Abstract
Alteration of regulatory DNA elements or their binding proteins may have drastic consequences for morphological evolution. Chromatin insulators are one example of such proteins and play a fundamental role in organizing gene expression. While a single insulator protein, CTCF (CCCTC-binding factor), is known in vertebrates, Drosophila melanogaster utilizes six additional factors. We studied the evolution of these proteins and show here that—in contrast to the bilaterian-wide distribution of CTCF—all other D. melanogaster insulators are restricted to arthropods. The full set is present exclusively in the genus Drosophila whereas only two insulators, Su(Hw) and CTCF, existed at the base of the arthropod clade and all additional factors have been acquired successively at later stages. Secondary loss of factors in some lineages further led to the presence of different insulator subsets in arthropods. Thus, the evolution of insulator proteins within arthropods is an ongoing and dynamic process that reshapes and supplements the ancient CTCF-based system common to bilaterians. Expansion of insulator systems may therefore be a general strategy to increase an organism’s gene regulatory repertoire and its potential for morphological plasticity.
Keywords: Adaptive evolution, barrier element, BEAF-32, CP190, GAGA factor, gene loss, lineage-specific genes, Mod(mdg4), Su(Hw), Zw5
Since more than a century, the molecular causes of morphological change are being examined in the fruit fly Drosophila melanogaster (Morgan 1911; Altenburg and Muller 1920). A well-studied example of morphological change are homeotic mutations. They alter the identity of particular body parts by transforming them into other parts (for review, see Lewis 1978). Genetic analysis of these mutations revealed that they often affect regulatory elements controlling the expression of an associated homeotic (Hox) gene (see Pfeifer et al. 1987, for examples). Misexpression of genes by regulatory mutations can therefore contribute to morphological change.
Comparative studies in additional arthropods demonstrated that differences in Hox gene expression are correlated with morphological differences across the phylum (e.g., Warren et al. 1994; Averof and Akam 1995; Averof and Patel 1997; Abzhanov and Kaufman 2000; Hughes and Kaufman 2002). Expression of the Hox gene ubx, for example, is linked to abdominal limb number (Warren et al. 1994; Averof and Patel 1997) and functional studies suggest that altered Hox gene expression is indeed a cause of morphological diversity (Lewis et al. 1999; Liubicich et al. 2009; Pavlopoulos et al. 2009). Also in other contexts, there is ample evidence that mutations in regulatory elements play an important role for the evolution of morphological traits (Jeong et al. 2008; Peter and Davidson 2011).
Regulatory elements, however, are abundant and can exert their function over a wide range of physical distances (Miele and Dekker 2008; Lieberman-Aiden et al. 2009). To protect genes from the inappropriate influence of these sequences, a process called chromatin insulation participates in the creation of independent chromatin domains (for review, see Wallace and Felsenfeld 2007; Yang and Corces 2012). As mediators of this kind of regulation, insulator proteins and the mechanisms by which they act have been studied intensely (see Van Bortle and Corces 2013, for a recent review].
On the basis of its ability to protect genes from position effects in transgenic flies, Suppressor of Hairy Wing [Su(Hw)] was the first insulator protein to be identified (Spana et al. 1988; Geyer and Corces 1992). To date, several additional proteins with insulator activity are known in D. melanogaster: Boundary Element Associated Factors (BEAF-32A and B), Zeste-white 5 (Zw5), GAGA Associated Factor (GAF), Modifier of mdg4 [Mod(mdg4)], Centrosomal Protein 190 (CP190), and dCTCF, the D. melanogaster ortholog of mammalian CCCTC-binding factor. Although insulator proteins have been described originally as enhancer blockers when positioned between a promoter and an enhancer, there is evidence for additional and more complex functions. An emerging role is their involvement in the spatiotemporal control of gene expression by modifying long-range chromosomal interactions, suggesting that they are key players in establishing an appropriate three-dimensional chromosome structure during cell differentiation and development (reviewed in Van Bortle and Corces 2013).
In agreement with this view, knockdown or mutation of insulator proteins and the sequences they bind has severe consequences. Impairment of dCTCF and the deletion of CTCF-binding sites, for example, eliminates boundary elements required for proper Hox gene expression and leads to homeotic transformations (Mohan et al. 2007; Iampietro et al. 2010); expressing dominant-negative BEAF-32 during embryogenesis is lethal (Gilbert et al. 2006) and disturbs Hox gene expression (Roy et al. 2011); mutations in trithorax-like, the gene encoding GAGA factor, display a maternal effect lethal phenotype and abnormalities in the expression of homeotic genes (Biggin and Tjian 1988; Bhat et al. 1996; Ohtsuki and Levine 1998; Belozerov et al. 2003). Recently, comparative ChIP-seq analysis in several Drosophila species revealed that CTCF and BEAF-32 are directly involved in the evolution of gene expression and genome organization through adaptive changes in their respective binding sites (Ni et al. 2012; Yang et al. 2012). Thus, insulator proteins regulate fundamental processes during Drosophila development, and evolutionary changes in the binding pattern of these factors have direct consequences for gene expression and phenotype.
The conservation of many D. melanogaster insulator binding sequences (Holohan et al. 2007; Negre et al. 2010; Ni et al. 2012) together with studies in other animals (e.g., Heger et al. 2012; Schmidt et al. 2012) indicates further that this fundamental aspect of genetic regulation may be relevant across different phyla. One would expect therefore that most eukaryots possess orthologous genes to implement insulator mechanisms. However, the phylogenetic distribution of only one factor, CTCF, has been investigated in detail (Heger et al. 2012). In this study, we examine the origin of the other known D. melanogaster insulator proteins.
Results
Seven proteins with insulator function have been described in D. melanogaster. In contrast, our previous work showed that orthologs of only one chromatin insulator, CTCF, can be found in nematodes (Heger et al. 2009) echoing the situation in vertebrates (Phillips and Corces 2009). Thus, the possession of additional insulator systems might be an arthropod-, insect-, or Drosophila-specific property. To investigate this idea, we searched the sequence databases at NCBI for putative orthologs of the seven D. melanogaster insulator proteins CTCF, Su(Hw), Zw5, CP190, Mod(mdg4), GAF, and BEAF-32. Using the respective D. melanogaster sequences as query, we performed within the arthropod phylum separate searches for each protein and retrieved > 14,000 candidates in total (Table 1). As many of these sequences (59.2%) were collected multiple times, we removed redundancy and retained 5810 unique sequences (Table 1). Subsequently, we performed in two parallel workflows clustering and phylogenetic analysis of the ZF (zinc finger) and of the BTB (broad complex, tramtrack, and bric-à-brac) domain containing subsets of insulator proteins.
Table 1.
Number of collected candidate sequences. Using BLAST searches with a specified cutoff (threshold), the number of arthropod candidate sequences retrieved in total, the number of unique sequences, and the number of sequences retained after clustering are shown. Candidates for a vertebrate GAF were collected from Danio rerio, Homo sapiens, and Strongylocentrotus purpuratus
| Insulator | Threshold | Total | Unique | After clustering |
|---|---|---|---|---|
| CTCF, Su(Hw), Zw5 | 10−05 | 8929 | 4245 | 587 |
| CP190, GAF, Mod(mdg4) | 10−14 | 5166 | 1501 | n. d./727 |
| BEAF-32 | ∞ | 130 | 64 | n. d. |
| Vertebrate GAF | 10−05 | 1135 | 161 | n. d. |
n. d., not determined.
The ZF Domain Containing Insulators: CTCF, Su(Hw), and Zw5
The insulator proteins Su(Hw), CTCF, and Zw5 are poly-ZF proteins with 12, 11, and eight C2H2 ZF domains, respectively (Fig. 1). The ZF domain constitutes an ancient DNA-binding motif present in all eukaryotes and also in some Archaea (Bouhouche et al. 2000) and the C2H2 ZF in particular is the most common DNA-binding motif of eukaryotic transcription factors (Clarke and Berg 1998; Tadepally et al. 2008). Thus, it is not surprising that our search recovered more than 4200 candidates (Table 1) belonging to 227 different arthropod species (Table S1).
Figure 1.
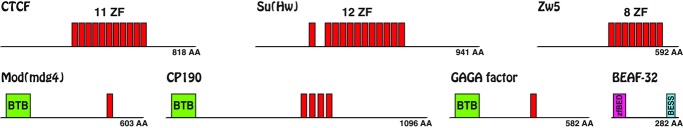
Domain structure of D. melanogaster insulator proteins. The domain composition of known D. melanogaster insulator proteins is drawn approximately to scale. Upper row: Factors with poly-zinc finger (ZF) domain. Individual ZFs (red) comprise about 29 AA and are regularly spaced, except for Su(Hw)’s first ZF. Lower row: Three insulator proteins with BTB (∼105 AA, green) s+ ZF domain (red) and BEAF-32 (with zf-BED and BESS domain).
To extract potential insulators from the 4245 candidates, we clustered the sequences according to their similarity to known insulators and obtained a set with 587 proteins. We next aligned these sequences and determined their orthology to given ZF insulators with phylogenetic methods. The resulting maximum likelihood tree displayed well-supported clusters for CTCF and Zw5, but low support for a Su(Hw) cluster (not shown). To prevent this problem, we extracted according to the maximum likelihood tree the members of potential insulator protein clusters and evaluated them separately in new experiments, thereby omitting the bulk of nonorthologous sequences. Using this strategy, we obtained high support for all three groups of ZF insulator proteins (CTCF, Su(Hw), and Zw5; Fig. S1).
To visualize the phylogenetic composition of these clusters, we mapped the source organism of the respective sequences to a consensus arthropod phylogeny (Figs. 2, S1). We found that CTCF orthologs are present in all arthropod groups with a sequenced genome and in many unsequenced species of the three arthropod subgroups (Fig. S1). These results emphasize the importance of this factor for arthropod biology and agree with a more general role of CTCF in bilaterians (Heger et al. 2012). In addition, they indicate that our strategy is able to detect orthologous sequences in arthropods with confidence.
Figure 2.
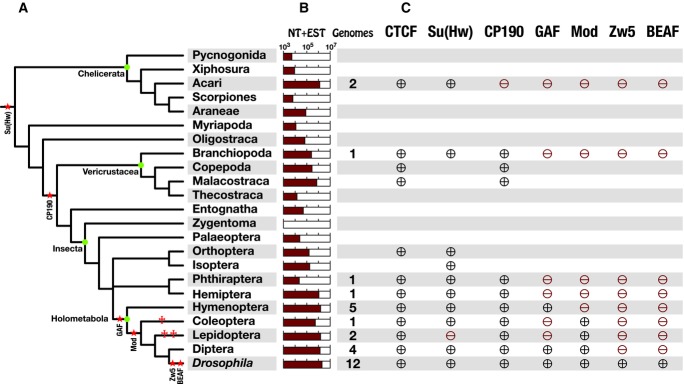
Successive gain of insulator proteins during arthropod evolution. (A) Schematic representation of arthropod relationships (after Regier et al. (2010); Simon et al. (2009); Wiegmann et al. (2009); Trautwein et al. (2012)). Green dots highlight the position of major arthropod groups. The birth (*) and loss (†) of each insulator protein is indicated in red. Several clades/orders are omitted for clarity. Ephemeroptera (mayflies) and Odonata (dragonflies) are combined in Palaeoptera. (B) For the taxa shown in A, the number of ESTs and nucleotide sequences deposited at NCBI (http://www.ncbi.nlm.nih.gov; reference day is May 7, 2012) is indicated as logarithmic bar graph, along with the number of available genome sequences (right column). Sequence data are biased toward holometabolous insects and particular crustacean and chelicerate lineages. Zygentoma have less than 1000 sequences (empty bar graph). (C) Phylogenetic mapping of insulator proteins. Presence of a particular insulator protein is indicated by ⊕, absence by ⊖. Absence is only indicated in orders with sufficient EST and genomic data. Results refer to the BLAST/phylogeny-based strategy and are identical for the HMM/OrthoMCL-based strategy, except some minor modifications (Table S4).
The phylogenetic distribution of Su(Hw) was similar to that of CTCF, with all three arthropod subgroups being represented in the respective cluster (Figs. 2, S1). As we could not find orthologs of Su(Hw) in nematodes, a sister phylum to arthropods, in a previous study (Heger et al. 2009), it is likely that this protein evolved at the base of arthropods or close to this base. Although this conclusion is derived from a modest number of sequences (two chelicerate, two crustacean) and the absence of a detectable ortholog in fully sequenced nematode genomes, the branching pattern and lineage-specific synapomorphies of the chelicerate and crustacean candidates argue for a common ancestry with Su(Hw) orthologs from insects.
We observed a misplacement of crustacean and chelicerate Su(Hw) and a split of dipteran sequences in a few experiments (Fig. S1 and data not shown). These inconsistencies with accepted arthropod relationships are likely a consequence of our dataset (single gene phylogeny with many short and incomplete sequences) and of the fast evolution in Drosophila (Savard et al. 2006) and also affect the CTCF and YY1 clusters (Fig. S1). Indeed, it is well known that gene trees and species trees do not necessarily agree if the number of analyzed loci is small (Pamilo and Nei 1998; Degnan and Rosenberg 2006). Even if our data are not sufficient to reconstruct the correct species genealogy in all detail, they offer substantial support for the existence of a distinct clade of Su(Hw) orthologs with representatives from all three arthropod subphyla.
Despite its broad distribution, Su(Hw) is not indispensable for arthropods. While orthologs to other ZF proteins such as CTCF or YY1 can be found in all arthropod lineages, there is no evidence for the presence of Su(Hw) in Lepidoptera (butterflies) although large amounts of expressed sequence tags (ESTs) and several genome sequences are available in this group (Figs. 2, S1).
When we analyzed the Zw5 cluster in preliminary experiments, we noticed that it seemed to contain exclusively sequences from the 12 Drosophila species we used to define this cluster. Occasionally however, some other arthropod sequences were positioned nearby. When we probed the reliability of this association in smaller datasets, the 12 Drosophila Zw5 orthologs alone gave rise to a distinct and highly supported cluster in all cases (Fig. S1). Thus, no sequences from other dipterans or more distantly related arthropods are orthologous to Drosophila Zw5 although, for example, more than 2000 ZF sequences from 57 non-Drosophila dipterans were present in the “unique” dataset (Table S1). These results indicate that the insulator protein Zw5 is specific for the genus Drosophila (Fig. 2) as it has been suggested in a previous study (Schoborg and Labrador 2010). To investigate whether Zw5 is a true synapomorphy of drosophilids, sequences from closely related brachyceran outgroups need to be analyzed.
The BTB Domain Containing Insulators: GAGA Factor, Mod(mdg4), and CP190
Like the C2H2 ZF domain, the BTB domain is ancient and found in all eukaryotes (Perez-Torrado et al. 2006). It consists of an N-terminal 105 amino acid motif that mediates homo- and heteromeric dimerization in a number of Drosophila transcriptional regulators, for example, broad complex, tramtrack, and bric-à-brac (Zollman et al. 1994). Searching for BTB domain containing insulators, we collected more than 1500 candidate sequences (Table 1) that belonged to 111 arthropod species (Table S2). To determine which of these sequences could be orthologous to a D. melanogaster insulator, we clustered them and obtained a dataset with 727 sequences. A maximum likelihood analysis of this dataset indicated high support for distinct CP190 and GAGA factor clusters. In contrast, the Mod(mdg4) cluster, containing more than 200 nearly identical sequences from Lepidoptera, was supported less well (not shown). We therefore extracted from the previous sequence set all potential orthologs of CP190, Mod(mdg4) (9/220 sequences), and GAF and determined their orthology to the Drosophila insulators in additional experiments.
These new analyses revealed that only two insect orders were represented within the GAF cluster: Hymenoptera and Diptera (Figs. 2, S2). We could not identify GAF orthologs in other arthropods despite the availability of substantial genomic and EST data (Fig. 2). This suggests that GAGA factor evolved in the last common ancestor of Hymenoptera and Diptera and has been lost at least twice during evolution of holometabolous insects, in Coleoptera (beetles) and Lepidoptera (butterflies).
In previous studies, putative vertebrate homologs of the GAGA factor have been reported (Matharu et al. 2010; Kumar 2011). As vertebrates and Drosophila share a common ancestor with all other bilaterians, these conclusions imply that GAGA factors should be present in other protostomes and deuterostomes. To investigate this assumption, we collected with a relaxed E-value of 10−05 more than 150 candidate sequences from three different deuterostome lineages (vertebrates: Danio rerio and Homo sapiens; echinoderms: Strongylocentrotus purpuratus), including four proposed vertebrate GAFs, and analyzed their relation to the insect GAF cluster. However, none of these sequences localized to the highly supported cluster (97% bootstrap support; Fig. S3). Rather, the proposed vertebrate GAFs formed a separate cluster, arguing for a common ancestry of these proteins in the vertebrate lineage.
To test whether this result is a consequence of insufficient BLAST sensitivity, we generated from the members of the insect GAGA cluster two representative HMM profiles (full length and BTB domain only) and performed a more sensitive profile–profile search on the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred). We retrieved from these searches 45 additional candidates from humans and mice and examined their relationship to the insect counterparts. As in the previous case, the resulting trees did not place any of the new candidates to the insect GAGA cluster (Fig. S4). Thus, none of the 200 analyzed deuterostome candidates is closer related to the insect GAGA cluster than any other candidate, strongly limiting the possibility that there is among them a GAGA ortholog, that is, a sequence that originated from the same last common ancestor than insect GAFs. Given that we could not identify GAF orthologs in arthropods preceding the Hymenoptera–Diptera split (Fig. S2) and in nematodes (Heger et al. 2009), these results indicate that the GAGA factor is unique to particular lineages of holometabolous insects and is related to the proposed homologs in other phyla by the presence of an ancient BTB domain. This conclusion contradicts Matharu et al. (2010) and Kumar (2011). However, Kumar (2011) presented his result on the basis of an HHpred search initiated with a single sequence and without verification in a phylogenetic context whereas Matharu et al. (2010) carried out a phylogenetic analysis without bootstrapping, also lacking phylogenetic implications. Our comprehensive survey of candidates within the whole range of arthropods, nematodes (Heger et al. 2009), and several deuterostomes, including those proposed by Matharu et al. (2010) and Kumar (2011), argues that GAGA factor originated in the ancestor of Hymenoptera and Diptera rather than in the ancestor of the Bilateria. A functional similarity that has been attributed to the proposed vertebrate GAGA factors (Matharu et al. 2010) therefore likely involves convergent evolution.
After removing most lepidopteran sequences (231/240) that clustered to D. melanogaster Mod(mdg4) in the 727 candidate set, we newly analyzed the candidates positioned within each supposed BTB cluster (108 sequences). Our results showed high bootstrap support (100%) for the presence of Mod(mdg4) orthologs in Drosophila, other Diptera, Lepidoptera (butterflies), and Coleoptera (beetles; Figs. 2, S2). Although we collected in the original dataset 277 sequences from hymenopterans, the next possible sister group, none of these were orthologous to Mod(mdg4). Taking into account the availability of five genome sequences, this indicates that Mod(mdg4) does not belong to the gene repertoire of hymenopterans (Fig. 2). These findings confirm previous reports of a mod(mdg4) locus in Lepidoptera (Dorn and Krauss 2003; Krauss and Dorn 2004) and establish the origin of this locus even earlier, in the common ancestor of Coleoptera, Lepidoptera, and Diptera.
In D. melanogaster, the mod(mdg4) locus gives rise to > 20 different isoforms that share an N-terminal 405 AA region containing the BTB domain (Buchner et al. 2000). A similarly complex organization was reported for the mod(mdg4) locus of lepidopterans (Shao et al. 2012). To find out whether this feature is also present in Coleoptera, we searched for Tribolium castaneum ESTs that share their 5′ region (with BTB domain), but have different 3′ ends. However, we could not find evidence for different isoforms in 12 ESTs that mapped in T. castaneum to the BTB domain region of the mod(mdg4) locus, as it is indicated by EST GI:189241700 (ChLG10:8,779,000–8,780,000; Fig. S2). Alignment of 18 sequences from the beetle Dendroctonus ponderosae that belonged to the mod(mdg4) cluster did not give evidence for the existence of separate isoformes either. Thus, currently available data from Coleoptera are not able to resolve the origins of the complex mod(mdg4) locus.
The D. melanogaster CP190 protein is an essential component of many insulator complexes organized by CTCF, Su(Hw), and BEAF-32 (Pai et al. 2004; Mohan et al. 2007; Bartkuhn et al. 2009; Negre et al. 2010; Van Bortle et al. 2012). Its N-terminal BTB domain is indispensable in providing this activity (Oliver et al. 2010). Our search for CP190 orthologs in arthropods revealed a distinct set of sequences clustering to D. melanogaster CP190 with high confidence. These sequences covered three crustacean branches and all insects with a sequenced genome, but not the Chelicerata although two genome sequences exist and 71 candidate sequences from different chelicerate lineages were present in the “unique” dataset (Figs. 2, S2). Thus, it is likely that CP190 originated in the ancestor of hexapods and crustaceans. As we could not observe a loss in any of the well-sampled taxa, the interaction between CP190 and CTCF/Su(Hw) insulator complexes could be a critical feature of all pancrustaceans (Crustacea plus Hexapoda; Regier et al. 2010).
BEAF-32
The insulator protein BEAF-32 exists in two isoforms and is associated with chromosomal domains (Zhao et al. 1995) and transcriptionally active regions in D. melanogaster (Jiang et al. 2009). It has an unusual N-terminal ZF, the BED finger (58 AA), and a C-terminal BESS domain (35 AA; Fig. 1). An extensive analysis of BEAF-32 has been performed by Schoborg and Labrador (2010), which suggested, on the basis of BLAST experiments, that BEAF-32 is specific to the Drosophila genus. We wanted to challenge this finding with a more powerful phylogenetic approach. Despite the relaxed threshold (E-value  ), we obtained with our search only 64 candidate sequences from the whole arthropod phylum (Table 1). None of these sequences was a reasonable BEAF-32 candidate (data not shown). In agreement with the previous study (Schoborg and Labrador 2010), we assume therefore that BEAF-32, like Zw5, is a Drosophila-specific insulator protein absent from other insects (Fig. 2).
), we obtained with our search only 64 candidate sequences from the whole arthropod phylum (Table 1). None of these sequences was a reasonable BEAF-32 candidate (data not shown). In agreement with the previous study (Schoborg and Labrador 2010), we assume therefore that BEAF-32, like Zw5, is a Drosophila-specific insulator protein absent from other insects (Fig. 2).
An HMM/OrthoMCL-Based Pipeline to Validate Our Conclusions
Our results suggest that individual insulator proteins have been acquired at different stages of arthropod evolution. To rule out that these observations suffer from a lack of sensitivity and do not reflect the underlying evolutionary history, we complemented the BLAST-based analysis with a more sensitive approach, a combination of HMM scans and OrthoMCL-clustering of candidates into orthologous groups, performed on all accessible sequence data from 26 species in all groups of protostomes available (Table S3). To achieve the best possible coverage, we translated every genome assembly and the unplaced reads data into six open reading frames (ORFs) and combined the resulting ORFeomes with their corresponding downloaded protein sets. With this approach, the potential failure to detect an ortholog cannot be attributed to the often incorrect or incomplete annotation of proteomes. To find insulator orthologs in this wealth of sequence data, we prepared from the clusters obtained in the previous approach HMM profiles specific for each insulator protein (except CTCF). Scanning the 26 datasets with all six insulator profiles yielded a total of 39,573 unique candidate sequences below the default threshold. We analyzed the orthology of these sequences to a given insulator protein family using a custom implementation of the OrthoMCL clustering pipeline (Li et al. 2003). All findings of the previous approach, which employed BLAST search and phylogenetic reconstruction, could be confirmed or further refined in this workflow (Table S4). Some important additional aspects shall be mentioned shortly: (1) The six insulator proteins Su(Hw), CP190, Mod(mdg4), Zw5, BEAF-32, and—notably—GAGA factor are specific for arthropods. In none of the seven protostome outgroup species could we find sequences orthologous to these proteins whereas the previously reported pattern of CTCF occurrence in annelids, molluscs, platyhelminthes, and nematodes (Heger et al. 2009, 2012) could be reproduced accurately. (2) Su(Hw) could not be found in an additional butterfly genome, confirming that it may have been lost in this insect order. (3) If GAGA factor disappeared secondarily in Lepidoptera and Coleoptera (Fig. 2 and Table S4), it should also be missing in Strepsiptera. Our results from the genome scan of Mengenilla moldrzyki are consistent with this assumption. (4) A sequence orthologous to Zw5 is present in Glossina morsitans, a dipteran closer to Drosophila than the nematoceran flies Aedes and Anopheles. As this sequence was generated during translation of the genome, it could not be detected with the BLAST-based strategy. This finding demonstrates the power of our methodology and slightly modifies the previous conclusion that Zw5 is Drosophila specific.
On the other hand, negative results for particular insulator proteins in some genomes where we expected them may indicate insufficient assembly quality or that the evolution of insulator proteins is more dynamic than anticipated (e.g., Su(Hw) not detected in Glossina and Lepeophtheirus; no CP190 in Heliconius; no GAGA factor in Acyrthosiphon; no Mod(mdg4) in Mengenilla). Further work is necessary to resolve these issues.
It is possible that a fragment of a particular ortholog is present in our dataset, but was not recognized as ortholog by OrthoMCL, for example, because of its shortness. Indeed, we observed in the HMM-derived candidate set some short open reading frames that belonged to insulator orthologs, but did not appear in the final clusters. For two reasons we think that this shortcoming does not confound our conclusions. First, the “twilight zone” is confined to ORFs in the range of approximately 30–75 AA (ORF minimum length—shortest ORF in an observed orthologous cluster). The majority of ORFs (84.5%) is larger. To completely miss an ortholog in our genome scan requires that all ORFs corresponding to that ortholog are within this range, a possible, but unlikely event. Second and more important, these limitations apply to the 26 genomes likewise. It is therefore implausible that deficiencies of our methodology generate the evolutionary patterns we report.
Finally, our results suggest that there is a single BEAF-32 ortholog outside Drosophila in the distantly related insect Pediculus humanus (Table S4). To explain this unexpected result, we analyzed the domain composition of the 95 BEAF-32 candidates from Table S4 and observed that a duplicated zfBED domain is present exclusively in this protein (ID: PHUM580690-PA). As the best BLAST hit of this sequence in D. melanogaster is not BEAF-32 either, we conclude that the domain duplication led to a false-positive signal and erroneously triggered its inclusion into the BEAF orthology group.
Discussion
Insulator proteins confer activity to insulators sequences, a class of functional elements with important roles in the regulation of chromosomal organization. In this study, we investigated the origin of the seven known proteins associated with insulator activity in D. melanogaster. To find potential orthologs of these proteins in other organisms, we followed two independent strategies. On the one hand, we performed BLAST searches and analyzed a large number of candidate sequences in phylogenetic experiments (Table 1; Figs. S1–S4). On the other hand, we combined HMM searches and MCL clustering to examine which of the candidates share orthology with a known Drosophila insulator (Table S4). With both methods we found robust evidence that the known Drosophila insulators (except CTCF) are restricted to arthropods and have been acquired successively during arthropod evolution (Fig. 2; Table S4). We draw these conclusions on the basis of several observations.
First, we did not find evidence for the existence of known Drosophila insulator proteins outside the arthropods by analyzing seven genomes from four protostome phyla (Annelida, Mollusca, Platyhelminthes, Nematoda). It is unlikely that a lack of sensitivity is responsible for this result as we find CTCF orthologs in the annelid Capitella spI, in the mollusc Lottia gigantea, and in the nematode Trichinella spiralis, faithfully replicating the results of previous work with an independent method (Heger et al. 2012). The detection of CTCF in all 19 analyzed arthropod genomes, including so far unknown orthologs (e.g., from the myriapod Strigamia maritima and the strepsipteran M. moldrzyki), further confirms the specificity and sensitivity of our approach.
Second, the ZF proteins CTCF and Su(Hw) are consistently present in arthropods from which genome sequences are available plus in some additional orders with a significant number of ESTs. In both cases, the presence of orthologous sequences in the three subgroups of arthropods indicates that the common ancestor of arthropods already had these proteins. Such an assumption is well supported for the CTCF protein that has been found in all bilaterians (Heger et al. 2012). The origin of the Su(Hw) protein is less clear. As it is absent in nematodes (Table S4; Heger et al. 2009), it could have evolved in the ancestor of arthropods or in the common ancestor of arthropods and a closely related ecdysozoan sister group, for example, tardigrades or onychophorans. The resolution of our study is not sufficient to answer this question.
Third, in contrast to CTCF and Su(Hw), the proteins Zw5 and BEAF-32 are restricted to a remarkably limited subset of arthropods. We obtained with both methods only few BEAF-32 candidates outside the Drosophila genus (64 and 95, respectively), and none of them could be placed into the Drosophila BEAF-32 orthology group. Although the domain composition of Zw5 issued a much higher number of candidates (28,431) across the 26 genomes (Table S4), we could only recover Zw5 orthologs in Drosophila and G. morsitans, another brachyceran fly. A Zw5 ortholog could not be found in nematoceran dipteres and other arthropods despite the existence of several sequenced genomes and large amounts of ESTs. These results are based on the most comprehensive study undertaken so far and provide consistent evidence that Zw5 and BEAF-32 likely emerged in or close to the common ancestor of the genus Drosophila. They are therefore the most recent additions to the group of insulator proteins.
Finally, our results with respect to the BTB domain containing insulators suggest that at least some of them have evolved at intermediate stages when compared with the “ubiquitous” proteins CTCF/Su(Hw) and the “restricted” factors Zw5/BEAF-32. The mapping of CP190, for example, shows that it is present in all insects with a sequenced genome and in three crustacean orders. The inability to detect this protein in a myriapod and two mite genomes and in a large amount of ESTs from chelicerates suggests that CP190 evolved in the ancestor of Pancrustacea.
GAGA factor, on the other hand, formed a highly supported cluster containing exclusively sequences from Diptera and Hymenoptera in phylogenetic experiments (Fig. S2). The HMM-based approach confirmed this result, but assigned in addition sequences from the hemipteran Rhodnius prolixus to the GAGA factor orthology group (Table S4). In all further insect, crustacean, and chelicerate genomes, orthologs were not detectable, indicating that this protein likely emerged in the common ancestor of Hemiptera, Hymenoptera, and Diptera. Importantly, these findings imply that GAGA factors do not exist outside the arthropod phylum. To elucidate the conflict of these findings with the proposed existence of vertebrate GAGA factors (Matharu et al. 2010; Kumar 2011), we performed phylogenetic analysis with two sets of candidates, acquired by BLAST and HMM searches. However, neither the four proposed vertebrate GAGA factors nor our additional candidates were placed to the insect orthology group (Figs. S3, S4), emphasizing the consistency of our results.
Although our study provides evidence for a consecutive gain of insulator proteins in arthropods, it also suggests that insulator evolution is dynamic in terms of losses. Although it is difficult to prove the absence of a gene in potentially inaccurate genome assemblies, our results indicate that inference of at least some secondary losses is reasonable. The two best examples are the repetitive loss of GAGA factor in Coleoptera/Strepsiptera and in Lepidoptera and the loss of Su(Hw) in Lepidoptera, each supported by the analysis of two genomes with both methods (Fig. 2, Table S4). In five additional cases, we were not able to discover an expected ortholog in a single genome (Table S4). This may be a consequence of incomplete genome assemblies, but could alternatively reflect a dynamic nature of insulator evolution in arthropods. Importantly, these patterns of change are confined to the arthropod-specific insulators. We did not observe a loss of the more ancient CTCF insulator in any arthropod genome. This is compatible with the idea that CTCF function is needed for fundamental processes in the Bilateria (Heger et al. 2012) and might have been supplemented and modified by additional components in the arthropod lineage.
Although our results have been established by two fairly independent methods, we cannot formally prove the absence of orthologous proteins from certain clades. With growing databases and dependent on the sensitivity of homology detection tools, the exact placement of the origin of some proteins may still change.
However, irrespective of uncertainties in the exact time of gain and loss, our observations reveal a consistent model for the evolution of the known D. melanogaster chromatin insulators through a series of successive acquisitions.
This result has several implications. It has been confirmed repeatedly that insulator proteins colocalize and interact with each other (Gerasimova et al. 1995; Melnikova et al. 2004; Pai et al. 2004; Gerasimova et al. 2007; Bartkuhn et al. 2009; Negre et al. 2010; Van Bortle et al. 2012), thereby creating a network of dependencies that is thought to contribute to cell-specific differences in nuclear organization and gene expression (Yang and Corces 2011). Moreover, there is recent evidence that each D. melanogaster insulator subclass, determined by the presence of Su(Hw), CTCF, BEAF, or GAF, shares the CP190 protein and possibly also Mod(mdg4) (Van Bortle et al. 2012; Yang and Corces 2012). Our findings point out that the mechanisms, interactions, and components of insulator complexes must be considerably different from Drosophila not only in the great majority of arthropods (that share two of the seven factors), but also in other bilaterian phyla that only have CTCF in common. According to our findings, the presence of different insulator systems and the complex interactions between their components seen in D. melanogaster today are a result of ongoing evolution and diversification. These processes started more than ∼ 600 million years ago in the ancestor of bilaterians (www.timetree.org) with CTCF, the oldest known chromatin insulator of multicellular animals (Heger et al. 2012). At or close to the root of arthropods, Su(Hw) emerged. These two basal systems experienced subsequent additions and modifications, with the gain of BEAF-32 in the genus Drosophila, ∼ 60 million years ago, being the most recent acquisition. To what extent the successive acquisition of new insulator genes is involved in adaptive processes, shall be the topic of future investigations.
Although we can describe in detail an expansion of insulator proteins only in the Drosophila history, this process is not necessarily confined to a single species. Millions of arthropod species trace back to the same common ancestor that was equipped with the CTCF and Su(Hw) insulators. It is therefore possible that other arthropods simultaneously increased their initial repertoire of insulator proteins during the past 600 million years, giving rise to a plethora of unexplored territories. Thus, the expansion of insulator mechanisms that happened in D. melanogaster history might in fact be a general characteristic of arthropods and other animals. The description of non-CTCF insulators in echinoderms exemplifies that such an expansion could indeed apply to a greater variety of animals (Yajima et al. 2012). If so, this characteristic could provide a mechanism to modulate an ancient insulator system and fine-tune gene expression in a lineage-specific way.
Methods Summary
Blast-Based Search for Candidate Insulator Proteins
With the known Drosophila insulator proteins as query (dCTCF, GI:21356747; Su(Hw), GI:33860216; CP190, GI:23171337; GAF, GI:83287912; Mod(mdg4), GI:158030328; Zw5, GI:45549097; BEAF, GI:17647187), standard BLASTX, BLASTP, or TBLASTN (Altschul et al. 1997) searches were conducted in publicly available sequence databases at NCBI (http://blast.ncbi.nlm.nih.gov/). To minimize the chance of missing an ortholog, we performed parallel searches in different databases (nucleotide, EST, and protein) and subdivided a search into smaller entities if a given taxonomic range reported > 500 hits below threshold (= download restriction at the NCBI BLAST web interface). We collected all ZF domain candidates below a relaxed BLAST expectation value of 10−05. We set the threshold for BTB domain protein candidates to 10−14 because in preliminary experiments this value effectively incorporated BTB domain proteins distinct from BTB domain containing insulators, for example, tramtrack or broad complex. Collected nucleotide sequences were translated to the appropriate reading frame using EMBOSS (Rice et al. 2000).
Clustering and Multiple Sequence Alignment
We obtained from the initial dataset a collection of unique sequences using the EMBOSS tool “skipredundant” (Rice et al. 2000). We added to this collection a set of reference sequences for each insulator protein. The references served as guide for the clustering step. All other candidates from the initial dataset were passed to the SiLiX clustering algorithm (Miele et al. 2011) as “partial” sequences (command line parameter “-p”). We ran SiLiX with default parameters (35% min. identity; 80% min. overlap; 100 nt min. length; 50% min. overlap of partial sequences). For subsequent analysis, we took all reference sequences plus candidates that clustered to one of the references. Multiple sequence alignments were performed using the Clustal Ω algorithm (Sievers et al. 2011). Alignments were viewed and manually edited using SeaView (Galtier et al. 1996).
Phylogenetic Analysis
For the Zw5, Su(Hw), and CTCF proteins, the described Drosophila orthologs served as positive control for cluster identification. Similarly, we included the known Drosophila orthologs of the BTB-domain proteins GAGA factor, CP190, and Mod(mdg4) to specify these clusters. Outgroup for the analysis of ZF insulators was the widely distributed ZF transcription factor YY1. For BTB domain insulators, we used as outgroup a set of lola-like orthologs from diverse arthropods. Phylogenetic trees resulting from the alignments were computed under the maximum likelihood criterion using parallel RAxML version 7.2.6 (Stamatakis 2006) with 100 bootstrap resamplings. As optimal models of sequence evolution we used the WAG+Γ or DCmut+Γ+F model (ZF domain proteins), the JTT+Γ+F model (BTB domain proteins), and the WAG+Γ+F model (vertebrate GAFs) as selected by ProtTest3 (Darriba et al. 2011). Likelihood trees were visualized and arranged with FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and then graphically edited with Adobe Illustrator software.
HMM-Based Candidate Search and MCL-Clustering of Orthologous Groups
To verify our results with an independent method, we combined a search on the basis of insulator-specific hidden Markov models (http://hmmer.org/) with the Markov cluster algorithm (van Dongen 2000). We downloaded from various sites (Table S3) genome and, if available, proteome and unplaced reads data of 19 different arthropod and seven outgroup species. The 26 selected species represent maximal diversity while avoiding over-representation of well-sampled groups such as dipterans. As subsequent comparisons relied on protein sequences, we translated the 26 genomes and the respective unplaced reads data into all six reading frames (>90 nucleotides) and combined the resulting open reading frames of each species with the corresponding protein set as offered by the sequencing center. To obtain specific HMMer profiles, we selected—with attention to maximal diversity and length—for each insulator protein 8–15 previously verified orthologs as representatives of a cluster. We then calculated and manually refined multiple alignments of these sequences using the MAFFT “einsi” algorithm (Katoh et al. 2005) and derived a representative full length HMMer profile for each insulator protein. All 26 ORF sets were scanned with the six custom made HMMer profiles and all sequences below the default inclusion threshold of HMMSEARCH (E-value < 0.01; 39,573 unique sequences) were fed to a dedicated OrthoMCL pipeline (Li et al. 2003) as described elsewhere (http://orthomcl.org/common/downloads/software/v2.0/UserGuide.txt). We used the recommended inflation parameter “1.5” for the MCL step. Before clustering, we removed duplicates and supplemented each of the 26 sequence collections with reference insulator sequences of that species (verified by phylogenetic experiments) to facilitate cluster recognition. Orthologous clusters that contained at least one reference sequence were analyzed further. For CTCF, Mod(mdg4), Zw5, and BEAF, we detected a single orthology group whereas Su(Hw) (2), CP190 (4), and GAF (3) orthologs split to more than one group.
HHpred-Based Search for GAGA Factor Orthologs
Like described above, we constructed two multiple sequence alignments from representative members of the GAGA cluster (full length and BTB domain only) and uploaded these alignments to HHpred (http://toolkit.tuebingen.mpg.de/hhpred) for highly sensitive profile–profile searches. We searched with default parameters in the proteomes of H. sapiens and Mus musculus, the only deuterostome datasets available. After removing duplicates, we analyzed the remaining 45 candidates with phylogenetic methods (see above).
Associate Editor: E. Abouheif
Acknowledgments
This research was supported by grants from the German Research Foundation to TW (DFG-SFB680). Phylogenies and parallel BLAST were computed on CHEOPS, the Cologne High Efficient Operating Platform for Science. The genome sequences of Capitella teleta and Lottia gigantea were produced by the U.S. Department of Energy Joint Genome Institute, http://www.jgi.doe.gov/, in collaboration with the user community. Preliminary sequence data for Schmidtea mediterranea were obtained from the Genome Institute at Washington University, St. Louis (http://genome.wustl.edu/pub/organism/Invertebrates/). Preliminary sequence data for Strigamia maritima were obtained from the Baylor College of Medicine Human Genome Sequencing Center website at http://www.hgsc.bcm.edu/. Preliminary sequence data for Ixodes scapularis, Rhodnius prolixus, and Glossina morsitans were obtained from http://www.vectorbase.org. We thank the Daphnia Genomics consortium for providing preliminary sequence data for Daphnia magna. These sequence data were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu. We further wish to thank E. Schierenberg for support in the early stages of this work and B. Marin for helpful discussion.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s website:
Figure S1. Phylogeny of ZF domain insulator proteins.
Figure S2. Phylogeny of BTB domain insulator proteins.
Figure S3. Absence of GAGA factor orthologs from vertebrates I.
Figure S4. Absence of GAGA factor orthologs from vertebrates II.
File S1. Multiple sequence alignment of ZF domain proteins.
File S2. Multiple sequence alignment of BTB domain proteins.
File S3. Multiple sequence alignment of vertebrate GAGA factor candidates (BLAST).
File S4. Multiple sequence alignment of vertebrate GAGA factor candidates (HMM).
Table S1. The top 40 arthropod species providing ZF candidate sequences.
Table S2. The top 40 arthropod species providing BTB candidate sequences.
Table S3. Download location of 26 genomes and proteomes used for a HMM/OrthoMCL-based search for insulator proteins.
Table S4. Summary of the HMM/OrthoMCL-based search for insulator proteins.
Literature Cited
- Abzhanov A. Kaufman TC. Crustacean (malacostracan) Hox genes and the evolution of the arthropod trunk. Development. 2000;127:2239–2249. doi: 10.1242/dev.127.11.2239. [DOI] [PubMed] [Google Scholar]
- Altenburg E. Muller HJ. The genetic basis of truncate wing—an inconstant and modifiable character in Drosophila. Genetics. 1920;5:1–59. doi: 10.1093/genetics/5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W. Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof M. Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- Averof M. Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB. Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P. Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J. Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- Biggin MD. Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Bouhouche N, Syvanen M. Kado CI. The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol. 2000;8:77–81. doi: 10.1016/s0966-842x(99)01679-0. [DOI] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G. Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ND. Berg JM. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R. Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH. Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R. Krauss V. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica. 2003;117:165–177. doi: 10.1023/a:1022983810016. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O. Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM. Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK. Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Gilbert MK, Tan YY. Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–1375. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B. Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol. Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E. Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. USA. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S. White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL. Kaufman TC. Exploring the myriapod body plan: expression patterns of the ten Hox genes in a centipede. Development. 2002;129:1225–1238. doi: 10.1242/dev.129.5.1225. [DOI] [PubMed] [Google Scholar]
- Iampietro C, Gummalla M, Mutero A, Karch F. Maeda RK. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 2010;6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J. Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O. Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H. Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V. Dorn R. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene. 2004;331:165–176. doi: 10.1016/j.gene.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Kumar S. Remote homologue identification of Drosophila GAGA factor in mouse. Bioinformation. 2011;7:29–32. doi: 10.6026/97320630007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis DL, DeCamillis MA, Brunetti CR, Halder G, Kassner VA, Selegue JE, Higgs S. Carroll SB. Ectopic gene expression and homeotic transformations in arthropods using recombinant Sindbis viruses. Curr. Biol. 1999;9:1279–1287. doi: 10.1016/s0960-9822(00)80049-4. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ., Jr Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liubicich DM, Serano JM, Pavlopoulos A, Kontarakis Z, Protas ME, Kwan E, Chatterjee S, Tran KD, Averof M. Patel NH. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc. Natl. Acad. Sci. USA. 2009;106:13892–13896. doi: 10.1073/pnas.0903105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu NK, Hussain T, Sankaranarayanan R. Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J. Mol. Biol. 2010;400:434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G. Georgiev P. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A. Dekker J. Long-range chromosomal interactions and gene regulation. Mol. Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Penel S. Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011;12:116. doi: 10.1186/1471-2105-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. The origin of nine wing mutations in Drosophila. Science. 1911;33:496–499. doi: 10.1126/science.33.848.496. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhang YE, Negre N, Chen S, Long M. White KP. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol. 2012;10:e1001420. doi: 10.1371/journal.pbio.1001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Hartig G, Grath S, Pohl H, Lehmann J, Tafer H, Donath A, Krauss V, Eisenhardt C, Hertel J, et al. Genomic and morphological evidence converge to resolve the enigma of Strepsiptera. Curr. Biol. 2012;22:1309–1313. doi: 10.1016/j.cub.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S. Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D, Sheehan B, South H, Akbari O. Pai CY. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 2010;11:101. doi: 10.1186/1471-2121-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D. Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pamilo P. Nei M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1998;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A, Kontarakis Z, Liubicich DM, Serano JM, Akam M, Patel NH. Averof M. Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc. Natl. Acad. Sci. USA. 2009;106:13897–13902. doi: 10.1073/pnas.0902804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D. Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Peter IS. Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Karch F. Bender W. The bithorax complex: control of segmental identity. Genes Dev. 1987;1:891–898. doi: 10.1101/gad.1.9.891. [DOI] [PubMed] [Google Scholar]
- Phillips JE. Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW. Cunningham CW. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I. Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roy S, Jiang N. Hart CM. Lack of the Drosophila BEAF insulator proteins alters regulation of genes in the Antennapedia complex. Mol. Genet. Genomics. 2011;285:113–123. doi: 10.1007/s00438-010-0591-y. [DOI] [PubMed] [Google Scholar]
- Savard J, Tautz D. Lercher MJ. Genome-wide acceleration of protein evolution in flies (Diptera) BMC Evol. Biol. 2006;6:7. doi: 10.1186/1471-2148-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P. Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg TA. Labrador M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J. Mol. Evol. 2010;70:74–84. doi: 10.1007/s00239-009-9310-x. [DOI] [PubMed] [Google Scholar]
- Shao W, Zhao QY, Wang XY, Xu XY, Tang Q, Li M, Li X. Xu YZ. Alternative splicing and trans-splicing events revealed by analysis of the Bombyx mori transcriptome. RNA. 2012;18:1395–1407. doi: 10.1261/rna.029751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Strauss S, von Haeseler A. Hadrys H. A phylogenomic approach to resolve the basal pterygote divergence. Mol. Biol. Evol. 2009;26:2719–2730. doi: 10.1093/molbev/msp191. [DOI] [PubMed] [Google Scholar]
- Spana C, Harrison DA. Corces VG. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tadepally HD, Burger G. Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol. Biol. 2008;8:176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein MD, Wiegmann BM, Beutel R, Kjer KM. Yeates DK. Advances in insect phylogeny at the dawn of the postgenomic era. Annu. Rev. Entomol. 2012;57:449–468. doi: 10.1146/annurev-ento-120710-100538. [DOI] [PubMed] [Google Scholar]
- Van Bortle K. Corces VG. The role of chromatin insulators in nuclear architecture and genome function. Curr. Opin. Genet. Dev. 2013;23:212–218. doi: 10.1016/j.gde.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE. Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012;22:2176–2187. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S. Utrecht: Netherlands University of Utrecht; 2000. Graph clustering by flow simulation. [Google Scholar]
- Wallace JA. Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RW, Nagy L, Selegue J, Gates J. Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Kim JW, Cassel BK, Bertone MA, Winterton SL. Yeates DK. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biol. 2009;7:34. doi: 10.1186/1741-7007-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Fairbrother WG. Wessel GM. ISWI contributes to ArsI insulator function in development of the sea urchin. Development. 2012;139:3613–3622. doi: 10.1242/dev.081828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Corces VG. Insulators, long-range interactions, and genome function. Curr. Opin. Genet. Dev. 2012;22:86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Corces VG. Chromatin insulators: a role in nuclear organization and gene expression. Adv. Cancer Res. 2011;110:43–76. doi: 10.1016/B978-0-12-386469-7.00003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ramos E. Corces VG. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 2012;22:2199–2207. doi: 10.1101/gr.142125.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM. Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL. Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogeny of ZF domain insulator proteins.
Figure S2. Phylogeny of BTB domain insulator proteins.
Figure S3. Absence of GAGA factor orthologs from vertebrates I.
Figure S4. Absence of GAGA factor orthologs from vertebrates II.
File S1. Multiple sequence alignment of ZF domain proteins.
File S2. Multiple sequence alignment of BTB domain proteins.
File S3. Multiple sequence alignment of vertebrate GAGA factor candidates (BLAST).
File S4. Multiple sequence alignment of vertebrate GAGA factor candidates (HMM).
Table S1. The top 40 arthropod species providing ZF candidate sequences.
Table S2. The top 40 arthropod species providing BTB candidate sequences.
Table S3. Download location of 26 genomes and proteomes used for a HMM/OrthoMCL-based search for insulator proteins.
Table S4. Summary of the HMM/OrthoMCL-based search for insulator proteins.


