication of P. falciparum infection that is characterized by adherence of parasitized RBC to the cerebral microvasculature. Analysis of the brain vessels from CM mouse models reveals Hz accumulation not only within parasites, but also free and within phagocytic cells (53). Further, examination of the cortex of postmortem CM victims shows slate-gray discoloration, commonly attributed to Hz deposition (54). Blood brain barrier (BBB) destruction is a major factor associated with CM (55). Matrix metalloproteinases (MMPs), secreted enzymes involved in ECM remodeling, are able to degrade basal lamina leading to BBB damage (56). Interestingly, Hz increases the transcription, translation, and activity of MMP9 in monocytes (57). Activation of MMP9 is also observed during P. falciparum infection (30) and may contribute to the disruption of endothelial basement membranes and extravasation of blood cells (57).
The activity of MMP9 is controlled by its cognate inhibitor, TIMP1. HNE exposure initially repressed Mmp9 expression, however, by 24 h the level of mRNA was significantly increased. Notably, both 6 h and steady-state mRNA levels of HNE treated samples indicate severely impaired Timp1 expression (−2000-fold at 24 h by qRT-PCR). Taken together, Mmp9 induction coupled with Timp1 repression indicates a steady state MMP9/TIMP1 imbalance that may lead to increased proteolysis of the ECM (Table 3) (57). Mmp9 expression can be regulated through a variety of signaling cascades including NF-κB, p38 MAPK, and ERK1/2 pathways (58–60). Given that HNE abrogates NF-κB mediated iNOS expression in both LPS stimulated- and serum deprived-RAW 264.7 cells, Mmp9 up-regulation in the present study is not a result of NF-κB activation (21, 50).
Active MMP9 is capable of pro-TNF-α cleavage which releases the active cytokine and promotes Mmp9 expression (61). Thus, the increased expression of Tnf discussed previously may enhance a positive feedback cycle in this study. Importantly, analyses of postmortem brain tissue of CM victims identified elevated TNF mRNA and protein (62), and immunostaining studies identified significant cerebrum, brainstem, and cerebellar localization (63).
CM victims possess several traits including obstructed microvascular flow, attributable to the sequestration of blood cells including parasitized RBC and leukocytes (64). Intercellular adhesion molecule 1 (ICAM1), one of the most important receptors involved in cytoadherence (65), is upregulated in naturally-acquired malaria and may contribute to ECM degradation (65, 66). In the current study, HNE up-regulated Icam1 expression at 6 and 24 h (Table 3). Through cell adhesion, ICAM1 aids ECM binding and may trigger macrophage accumulation and localized MMP9 activity.
Dyserythropoiesis
The specific mechanism(s) leading to malarial anemia have not been clearly defined, but several factors including dyserythropoiesis are thought to play a role (67). Casals-Pascual et al. provide evidence correlating dyserythropoiesis with Hz (68). Further, HNE and the supernatant of native Hz-fed monocytes have both been shown to dose-dependently inhibit erythroid-progenitor growth in culture (69). Repressed CCL5 has been correlated with dyserythropoiesis and may be a contributing factor (70). Interestingly, CM victims, which have been found with significant Hz accumulation in their brains (54), exhibit decreased levels of CCL5 (71). The significant repression of Ccl5 expression by HNE at 24 h suggests a potential role in dyserythropoiesis. Moreover, impaired expression of two upstream regulators of Ccl5, namely Traf3 and Tsc22d3, may be directly involved in Ccl5 repression in the current study (Table 3).
Conclusion
The host immune response to malarial infection is multifactorial, including complex innate and adaptive immune responses to the parasites, composite native Hz, Hz-derived lipid peroxidation products, and other cellular debris. Not unexpectedly, countless interactions between an array of malaria toxins and host cells result in adverse biological effects, prompting a reductionist examination of such complex systems. In the current study, gene expression analysis of HNE-treated cells supports some role for HNE in malaria pathogenesis. A wide range of substantive modulations occurred in activated cells following HNE treatment at both 6 and 24 h. The early response predominantly involves an oxidative stress activity that involves induction of ARE and glutathione metabolism genes. Steady state gene expression changes are associated with a variety of documented malaria responses such as macrophage activation, immune and inflammatory responses, NF-κB signal transduction, ECM degradation, and dyserythropoiesis. The modest, primarily phagocytic, response to BH supports the hypothesis that Hz predominantly functions as a vehicle to generate and introduce toxic mediators (e.g., HNE, hydroxylated fatty acids) that are closely associated with the biomineral into phagocytic cells (35). Further, the gene expression changes induced by HNE may be illustrative of other reactive lipid oxidation products generated by Hz. Although valuable for exploring the interactions between Hz components and immune active cells, it must be kept in perspective that there are limitations to using any model system. It is possible that stimulation antagonizes expression changes mediated by BH or HNE. However, activated immune cells are physiologically relevant in the pathology of malaria. Future studies will be aimed at comparing BH- and HNE-mediated gene expression changes with those resulting from other biologically active Hz-associated components, and confirming expression changes in primary human macrophages and monocytes.
Supplementary Material
Acknowledgment
Financial support for this work was provided by NIH (NIAID) grant R03AI060827. The Vanderbilt Microarray Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). We thank M. F. Richards for editorial assistance.
Abbreviations
- Hz
hemozoin
- HNE
4-hydroxy-2-nonenal
- BH
β-hematin
- ECM
extracellular matrix
- HETE
hydroxyeicosatetraenoic acid
- HODE
hydroxyoctadecadienoic acid
- RBC
red blood cell
- LPS
lipopolysaccharide
- PMA
phorbol-12-myristate-13-acetate
- iNOS
inducible nitric oxide synthase
- IPA
Ingenuity Pathway Analysis
- qRT-PCR
quantitative real-time reverse transcription polymerase chain reaction
- MHC
major histocompatibility complex
- NO
nitric oxide
- PM
peritoneal macrophage
- CM
cerebral malaria
- BBB
blood brain barrier
Footnotes
Supporting Information Available: Representative SEM image of BH and histogram showing the length distribution of particles (S1) and flow cytometric analyses of latex bead phagocytosis (S2), BH phagocytosis (S3), and potential cell death mediated by HNE (S4). Selected genes that are associated with (A) specific genes or gene products correlated to malarial infection or (B) genes that are classified under specific over-expressed biological processes in malaria models that are modulated ≥ 1.8 fold by 35 µM HNE or 0.1 mg/mL BH at 6 or 24 h are shown in Supplementary Tables 1–8. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Greenwood B, Mutabingwa T. Malaria in 2002. Nature. 2002;415:670–672. doi: 10.1038/415670a. [DOI] [PubMed] [Google Scholar]
- 2.Urquhart AD. Putative pathophysiological interactions of cytokines and phagocytic cells in severe human falciparum malaria. Clin. Infect. Dis. 1994;19:117–131. doi: 10.1093/clinids/19.1.117. [DOI] [PubMed] [Google Scholar]
- 3.Boutlis CS, Yeo TW, Anstey NM. Malaria tolerance - for whom the cell tolls? Trends Parasitol. 2006;22:371–377. doi: 10.1016/j.pt.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagola S, Stephens PW, Bohle DS, Kosar AD, Madsen SK. The structure of malaria pigment β-haematin. Nature. 2000;404:307–310. doi: 10.1038/35005132. [DOI] [PubMed] [Google Scholar]
- 5.Goldie P, Roth EF, Jr, Oppenheim J, Vanderberg JP. Biochemical characterization of Plasmodium falciparum hemozoin. Am. J. Trop. Med. Hyg. 1990;43:584–596. doi: 10.4269/ajtmh.1990.43.584. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzer E, Kuhn H, Valente E, Arese P. Malaria-parasitized erythrocytes and hemozoin nonenzymatically generate large amounts of hydroxy fatty acids that inhibit monocyte functions. Blood. 2003;101:722–728. doi: 10.1182/blood-2002-03-0979. [DOI] [PubMed] [Google Scholar]
- 7.Schwarzer E, Muller O, Arese P, Siems WG, Grune T. Increased Levels of 4-hydroxynonenal in human monocytes fed with malarial pigment hemozoin. FEBS Lett. 1996;338:119–122. doi: 10.1016/0014-5793(96)00523-6. [DOI] [PubMed] [Google Scholar]
- 8.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-Hydroxynonenal: A membrane lipid oxidation product of medicinal interest. Medicinal Research Reviews. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 9.Sherry BA, Alava G, Tracey KJ, Martiney J, Cerami A, Slater AF. Malaria-specific metabolite hemozoin mediates the release of several potent endogenous pyrogens (TNF, MIP-1 alpha, and MIP-1 beta) in vitro, and altered thermoregulation in vivo. J Inflamm. 1995;45:85–96. [PubMed] [Google Scholar]
- 10.Prada J, Malinowski J, Muller S, Bienzle U, Kremsner PG. Hemozoin differentially modulates the production of interleukin 6 and tumor necrosis factor in murine malaria. Eur. Cytokine Network. 1995;6:109–112. [PubMed] [Google Scholar]
- 11.Ochiel DO, Awandare GA, Keller CC, Hittner JB, Kremsner PG, Weinberg JB, Perkins DJ. Differential regulation of B-chemokines in children with Plasmodium falciparum malaria. Infect. Immun. 2005;73:4190–4197. doi: 10.1128/IAI.73.7.4190-4197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzer E, Turrini F, Ulliers D, Giribaldi G, Ginsburg H, Arese P. Impairment of macrophage functions after ingestion of Plasmodium falciparum-infected erythrocytes or isolated malarial pigment. J. Exp. Med. 1992;176:1033–1041. doi: 10.1084/jem.176.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarzer E, Arese P. Phagocytosis of malarial pigment hemozoin inhibits NADPH-oxidase activity in human monocyte-derived macrophages. Biochim. Biophys. Acta. 1996;1316:169–175. doi: 10.1016/0925-4439(96)00021-x. [DOI] [PubMed] [Google Scholar]
- 14.Skorokhod OA, Alessio M, Mordmuller B, Arese P, Schwarzer E. Hemozoin (malarial pigment) inhibits differentiation and maturation of human monocyte-derived dendritic cells: a peroxisome proliferator-activated receptor-γ-mediated effect. J. Immunol. 2004;173:4066–4074. doi: 10.4049/jimmunol.173.6.4066. [DOI] [PubMed] [Google Scholar]
- 15.West JD, Marnett LJ. Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2005;18:1642–1653. doi: 10.1021/tx050211n. [DOI] [PubMed] [Google Scholar]
- 16.Weigel AL, Handa JT, Hjelmeland LM. Microarray analysis of H2O2-, HNE, or tBH-treated Arpe-19 cells. Free Radical Biol. Med. 2002;33:1419–1432. doi: 10.1016/s0891-5849(02)01082-1. [DOI] [PubMed] [Google Scholar]
- 17.Green MD, Xiao L, Lal AA. Formation of hydroxyeicosatetraenoic acids from hemozoin-catalyzed oxidation of arachidonic acid. Mol. Biochem. Parasitol. 1996;83:183–188. doi: 10.1016/s0166-6851(96)02769-7. [DOI] [PubMed] [Google Scholar]
- 18.Miller CM, Carney CK, Schrimpe AC, Wright DW. B-hematin (hemozoin) mediated decomposition of polyunsaturated fatty acids to 4-hydroxy-2-nonenal. Inorg. Chem. 2005;44:2134–2136. doi: 10.1021/ic048821i. [DOI] [PubMed] [Google Scholar]
- 19.Carter MD, Reese Harry S, Wright DW. Identification of hydroxyeicosatetraenoic acid components of schistosomal hemozoin. Biochem. Biophys. Res. Commun. 2007;363:867–872. doi: 10.1016/j.bbrc.2007.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohle DS, Helms JB. Synthesis of β-hematin by dehydrohalogenation of hemin. Biochem. Biophys. Res. Commun. 1993;193:504–508. doi: 10.1006/bbrc.1993.1652. [DOI] [PubMed] [Google Scholar]
- 21.Carney CK, Schrimpe AC, Halfpenny K, Harry RS, Miller CM, Broncel M, Sewell SL, Schaff JE, Deol R, Carter MD, Wright DW. The basis of the immunomodulatory activity of malaria pigment (hemozoin) J. Biol. Inorg. Chem. 2006;11:917–929. doi: 10.1007/s00775-006-0147-0. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzer E, Alessio M, Ulliers D, Arese P. Phagocytosis of the malarial pigment, hemozoin, impairs expression of major histocompatibility complex class II antigen, CD54, and CD11c in human monocytes. Infect. Immun. 1998;66:1601–1606. doi: 10.1128/iai.66.4.1601-1606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huy NT, Trang DTX, Kariu T, Sasai M, Saida K, Harada S, Kamei K. Leukocyte activation by malarial pigment. Parasit. Int. 2006;55:75–81. doi: 10.1016/j.parint.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Krämer B, Grobusch MP, Suttorp N, Neukammer J, Rinneberg H. Relative frequency of malaria pigment-carrying monocytes of nonimmune and semi-immune patients from flow cytometric depolarized side scatter. Cytometry. 2001;45:133–140. doi: 10.1002/1097-0320(20011001)45:2<133::aid-cyto1155>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Henderson GI, Freeman GL. Role of 4-Hydroxynonenal in modification of cytochrome C oxidase in ischemia/reperfused rat heart. J. Mol. Cell. Cardiol. 2001;33:1919–1927. doi: 10.1006/jmcc.2001.1454. [DOI] [PubMed] [Google Scholar]
- 26.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 27.Crabb JW, O'Neil J, Miyagi M, West K, Hoff HF. Hydroxynonenal inactivates cathepsin B by forming Michael adducts with active site residues. Protein Sci. 2002;11:831–840. doi: 10.1110/ps.4400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji C, Kozak KR, Marnett LJ. Ikappa B kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J. Biol. Chem. 2001;276:18223–18228. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 29.Szweda LI, Uchida K, Tsai L, Stadtman ER. Inactivation of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Selective modification of an active-site lysine. J. Biol. Chem. 1993;268:3342–3347. [PubMed] [Google Scholar]
- 30.Griffiths MJ, Mohammed JS, Popper SJ, Hemingway CA, Kortok MM, Wathen A, Rockett KA, Mott R, Levin M, Newton CR, Marsh K, Relman DA, Kwiatkowski DP. Genomewide analysis of the host response to malaria in Kenyan children. J. Infect. Dis. 2005;191:1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 31.Muehlenbachs A, Fried M, Lachowitzer J, Mutabingwa TK, Duffy PE. Genome-wide expression analysis of placental malaria reveals features of lymphoid neogenesis during chronic infection. J. Immunol. 2007;179:557–565. doi: 10.4049/jimmunol.179.1.557. [DOI] [PubMed] [Google Scholar]
- 32.Sexton AC, Good RT, Hansen DS, D'Ombrain MC, Buckingham L, Simpson K, Schofield L. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J. Infect. Dis. 2004;189:1245–1256. doi: 10.1086/382596. [DOI] [PubMed] [Google Scholar]
- 33.Schaecher K, Kumar S, Yadava A, Vahey M, Ockenhouse CF. Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes. Infect. Immun. 2005;73:6091–6100. doi: 10.1128/IAI.73.9.6091-6100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ockenhouse CF, Hu W-c, Kester KE, Cummings JF, Stewart A, Heppner DG, Jedlicka AE, Scott AL, Wolfe ND, Vahey M, Burke DS. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect. Immun. 2006;74:5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen PH, Day N, Pram TD, Ferguson DJ, White NJ. Intraleucocytic malaria pigment and prognosis in severe malaria. Trans. R. Soc. Trop. Med. Hyg. 1995;89:200–204. doi: 10.1016/0035-9203(95)90496-4. [DOI] [PubMed] [Google Scholar]
- 36.Jaramillo M, Plante I, Ouellet N, Vandal K, Tessier PA, Olivier M. Hemozoin-inducible proinflammatory events in vivo: potential role in malaria infection. J. Immunol. 2004;172:3101–3110. doi: 10.4049/jimmunol.172.5.3101. [DOI] [PubMed] [Google Scholar]
- 37.Taramelli D, Basilico N, Pagani E, Grande R, Monti D, Ghione M, Olliaro P. The heme moiety of malaria pigment (β-hematin) mediates the inhibition of nitric oxide and tumor necrosis factor-α production by lipopolysaccharide-stimulated macrophages. Exp. Parasitol. 1995;81:501–511. doi: 10.1006/expr.1995.1143. [DOI] [PubMed] [Google Scholar]
- 38.Jaramillo M, Godbout M, Olivier M. Hemozoin induces macrophage chemokine expression through oxidative stress-dependent and -independent mechanisms. J. Immunol. 2005;174:475–484. doi: 10.4049/jimmunol.174.1.475. [DOI] [PubMed] [Google Scholar]
- 39.Biswas S, Karmarkar MG, Sharma YD. Antibodies detected against Plasmodium falciparum haemozoin with inhibitory properties to cytokine production. FEMS Microbiol. Lett. 2001;194:175–179. doi: 10.1111/j.1574-6968.2001.tb09465.x. [DOI] [PubMed] [Google Scholar]
- 40.Jaramillo M, Gowda DC, Radzioch D, Olivier M. Hemozoin increases IFN-gamma-inducible macrophage nitric oxide generation through extracellular signal-regulated kinase- and NF-kappa B-dependent pathways. J. Immunol. 2003;171:4243–4253. doi: 10.4049/jimmunol.171.8.4243. [DOI] [PubMed] [Google Scholar]
- 41.Slater AF, Swiggard WJ, Orton BR, Flitter WD, Goldberg DE, Cerami A, Henderson GB. An iron-carboxylate bond links the heme units of malaria pigment. Proc Natl Acad Sci USA. 1991;88:325–329. doi: 10.1073/pnas.88.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blauer G, Akkawi M. Investigations of B- and β-Hematin. J. Inorg. Biochem. 1997;66:145–152. doi: 10.1016/s0162-0134(96)00200-0. [DOI] [PubMed] [Google Scholar]
- 43.Taramelli D, Recalcati S, Basilico N, Olliaro P, Cairo G. Macrophage preconditioning with synthetic malaria pigment reduces cytokine production via heme iron-dependent oxidative stress. Lab. Invest. 2000;80:1781–1788. doi: 10.1038/labinvest.3780189. [DOI] [PubMed] [Google Scholar]
- 44.Douki T, Odin F, Caillat S, Favier A, Cadet J. Predominance of the 1,N2-propano 2'-deoxyguanosine adduct among 4-hydroxy-2-nonenal-induced DNA lesions. Free Radical Biol. Med. 2004;37:62–70. doi: 10.1016/j.freeradbiomed.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Pichyangkul S, Saengkrai P, Webster HK. Plasmodium falciparum pigment induces monocytes to release high levels of tumor necrosis factor-α and interleukin-1β. Am. J. Trop. Med. Hyg. 1994;51:430–435. [PubMed] [Google Scholar]
- 46.Sinniah R, Rui-Mei L, Kara A. Up-regulation of cytokines in glomerulonephritis associated with murine malaria infection. Int. J. Exp. Pathol. 1999;80:87–95. doi: 10.1046/j.1365-2613.1999.00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlgren M, Abrams JS, Fernandez V, Bejarano MT, Azuma M, Torii M, Aikawa M, Howard RJ. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells and secretion of cytokines (IL-1-beta, IL-1RA, IL-6, IL-8, IL-10, TGF beta, TNF alpha, G-CSF, GM-CSF. Scand. J. Immunol. 1995;42:626–636. doi: 10.1111/j.1365-3083.1995.tb03705.x. [DOI] [PubMed] [Google Scholar]
- 48.Millington OR, Lorenzo CD, Phillips S, Garside P, Brewer JM. Suppression of adaptive immunity to heterologous antigens during Plasmodium infection through hemozoin-induced failure of dendritic cell function. J. Biol. 2006;5:5. doi: 10.1186/jbiol34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ocaña-Morgner C, Wong, Kurt A, Lega F, Dotor J, Borras-Cuesta F, Rodriguez A. Role of TGF-beta and PGE2 in T cell responses during Plasmodium yoelii infection. Eur. J. Immunol. 2007;37:1562–1574. doi: 10.1002/eji.200737068. [DOI] [PubMed] [Google Scholar]
- 50.Liu W, Kato M, Itoigawa M, Murakami H, Yajima M, Wu J, Ishikawa N, Nakashima I. Distinct involvement of NF-κB and p38 mitogen-activated protein kinase pathways in serum deprivation-mediated stimulation of inducible nitric oxide synthase and its inhibition by 4-hydroxynonenal. J. Cell. Biochem. 2001;83:271–280. doi: 10.1002/jcb.1234. [DOI] [PubMed] [Google Scholar]
- 51.Skorokhod OA, Schwarzer E, Ceretto M, Arese P. Malarial pigment haemozoin, IFN-gamma, TNF-alpha, IL-1beta and LPS do not stimulate expression of inducible nitric oxide synthase and production of nitric oxide in immuno-purified human monocytes. Malar. J. 2007;6:73–80. doi: 10.1186/1475-2875-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prada J, Malinowsky J, Muller S, Bienzle U, Kremsner PG. Effects of Plasmodium vinckei hemozoin on the production of oxygen radicals and nitrogen oxides in murine macrophages. Am. J. Trop. Med. Hyg. 1996;54:620–624. doi: 10.4269/ajtmh.1996.54.620. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan AD, Ittarat I, Meshnick SR. Patterns of haemozoin accumulation in tissue. Parasitology. 1996;112:285–294. doi: 10.1017/s003118200006580x. [DOI] [PubMed] [Google Scholar]
- 54.Newton CR, Taylor TE, Whitten RO. Pathophysiology of fatal falciparum malaria in African children. Am. J. Trop. Med. Hyg. 1998;58:673–683. doi: 10.4269/ajtmh.1998.58.673. [DOI] [PubMed] [Google Scholar]
- 55.Adams S, Brown H, Turner G. Breaking down the blood-brain barrier: signaling a path to cerebral malaria? Trends Parasitol. 2002;18:360–366. doi: 10.1016/s1471-4922(02)02353-x. [DOI] [PubMed] [Google Scholar]
- 56.Mandal M, Mandal A, Das S, Chakraborti T, Chakraborti S. Clinical implications of matrix metalloproteinases. Mol. Cell. Biochem. 2003;252:305–329. doi: 10.1023/a:1025526424637. [DOI] [PubMed] [Google Scholar]
- 57.Prato M, Giribaldi G, Polimeni M, Gallo V, Arese P. Phagocytosis of hemozoin enhances matrix metalloproteinase-9 activity and TNF-alpha production in human monocytes: role of matrix metalloproteinases in the pathogenesis of falciparum malaria. J Immunol. 2005;175:6436–6442. doi: 10.4049/jimmunol.175.10.6436. [DOI] [PubMed] [Google Scholar]
- 58.Woo C-H, Lim J-H, Kim J-H. Lipopolysaccharide induces matrix metalloproteinase-9 expression via a mitochondrial reactive oxygen species-p38 kinase-activator protein-1 pathway in RAW 264.7 Cells. J Immunol. 2004;173:6973–6980. doi: 10.4049/jimmunol.173.11.6973. [DOI] [PubMed] [Google Scholar]
- 59.Lai W-C, Zhou M, Shankavaram U, Peng G, Wahl LM. Differential regulation of lipopolysaccharide-induced monocyte matrix metalloproteinase (MMP)-1 and MMP-9 by p38 and extracellular signal-regulated kinase 1/2 mitogen-activated protein kinases. J Immunol. 2003;170:6244–6249. doi: 10.4049/jimmunol.170.12.6244. [DOI] [PubMed] [Google Scholar]
- 60.Rhee JW, Lee KW, Kim D, Lee Y, Jeon OH, Kwon HJ, Kim DS. NF-kappaB-dependent regulation of matrix metalloproteinase-9 gene expression by lipopolysaccharide in a macrophage cell line RAW 264.7. J Biochem Mol Biol. 2007;40:88–94. doi: 10.5483/bmbrep.2007.40.1.088. [DOI] [PubMed] [Google Scholar]
- 61.Gearing AJH, Beckett P, Christodoulou M, Churchill M, Clements J, Davidson AH, Drummond AH, Galloway WA, Gilbert R, Gordon JL, Leber TM, Mangan M, Miller K, Nayee P, Owen K, Patel S, Thomas W, Wells G, Wood LM, Woolley K. Processing of tumour necrosis factor-α precursor by metalloproteinases. Nature. 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- 62.Brown H, Turner G, Rogerson S, Tembo M, Mwenechanya J, Molyneux M, Taylor T. Cytokine expression in the brain in human cerebral malaria. The Journal of Infectious Diseases. 1999;180:1742–1746. doi: 10.1086/315078. [DOI] [PubMed] [Google Scholar]
- 63.Armah H, Wiredu EK, Dodoo AK, Adjei AA, Tettey Y, Gyasi R. Cytokines and adhesion molecules expression in the brain in human cerebral malaria. Int. J. Environ. Res. Public Health. 2005;2:123–131. doi: 10.3390/ijerph2005010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wassmer SC, Combes V, Candal FJ, Juhan-Vague I, Grau GE. Platelets potentiate brain endothelial alterations induced by Plasmodium falciparum. Infect. Immun. 2006;74:645–653. doi: 10.1128/IAI.74.1.645-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner GDH, Morrison H, Jones M, Davis TME, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B, White NJ, Berendt AR. An immunohistochemical study of the pathology of fatal malaria: evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 66.Silamut K, Phu NH, Whitty C, Turner GDH, Louwrier K, Mai NTH, Simpson JA, Hien TT, White NJ. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am. J. Pathol. 1999;155:395–410. doi: 10.1016/S0002-9440(10)65136-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohan and Stevenson Dyserythropoiesis and severe anaemia associated with malaria correlate with deficient interleukin-12 production. Br. J. Haematol. 1998;103:942–949. doi: 10.1046/j.1365-2141.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 68.Casals-Pascual C, Kai O, Cheung JOP, Williams S, Lowe B, Nyanoti M, Williams TN, Maitland K, Molyneux M, Newton CRJC, Peshu N, Watt SM, Roberts DJ. Suppression of erythropoiesis in malarial anemia is associated with hemozoin in vitro and in vivo. Blood. 2006;108:2569–2577. doi: 10.1182/blood-2006-05-018697. [DOI] [PubMed] [Google Scholar]
- 69.Giribaldi G, Ulliers D, Schwarzer E, Roberts I, Piacibello W, Arese P. Hemozoin- and 4-hydroxynonenal-mediated inhibition of erythropoiesis. Possible role in malarial dyserythropoiesis and anemia. Haematologica. 2004;89:492–493. [PubMed] [Google Scholar]
- 70.Were T, Hittner JB, Ouma C, Otieno RO, Orago AS, Ong'echa JM, Vulule JM, Keller CC, Perkins DJ. Suppression of RANTES in children with Plasmodium falciparum malaria. Haematologica. 2006;91:1396–1399. [PubMed] [Google Scholar]
- 71.John CC, Opika-Opoka R, Byarugaba J, Idro R, Boivin MJ. Low levels of RANTES are associated with mortality in children with cerebral malaria. J. Infect. Dis. 2006;194:837–845. doi: 10.1086/506623. [DOI] [PubMed] [Google Scholar]
- 72.Lovegrove FE, Pena-Castillo L, Mohammad N, Liles C, Hughes TR, Kain KC. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295–311. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.