Abstract
Apoptosis in response to the ligand CD95L (also known as Fas ligand) is initiated by caspase-8, which is activated by dimerization and self-cleavage at death-inducing signaling complexes (DISCs). Previous work indicated that the degree of substrate cleavage by caspase-8 determines whether a cell dies or survives in response to a death stimulus. To determine how a death ligand stimulus is effectively translated into caspase-8 activity, we assessed this activity over time in single cells with compartmentalized probes that are cleaved by caspase-8, and used multiscale modeling to simultaneously describe single-cell and population data with an ensemble of single-cell models. We derived and experimentally validated a minimal model in which cleavage of caspase-8 in the enzymatic domain occurs in an interdimeric manner through interaction between DISCs, whereas prodomain cleavage sites are cleaved in an intradimeric manner within DISCs. Modeling indicated that sustained membrane-bound caspase-8 activity is followed by transient cytosolic activity, which can be interpreted as a molecular timer mechanism reflected by a limited lifetime of active caspase-8. The activation of caspase-8 by combined intra- and interdimeric cleavage ensures weak signaling at low concentrations of CD95L and strongly accelerated activation at higher ligand concentrations, thereby contributing to precise control of apoptosis.
INTRODUCTION
Extrinsic apoptosis is initiated by extracellular death ligands, such as CD95L (also known as Fas ligand) or TRAIL, and by formation of the death-inducing signaling complex (DISC) (1), which serves as a platform for the activation of initiator caspases, caspase-8 and caspase-10. These enzymes cleave and activate effector caspases, caspase-3 and caspase-7, and cleave the proapoptotic Bcl-2 family member BID into tBID, which induces mitochondrial outer membrane permeabilization (MOMP). MOMP irreversibly triggers effector caspase activation by releasing further proapoptotic proteins. In type I cells, the activity of initiator caspases is sufficient for direct activation of effector caspases, whereas type II cells require indirect activation mediated by BID cleavage and MOMP (2, 3). Either type of cells can survive exposure to death ligand, if the activity of initiator caspases is not sufficient to cleave enough substrates. Despite extensive characterization of caspase-8 and caspase-10 activation, cleavage, and other posttranslational modifications, little is known regarding how their cellular activity is effectively generated and controlled over time and how the activity of these proteases allows cells to decide between death and survival.
DISCs initiated by the CD95 receptor (CD95R; also known as Fas) contain the clustered receptors bound to the adaptor protein Fas-associated death domain protein (FADD) on their cytosolic domain, on which dimers of procaspase-8 are assembled (4, 5). The two main procaspase-8 isoforms, procaspase-8a and procaspase-8b (p55 and p53), consist of a prodomain, which interacts with FADD, and an enzymatic domain with two active subdomains. The prodomain and the enzymatic subdomains are connected with linkers that can be cleaved by caspase-8 itself. Cleavage of procaspase-8 at Asp374 and Asp384 between the catalytic subdomains generates procaspase-8 fragments known as p43 (or p41 for the b isoform) and p10, which typically appear first after activation (fig. S1) (6, 7). Cleavage of procaspase-8 at Asp210 and Asp216, between the prodomain and the catalytic subunits, leads to the formation of p26 (or p24 for the b isoform) and p30 (8). Further cleavage events occur on the preprocessed procaspase-8 fragments p43 and p30; the cleavage of p43 (or p41 of the b isoform) at Asp210 and Asp216 produces more p26 (or p24 for the b isoform) and p18, and the cleavage of p30 at Asp374 and Asp384 leads to the formation of p18 and p10 fragments (8). Fully cleaved caspase-8 is released from the DISC to the cytosol as the heterotetramer (p18)2(p10)2, which we refer to for simplicity as p18.
Uncleaved procaspase-8 dimers can cleave themselves and a restricted group of local DISC-bound proteins (9, 10), whereas cleavage to p43 (or p41 for the b isoform) leads to a “substrate switch” enabling the cleavage of such downstream effectors of apoptosis as BID or procaspase-3 (11). Proximity-induced activation of caspase-8 is attributed to dimerization, whereas cleavage of the linker between enzymatic subdomains in procaspase-8 dimers is thought to stabilize the dimeric conformation (12, 13). Cleavage of this linker is required for the caspase-8 substrate switch toward downstream substrates (11, 14). Thus, two fully active caspase-8 pools are constituted from this activation process, both of which are processed in the linker between the enzymatic subdomains: a membrane-associated pool represented by p43 (and p41) and a cytosolic pool of p18, which is commonly referred to as caspase-8 and is the heterotetramer.
Autoprocessing of procaspase-8 can occur in three ways: (i) by an intramolecular process within the same molecule, (ii) by an intermolecular reaction between procaspase-8 molecules colocalized in the same DISC (intradimer processing), or (iii) by an intermolecular reaction between procaspase-8 molecules located in different DISCs (interdimeric processing). The relative contribution of these autocleavage mechanisms is expected to affect the dynamics of caspase-8 activation: Intramolecular and intradimeric processing occurs within the same molecular complex and can thus be described as a first-order process with kinetics not depending on the stimulus strength. In contrast, interdimeric processing involves a bimolecular reaction and would require transient interaction of receptor complexes in the plasma membrane. Therefore, interdimeric processing would result in accelerated caspase-8 autoprocessing with increasing stimulus strength. Complementary results have been obtained with regard to the intramolecular and intermolecular cleavage of caspase-8. Chen et al. (15) proposed that the first cleavage event in caspase-8 processing in the enzymatic domain occurs by an intramolecular process. Structural studies of dimeric procaspase-8 indicated that the cleavage sites in the enzymatic domain may access the active site of the attached protomer, thus favoring an intradimer processing mechanism (16, 17). Furthermore, active, processed caspase-8 can cleave dimeric procaspase-8 through an interdimeric process (6, 18). Here, we investigated the relative contribution of the experimentally observed cleavage mechanisms to caspase-8 activation and apoptosis induction in living cells.
To understand how caspase-8 activation is effectively regulated to produce the appropriate cellular response, we developed a mathematical model reflecting the mechanism of caspase-8 cleavage and activity. We used a modeling approach, in which an ensemble of single-cell models was fitted to single-cell observables in combination with population observables. Subsequently, cell-to-cell variability in apoptosis kinetics was explained by sets of initial protein concentrations estimated for each cell. We identified a minimal model variant that was experimentally validated; prodomain cleavage sites were cleaved within dimers, and enzymatic domain sites were cleaved between dimers. By model analysis, we showed how the auto-processing mechanism of caspase-8 and the concentrations of signaling proteins along the cell death pathway control the dynamics of apoptosis induction.
RESULTS
Using cleavage probes to obtain single-cell estimates for caspase-8 activity at the membrane and in the cytosol
Caspase-8 substrates, such as caspase-3 and BID, can be cleaved by the caspase-8 intermediate p43 or p41 (subsequently, we refer only to the p43 of the a isoform, but this represents both isoforms) at the plasma membrane and its final product (p18)2(p10)2 (referred to as p18), which is released to the cytosol. Neither p55 nor p53 (later referred to as p55) nor p30 cleaves downstream caspase-8 substrates, such as BID and procaspase-3 (11, 14). We developed a technique for quantifying the cleavage activities of the p43 (membrane-bound) and p18 (cytosolic) intermediates in single cells, using two compartmentalized caspase-8–specific cleavage probes (Fig. 1A). These cleavage probes are targeted to different cellular compartments by fusing fluorescence proteins to localization domains through the cleavage sequence Ile-Glu-Thr-Asp (IETD). Upon caspase-8 activation, the linker is cleaved and the fluorescent protein is released from its localization domain, allowing the fluorescent proteins to equilibrate over the entire cell. Dynamic changes in nuclear intensity, measured by time-resolved confocal microscopy imaging, served as the readout for enzymatic activity (19).
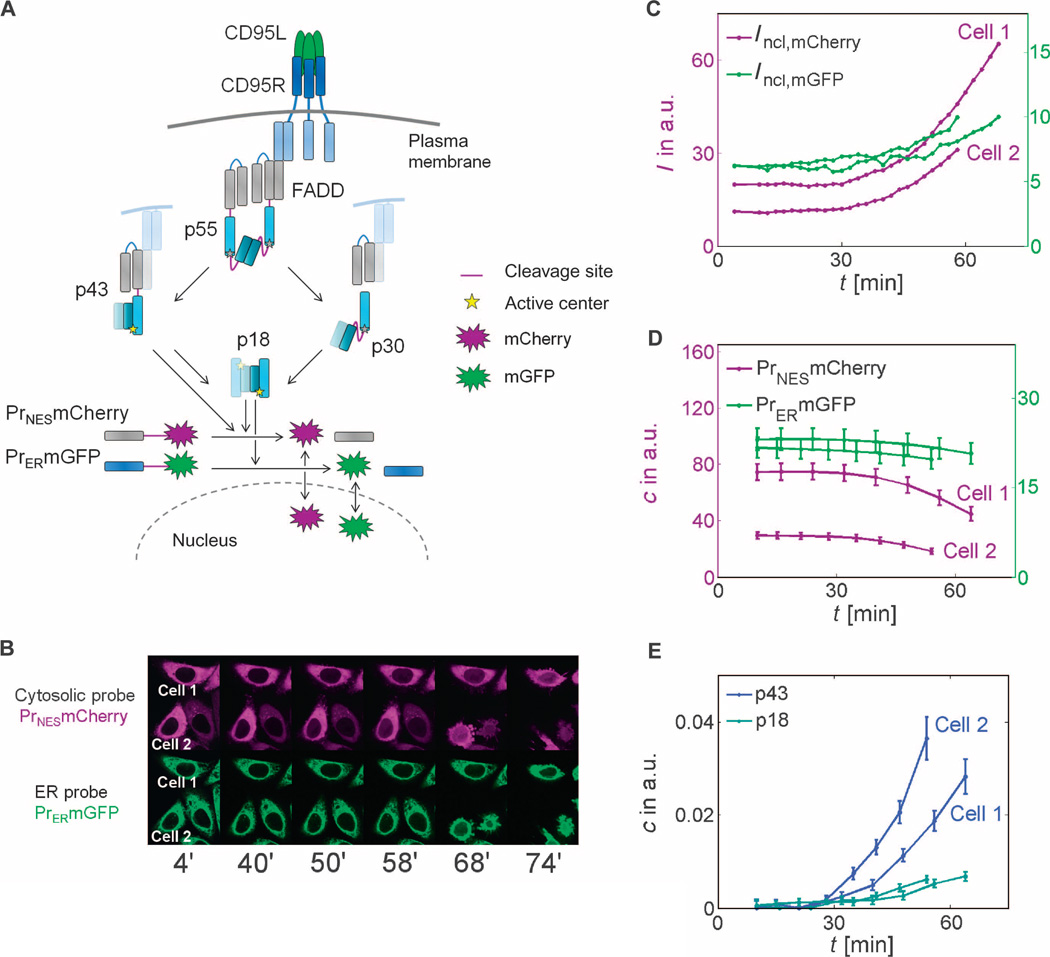
Fig. 1. Observation of single-cell caspase-8 trajectories with cleavage probes.
(A) Schematic representation of CD95 signaling. Subsequent to CD95L binding to CD95R and FADD binding, dimers of procaspase-8 (p55) are assembled and can be cut at two cleavage sites (drawn in purple). Cleavage in the catalytic domain (dark and light blue segments represent the p10 and p18 subdomains) generates p43, which is further processed in the prodomain (gray segments represent the two death domains) to p18, which enters the cytosol. Prodomain linker cleavage in p55 produces catalytically inactive p30. Whereas the cytoplasmic probe can be cleaved by p43 and p18, the ER probe is cleaved by p18 only. (B) Live-cell imaging of probe cleavage (red channel: mCherry/cytosolic probe; green channel: mGFP/ER probe). (C to E) In two marked cells, time series of nuclear fluorescence intensities (C) were measured and transformed to concentrations of uncleaved probes (D) or p43 and p18 (E) using Eqs. 2 and 3. (B to E) Quantification of enzyme activities in two exemplary CD95-HeLa cells. I, intensity; c, concentration; a.u., arbitrary units.
To distinguish the cleavage activity of caspase-8 at the plasma membrane and within the cytosol, we fused calnexin, which is localized at the surface of the endoplasmic reticulum (ER), to monomeric green fluorescent protein (mGFP) through IETD. Cleavage of this probe, which is catalyzed by the cytosolic form of caspase-8 (p18), allowed mGFP to enter the nucleus (Fig. 1, A to C). The other cleavage probe uses a nuclear export signal (NES) fused to the red fluorescent protein mCherry through IETD and thus before cleavage is present throughout the cytoplasm where it can be cleaved by caspase-8 both at the plasma membrane (p43) and within the cytosol (p18). Upon cleavage, mCherry can enter the nucleus (Fig. 1, A to C).
We then developed a mathematical model to derive the enzymatic activities of p18 and p43 from the imaging-based cleavage data (Fig. 1B). Rearranging the probe cleavage rate equation yields an expression for the total caspase-8 enzymatic activity, that is, the caspase-8 concentration times the rate constant of probe cleavage kprobe,
| (1) |
[PrF] and d[Pr]/dt are the uncleaved probe concentration and the temporal change of cleaved probe concentration, respectively. We neglected saturation in caspase-8–mediated probe cleavage because caspase-8 shows high KM values in the range of several micromolar for the cleavage sequence IETD (20, 21). We observed that the kinetics of cell death were independent of the probe concentration within the cell, further suggesting that caspase-8 saturation and substrate competition were negligible (fig. S2). Equation 1 was reformulated as a function of average probe intensities in the nucleus (IER,ncl and INES,ncl) and in the total cell (IER,tot and INES,tot), and their slopes were derived from live-cell imaging data (for details, see text S1: Data analysis). We used the ER probe intensities to determine p18 enzymatic activity
| (2) |
To estimate the enzymatic activity of p43, it is necessary to subtract the p18 activity (Eq. 2) from the term for NES probe cleavage
| (3) |
For simplicity, we assumed that p43 and p18 cleaved the probes equally well. This assumption is valid because the critical requirements for generating cleavage activity toward downstream substrates are equally present in p43 and p18: In both forms, the enzymatic subdomains are in dimeric conformation and the dimer is stabilized owing to cleavage of the linker between the enzymatic subdomains (16, 17). The presence of the prodomain in p43 (but not in p18) is unlikely to affect the catalytic activity toward the probes, because the crystal structure of caspase-8 dimers shows that the linkers between the dimerized enzymatic domains and the prodomain parts are distant from the active centers (20, 22). This suggests that probe cleavage by these two active caspase-8 forms can be described with the same kinetic constant kprobe. Caspase-8 activation is enhanced by caspase-3–mediated feedback cleavage, which is activated after MOMP in the last 6 to 8 min before apoptosis (23). Before MOMP, caspase-8 and not caspase-3 contributes to probe cleavage (19). Because we here focused on the initial dynamics of caspase-8 autoprocessing, we excluded the last 10 min before bleb appearance from analysis. We recorded the kinetics of probe cleavage in wild-type HeLa cells and in CD95-HeLa cells, which stably overexpress CD95 10-fold compared to the wild-type cells (24, 25). For each ligand concentration, more than 100 cells were analyzed for each cell line. Time of death, visually defined as the time of apoptotic membrane blebbing, was between 1 hour and more than 12 hours for wild-type HeLa cells and between 40 and 120 min for CD95-HeLa cells (fig. S3).
We used the probe localization to quantitatively estimate caspase-8 activity using our simple model of probe cleavage (Eqs. 1 to 3) (Fig. 1, B to E). The rate of probe cleavage was generally low in the beginning (around the first 30 min) and then was strongly enhanced (exemplary trajectories of two CD95-HeLa cells are shown in Fig. 1C; other data in fig. S3). Temporal profiles were confirmed with fluorescence resonance energy transfer (FRET) cleavage probes (23) (fig. S4). Fluorophore intensities were processed such that the amount of uncleaved probe was directly comparable (Fig. 1D). The similarity in calculated enzyme activity trajectories (Fig. 1E) between individual cells indicated that the method was robust. The activity of p43 (membrane-associated activity) exhibited a steep increase after the latency period, and this membrane-associated activity exceeded the activity of cytosolic p18, which showed a delayed start and a more gradual increase in activity than the p43 activity.
Kinetic modeling of receptor oligomerization explaining the inverse bell–shaped dose response of caspase-8 activity
To investigate the quantitative dynamics of caspase-8 activation, we developed kinetic models comprising two parts: one described receptor oligomerization and one described subsequent caspase-8 activation mechanisms. To characterize caspase-8 processing and identify potential nonlinearities, we performed a dose-response analysis (50 ng/ml to 10 µg/ml) with soluble CD95L fused to the T4-Foldon motif from the fibritin of the bacteriophage T4 (T4-CD95L), which stabilizes the ligand in its trimeric form (26). For wild-type HeLa cells, the rate of time to apoptosis was slower at high concentrations of T4-CD95L compared to the rate to apoptosis of cells exposed to intermediate concentrations (Fig. 2A). Because we observed a similar inverse bell–shaped dose-response behavior for single-cell caspase-8 activities (fig. S3; decreasing caspase-8 activity and delayed apoptosis at high CD95 concentration in wild-type HeLa cells and, to a lesser extent, in CD95-HeLa cells), we hypothesized that this characteristic behavior emerges early in the signaling cascade, upstream of caspase-8 activation, possibly at the receptor level.
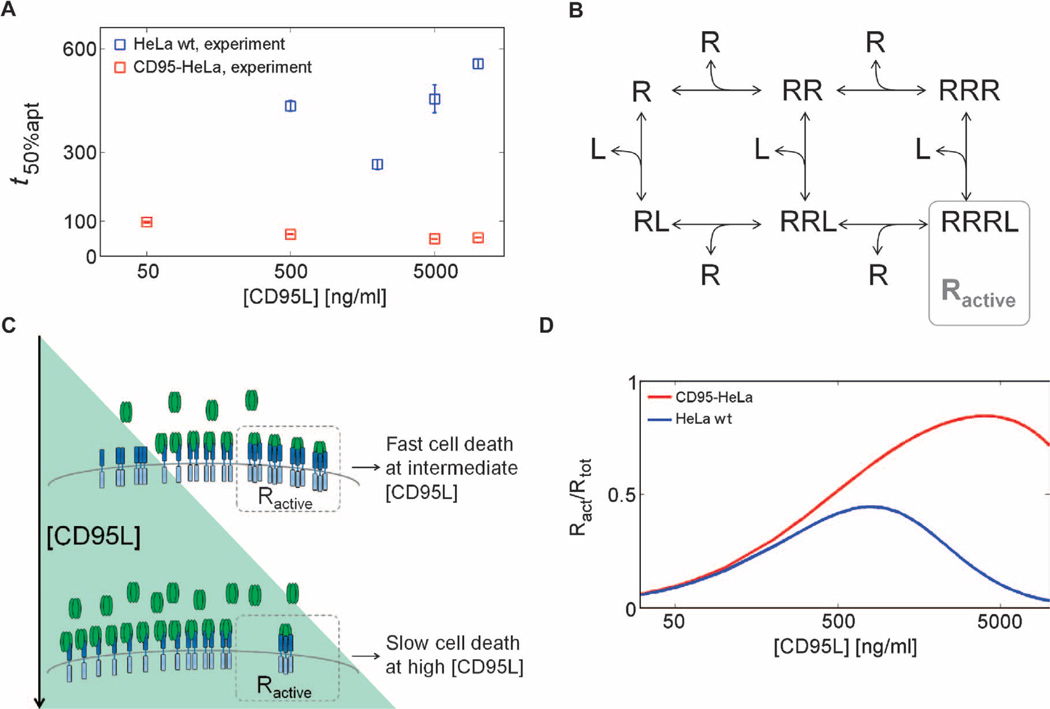
Fig. 2. Inverse bell–shaped dose-response profile of cell death.
(A) Median cell death times (squares) for wild-type HeLa (HeLa wt) and CD95-HeLa cells. Error bars represent SDs from medians in different microscopic fields. (B) Receptor oligomerization model topology, in which receptor (R) trimers bound to ligands (L) represent active receptors Ractive. (C) Mechanism of inverse bell–shaped behavior. At high ligand concentrations, most bound ligands are connected to receptors in a 1:1 stoichiometry, resulting in a low number of oligomerized receptors capable of caspase-8 activation. (D) Simulations for active receptor fractions for CD95-HeLa and HeLa wt cells calculated with kinetic parameters from model fitting. Ract, Ractive.
We used a mathematical model of ordinary differential equations (ODEs) based on known features of CD95R-CD95L binding to analyze the mechanisms underlying the inverse bell–shaped dose-response profile in the wild-type HeLa cells (Fig. 2B and text S2: Model implementation and fitting). The monomeric receptor R reversibly homotrimerizes and interacts with the trimeric ligand L to form what we assume is the biologically active ligand-bound receptor trimer RRRL. Because T4-CD95 ligand is a pure and stable trimer (26), we hypothesized that very high ligand concentrations inhibit signaling because each ligand molecule may interact with a single monomeric receptor, thereby preventing the formation of the active RRRL complex (Fig. 2C). Corresponding model simulations showed a peak of active receptor at intermediate ligand concentrations (Fig. 2D), which corresponds to the fast apoptosis kinetics measured at intermediate ligand concentrations. This inhibition at the high concentrations we used was not readily detected in the CD95-HeLa cells (Fig. 2A), which likely reflects that these cells have more receptor and thus would require even higher concentrations to achieve the inverse bell shape apoptotic kinetics. The simulations were consistent with this and showed only a decrease in apoptotic kinetics at the highest concentrations analyzed (Fig. 2D). To conclude, our results indicated that the observed inverse bell shape in caspase-8 activity can be explained on the basis of the dynamics of receptor oligomerization.
Kinetic model of caspase-8 activation by cis or trans cleavage mechanisms
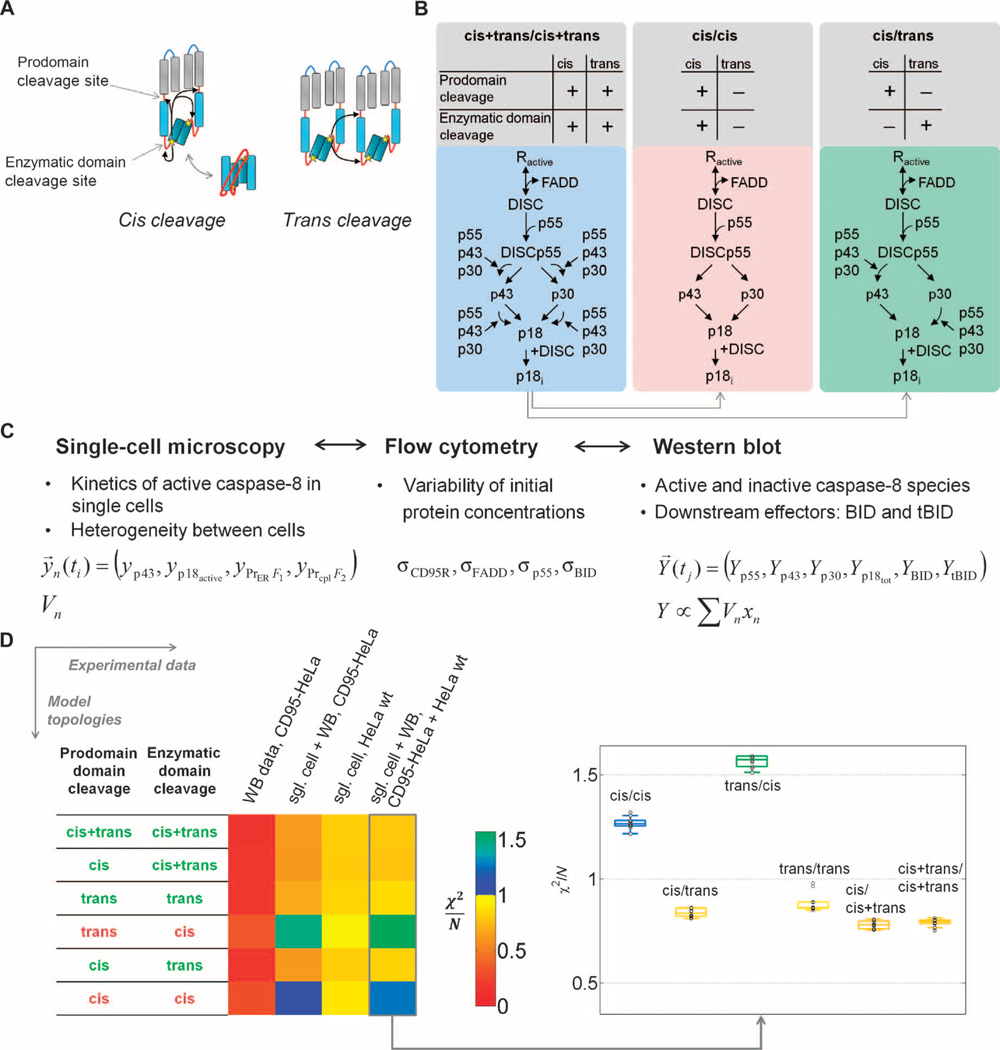
Several mechanisms have been proposed for caspase-8 autocleavage (27). Two classes of mechanisms could lead to different response characteristics: The ones that result in apparent intradimeric (“cis”) mechanisms would be independent of input signal strength, and the ones that cause apparent interdimeric (“trans”) mechanisms would be altered by input signal strength (Fig. 3A). Cis mechanisms can be seen as unimolecular reactions, and thus, the kinetics would be independent from input strength. In contrast, trans mechanisms introduce bimolecular reactions, implying accelerated activation rates with increased input strength. We formulated alternative model variants of caspase-8 autoprocessing that included the above described receptor activation model (Fig. 2B) and discriminated between intra- and interdimeric processing modes by fitting to experimental data (Fig. 3B). Here, we give a general outline of the model (for additional assumptions see text S2: Model implementation and fitting). We used mass action–based ODEs to describe DISC formation; p55 binding; the activation, release, and deactivation of caspase-8 (p18); as well as substrate cleavage (Fig. 3B). In the model, DISC formation involved ligand, receptor, FADD, and procaspase-8. c-FLIP proteins can influence caspase-8 activity: At low concentrations, c-FLIP inhibits apoptosis (24, 25), but at moderate to high concentrations, c-FLIP can promote apoptosis (25). We did not explicitly include c-FLIP in the model. Neglecting c-FLIP could mean that we have slightly over- or underestimated some of the caspase-8 kinetic parameters, but because c-FLIP abundance was not perturbed in our experiments and was low, in the range of a few hundred compared to 3 × 105 procaspase-8 molecules per cell (24, 25), we think that any effect is minor. Procaspase-8 autoprocessing can only occur in a dimer configuration where two (pro)caspase-8 molecules are in close proximity. The trimeric CD95 receptor must therefore recruit two or more FADD and procaspase-8 molecules, which then need to remain bound until auto-processing is completed. Procaspase-8 autocleavage is to some extent a random-ordered process and may thus proceed in two alternative routes. In processing of p55 through p43 to p18, cleavage between the enzymatic subdomains precedes cleavage between prodomain and enzymatic domain. In processing of p55 through p30 to p18, cleavage between the prodomain and enzymatic domain precedes cleavage between the enzymatic sub-domains (fig. S1). We confirmed the existence of two competing pathways by analyzing a caspase-8 mutant lacking the enzymatic domain cleavage sites (fig. S5). To avoid combinatorial complexity, we made several assumptions for describing the dimer dynamics by a simplified reaction scheme with monomeric reaction steps (Fig. 3B; for details, see text S2: Model implementation and fitting).
Fig. 3. Model-based discrimination of caspase-8 activation mechanisms.
(A) Intra- versus interdimeric cleavage mechanisms: Active centers (stars) in procaspase-8 can cleave domain linkers (red lines) within procaspase-8 dimers (cis) or between neighboring dimers (trans). (B) Schematic representation of model variants. The enzymatic domain sites and the prodomain sites are potentially cleaved in cis, in trans, or in both cis and trans. Here, the maximal variant cis+trans/cis+trans and two minimal variants are symbolized: a pure cis and the cis/trans topology. See fig. S6B for the other models. (C) Representation of data from different experimental techniques in the model. Single-cell models were fitted to single-cell measurements yn. Population- based measurements Y for caspase-8 intermediates, BID, and tBID are represented by sums of single-cell variables xn weighted by cellular volumes Vn. Distribution parameters from flow cytometry and mean initial protein concentrations from Western blot were not directly fitted but used to constrain parameter limits. (D) Model discrimination between cis and trans cleavage variants. Color-coded χ2/N values, indicating goodness of fit, for the 1% best of n = 1000 fits are shown for model topologies that were fit to experimental data of increasing complexity, and box plots for fits to the full experimental data set, in which box colors indicate median χ2/N values. Models with χ2/N > 1 were regarded as unable to explain experimental data.
In the following, model variants are abbreviated by the cleavage modes—cis, trans, or cis+trans—of the two linkers. For example, cis/trans means that the prodomain linker cleavage can be described as unimolecular, and the catalytic linker can be described as a bimolecular trans reaction. Because the cis and trans mechanisms of each linker cleavage are not mutually exclusive, three topologies are possible for each linker, generating a total of nine different topologies (fig. S6; three examples are shown in Fig. 3B). Together with cleavage reactions for the cytosolic probe, the ER-anchored probe, and BID, the models contain between 13 and 25 reactions (table S1), 16 species, and 11 to 16 kinetic parameters (tables S2 and S3). Model parameters were unknown and determined by fitting to experimental data as described in the following.
Integrating single-cell and population data with an ensemble of single-cell models
Mathematical models of signal transduction are typically fitted to cell population data that have two main disadvantages: They hide cell-to-cell variability and smoothen kinetics that would be seen in individual cells. Here, we simultaneously fitted an ensemble of single-cell models to single-cell trajectories and population data. This cell ensemble modeling approach enabled description of the variability in a heterogeneous population of cells and also consideration of the dynamics of species that can be only measured on the population level. Combining single-cell data with population data further constrained the model and increased parameter identifiability.
Our single-cell measurements based on cleavage probes are restricted to catalytically active caspase-8 forms. We enriched our data set by population-based Western blot measurements of p55, p43, p30, p18, BID, and tBID in CD95-HeLa cells at 50 and 500 ng/ml (figs. S7 and fig. S8). We integrated experimental data obtained by several techniques, providing information of different qualities, in a multiscale modeling approach (Fig. 3C). To describe a heterogeneous cell population, we constructed an ensemble of single-cell models, in which each cell is described with the same sets of ODEs and the same kinetic parameters, but by different initial protein concentrations, thus introducing cell-to-cell variability. The ensemble of single-cell models was fitted to single-cell trajectories for the concentrations of the uncleaved probes, [PrER-mGFP] and [PrNES-mCherry], as well as the concentrations of [p43] and [p18], from an ensemble of cells. Along with fitting of individual parameters, ensemble averages were fitted to Western blot data, which were represented in the model by averages of the corresponding single-cell model variables (Fig. 3C; for details, see text S2: Model implementation and fitting).
To restrict fitting to relevant initial concentrations, we used GFP fusions to determine average molecule numbers of FADD, p55, and BID per cell by calibrated Western blot (table S4). For calibration, GFP fusions were blotted together with known amounts of recombinant GFP, and GFP fusions were blotted together with wild-type HeLa and CD95-HeLa lysates (figs. S9 to S11). Calibrated beads were used to determine average amounts of CD95R in the plasma membrane detected by flow cytometry (fig. S12 and table S5). We additionally restricted the cell-to-cell variability of signaling protein concentrations to a physiologically relevant range using flow cytometric measurements (figs. S12 and S13 and tables S6 and S7).
Model prediction of a combination of cis and trans cleavage for caspase-8 activation
Ensemble models of different caspase-8 autoprocessing scenarios were fitted to experimental data to discriminate between alternative hypotheses on cis or trans mechanisms. We first compared the capacity of the four smallest models—cis/trans, cis/cis, trans/cis, and trans/trans—to fit the data (Fig. 3B). These models are smaller compared with the model that contains the highest degrees of freedom, namely, cis+trans/cis+trans (fig. S6).
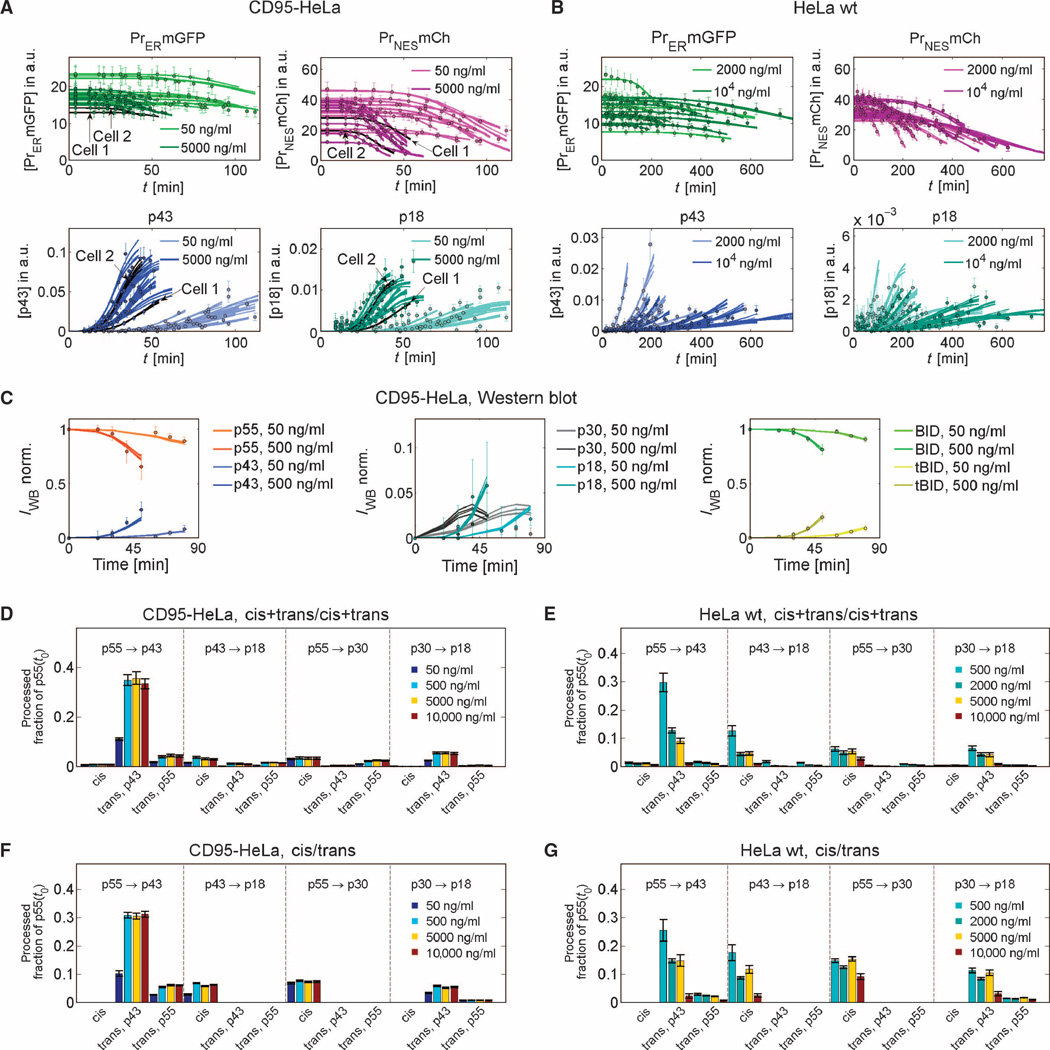
Strikingly, the two minimal models that constrain the cleavage in the catalytic domain to a cis mechanism could not reproduce the data (Fig. 3D). In contrast, the variants cis/trans and trans/trans could fit the data (χ2min,cis/trans/N = 0.81; χ2min,trans/trans/N = 0.85). Moreover, fitting with extended model variants did not significantly improve the fit (fig. S14). As an example, Fig. 4, A to C, illustrates the fitting of the cis/trans variant to single-cell data and Western blot population data for two of four concentrations of CD95L. Thus, the minimal models cis/trans and trans/trans were sufficient to explain the data (Fig. 3D). Analysis of reaction fluxes in different cleavage reactions further showed that the best fit of the full cis+trans/cis+trans version behaved similarly to the minimal cis/trans variant that explained the data: Most of the cleavage of prodomain linker in the cis+trans/cis+trans model originated from the cis mechanism, and most of the cleavage of the catalytic subunits originated from the trans mechanism (Fig. 4, D to G; for details, see text S3: Model analysis and verification, and figs. S15 to S17).
Fig. 4. Cis/trans model fits and flux analysis.
(A to C) Fits of the ensemble of single-cell cis/trans models to single-cell data of 80 cells (2 cell lines × 4 ligand concentrations × 10 cells per condition) or Western blot data (best 0.5% of n = 1000 fits). For clarity, only fits and data at two of four concentrations are shown. Experimental data (points) and model fits (lines) for cleavage probes, p43, and p18 for CD95-HeLa (A) and HeLa wt (B) cells. (C) Western blot data for CD95-HeLa cells for caspase-8 intermediates, BID, and tBID and model fits. (D to G) Flux analysis for different procaspase-8 cleavage reactions shows that the cis+trans/cis+trans model essentially behaves like the cis/trans variant. Bars indicate fractions of the initial p55 pool that are processed until median apoptosis times via different cleavage reactions at different ligand concentrations. Substrates and products of the reactions are indicated on top of the bars. For trans reactions, the enzymes are indicated below the bars. (D) and (E) show predictions of the cis+trans/cis+trans model, and (F) and (G) of the smaller cis/trans variant, for CD95-HeLa (D and F) or HeLa wt cells (E and G). Essentially, the cis+trans/cis+trans model behaves like the cis/trans version because fluxes in trans-cleavage reactions of the enzymatic domain cleavage sites (p55 to p43 and p30 to p18) and fluxes in cis-cleavage reactions of the prodomain cleavage sites (p43 to p18 and p55 to p30) are small and can be neglected. Error bars represent SDs for the best 1% of n = 1000 fits.
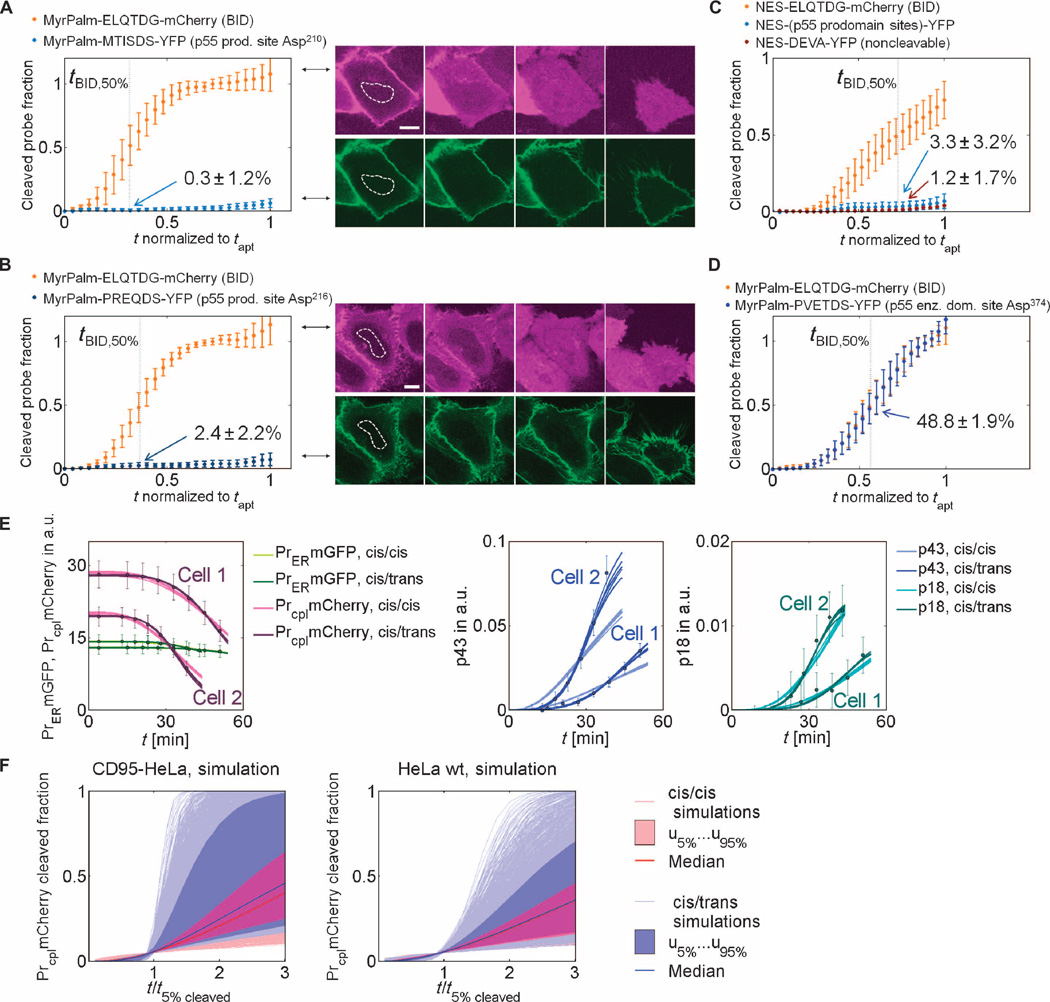
We sought to confirm the predicted cis/trans mechanism by an independent set of experimental data not used for model calibration. Therefore, we generated cleavage probes containing different caspase-8 autoprocessing sites and analyzed their cleavage dynamics. Cleavage probes containing the prodomain linker sequence Met-Thr-Ile-Ser-Asp-Ser (MTISDS) for prodomain cleavage site Asp210 or Pro-Arg-Glu-Gln-Asp-Ser (PREQDS) for prodomain cleavage site Asp216 require a bimolecular reaction step for cleavage by caspase-8. By localizing probes with a myristoylation-palmitoylation (MyrPalm) domain on the plasma membrane, we mimicked a caspase-8 trans autocleavage scenario (Asp210: MyrPalm-MTISDS-YFP or Asp216: MyrPalm-PREQDS-YFP; for details, see text S3: Model analysis and verification). Consistent with the predicted cis mechanism, we found that probes with MTISDS or PREQDS as the cleavage site in the linker showed no substantive cleavage before cell death, compared to a probe with the cleavage sequence of BID (MyrPalm-ELQTDG-mCherry, ELQTDG: Glu-Leu-Gln-Thr-Asp-Gly) (Fig. 5, A and B). Similar results were obtained with probes composed of the p55 prodomain sites representing a more complete caspase-8 prodomain linker containing Asp210, Asp216, and the less relevant cleavage site Asp223 (Fig. 5C). In contrast, the model predicted trans cleavage of the enzymatic domain linker. We therefore expected efficient cleavage of a probe with PVETDS, corresponding to the enzymatic domain site Asp374, as the cleavage site in the linker. As predicted, the PVETDS probe was fully cleaved at the time of death, comparable to an ELQTDG probe containing the cleavage sequence of BID (Fig. 5D). Together, these data provided independent support for the model-predicted cis/trans cleavage mechanism of caspase-8 autoprocessing.
Fig. 5. Experimental validation of model predictions and Monte Carlo simulations.
(A) Averaged normalized intensities from nuclear regions of interest (ROIs; white dotted lines) recorded in 21 HeLa wt cells expressing MyrPalm-ELQTDG-mCherry (top image row) and MyrPalm-MTISDS-YFP (bottom image row) probes. (B) Averaged nuclear intensities from 16 HeLa wt cells expressing MyrPalm-ELQTDG-mCherry (top image row) and MyrPalm-PREQDS-YFP (bottom image row) probes. The amount of the prodomain site cleavage was calculated from the trajectories in (A) and (B), and the percentage cleaved when half of the BID probes was cleaved (tBID,50%) is shown for each graph. Error bars, SDs; scale bars, 10 µm; p55 prod. site, p55 prodomain cleavage site. (C) Averaged nuclear intensities from 39 HeLa wt cells expressing NES-ELQTDG-mCherry probes, 16 HeLa wt cells expressing NES-(p55 prodomain sites)-YFP probes, and 23 cells expressing NES-DEVA-YFP probes, with a control sequence that cannot be cleaved. The amounts of probe cleavage at tBID,50% for the probe with prodomain cleavage sites and for the DEVA probe are shown in the graph. (D) Enzymatic domain cleavage can occur by a trans mechanism. Averaged normalized intensities from nuclear ROIs recorded in 20 HeLa wt cells expressing the two probes, MyrPalm-ELQTDG-mCherry and MyrPalm-PVETDS-YFP. The amount of the probe cleavage at tBID,50% for each probe was about the same and is shown in the graph. p55 enz. dom. site, p55 enzymatic domain cleavage site. (E) Model fits to two exemplary CD95-HeLa trajectories, indicated in Fig. 4A, at [CD95L] = 5000 ng/ml. Left graph: model fits to cytosolic and ER probe concentrations; middle graph: fits to the p43 cleavage activities; right graph: fits to the p18 cleavage activities. (F) Cytosolic probe cleavage trajectories from random parameter (Monte Carlo) simulations for CD95-HeLa (left plot) and HeLa wt cells (right plot). Trajectories were obtained by randomly sampling kinetic rate constants (Nsamples = 104) from large parameter intervals, temporally normalized by time t5% when 5% of the probes are cleaved, and normalized in amplitude by the total cytosolic NES-probe concentrations [shaded areas: space between 5% and 95% percentiles (u5% and u95%); solid lines: medians].
Typically, previous mathematical models were fitted to population data only. To investigate whether our ensemble modeling approach combining single-cell and population data enabled more reliable modeling of cellular systems, we tested if reduced amounts of experimental data would also be sufficient for model discrimination. For the wild-type HeLa cells, neither the Western blot (population) data alone nor the single-cell data alone were sufficient to discriminate among the models (Fig. 3D). However, when using data sets of combined single-cell and population data from CD95-HeLa, or from both wild-type HeLa and CD95-HeLa, we obtained clear and reproducible model discrimination.
We investigated why a trans cleavage of the enzymatic domain was required in the model to explain the experimental data. A cis mechanism for the cleavage of the catalytic domain linker failed to reproduce the steep increase in the p43 trajectories after a certain period of low enzymatic activity (Fig. 5E). A similar conclusion could be drawn when analyzing the qualitative features of cis/cis and cis/trans models using simulations with randomly chosen kinetic parameters in a Monte Carlo approach (Fig. 5F and fig. S18; for details, see text S3: Model analysis and verification). In contrast to the cis/cis models, the cis/trans models generated switch-like trajectories, especially in CD95-HeLa cells. Therefore, our modeling indicated that a trans mechanism is required to explain the experimental data and to establish the switch-like initiation of apoptosis.
p43 generation by itself through a positive feedback mechanism and function as a molecular timer
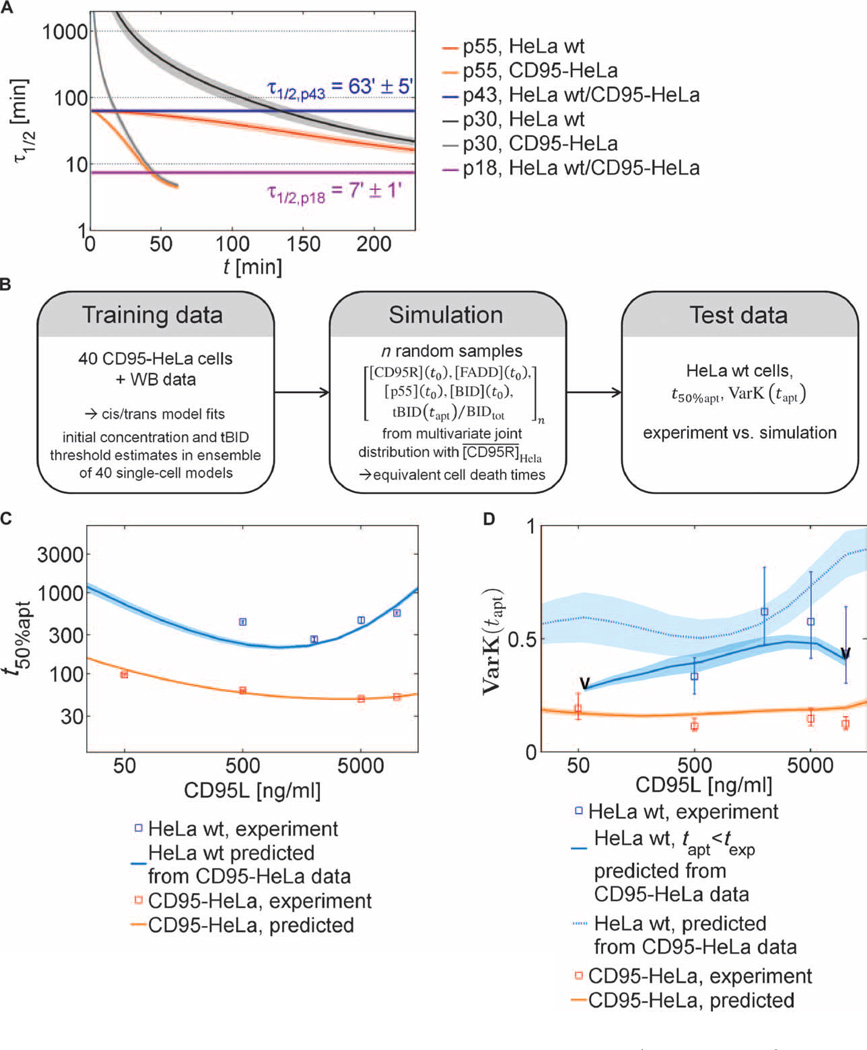
Combining single-cell and Western blot data, we concluded that modeling the prodomain linker cleavage as a cis mechanism and the catalytic domain linker cleavage as a trans mechanism could best reproduce the experimental results. The positive feedback of p43 on its own generation is mechanistically important because the estimated parameters showed about eight times higher trans-cleavage activity for p43 than for DISC-bound p55 or p30 (table S8). To further elucidate these mechanisms, we determined the half-life of the different caspase-8 cleavage products during the time course of apoptosis (Fig. 6A). p55 cleavage mostly generates p43. Because p43 is the main form that cleaves p55, this process can be considered a positive feedback that induces a decrease of the half-life of p55 over time. p43 is processed to p18 through an apparent cis mechanism, implying a constant half-life of p43. According to the best fits of the model, this half-life was around 1 hour (Fig. 6A). In contrast, the same fits predicted that p18 activity only remains for 7 min in the cytosol, which indicates that p43 is responsible for most of the caspase-8 activity in the cell. Thus, caspase-8 has an intrinsic molecular timer that determines the duration of its activity by the combination of the low cleavage constant for prodomain site cleavage in the reaction from p43 to p18 and the rapid in-activation of p18. Furthermore, because p43 is restricted to the DISC, the data indicated that most caspase-8 activity is spatially restricted to this complex and does not substantially propagate to the cytosol.
Fig. 6. Stability of procaspase-8 cleavage products and quantitative description of cell death kinetics based on a tBID threshold model.
(A) Half-life for caspase-8 intermediates. Lines show means, and areas indicate SDs for the best 1% best of n = 1000 cis/trans model fits, simulated at [CD95L] = 500 ng/ml. (B) Derivation of a cell death model. The cis/trans model was extended to describe cell death by assuming that a cell dies once the tBID concentration exceeds a certain threshold. The model was trained using CD95-HeLa cell data, then simulations were run to predict the response of the HeLa wt cells (after reducing the median CD95R number by 10-fold to account for the reduced number of receptors in HeLa wt compared to CD95-HeLa cells), and finally the amount of cell death of the HeLa wt cells under various conditions was compared to the results of the simulations. (C) Experimental values (squares) and predictions (lines and shaded areas) of median cell death times for HeLa wt and CD95-HeLa cells. For validation, model fits to CD95-HeLa cells were used to predict the kinetics of HeLa wt apoptosis. Error bars represent SDs between cell death time medians in different fields of view in a Lab-Tek chamber (solid lines: means; shaded areas: SDs; best 1% of n = 1000 fits). (D) Coefficients of variation for cell death times in the range of 0.5 to 0.6 for HeLa wt cells and in the range of 0.1 to 0.2 for CD95-HeLa cells (squares with error bars estimated by bootstrapping) and from cis/trans model simulations, based on fits to CD95-HeLa data. Lines and shaded areas correspond to SDs as in (C). Predicted coefficients of variation (VarK) were calculated at ligand concentrations at which at least 50% of the simulated cells underwent apoptosis during texp (tapt < texp, limits marked by “V”). VarK estimates including cells undergoing apoptosis later than texp are shown as a dotted line.
A quantitative description of cell death kinetics based on a tBID threshold model
To further validate the cis/trans model, we tested its capacity to predict cell death kinetics and the cell-to-cell variability in cell death times and conducted a systematic sensitivity analysis to study the functional roles of signaling proteins controlling cell death.
To link our cis/trans model of caspase-8 activation to cell death, we used tBID as molecular marker for cell death (23, 28, 29). Specifically, a cell was assumed to die if the tBID concentration in the model exceeded a certain threshold. In simulations, cells in which tBID did not reach this threshold were regarded as survivors. For model validation, we split our experimental data set into the subset of data from CD95-HeLa cells and the subset of data from wild-type HeLa cells. The first subset comprised single-cell and population data from CD95-HeLa cells and was used for model fitting (Fig. 6B). Then, the cis/trans model that was calibrated with only this training data set was used to generate predictions for wild-type HeLa cells. For this purpose, a multivariate log-normal joint distribution was defined with estimated initial concentrations for FADD, p55, and BID and with tBID thresholds from fits to the training data set of CD95-HeLa cells in combination with the experimentally determined mean receptor concentration of wild-type HeLa cells (for details, see text S3: Model analysis and verification). Cell death was simulated in wild-type HeLa cells by randomly sampling vectors of initial concentrations and tBID thresholds from the multivariate distribution and integrating the cis/trans model. Simulated wild-type HeLa cells were assumed to die if the tBID threshold was exceeded. Then, the second subset of wild-type HeLa cells was used to validate these predictions.
Predicted median cell death times in wild-type HeLa cells were in good accordance with experimental measurements, except for one data point for the ligand concentration of 500 ng/ml (Fig. 6C). The model trained on CD95-HeLa cells was further used to predict the cell-to-cell variability of wild-type HeLa cells. We used the coefficient of variation as a measure of variability for time to cell death. The model predicted a much larger variation for wild-type HeLa cells than for CD95-HeLa cells, which is in good agreement with experimental observations (Fig. 6D).
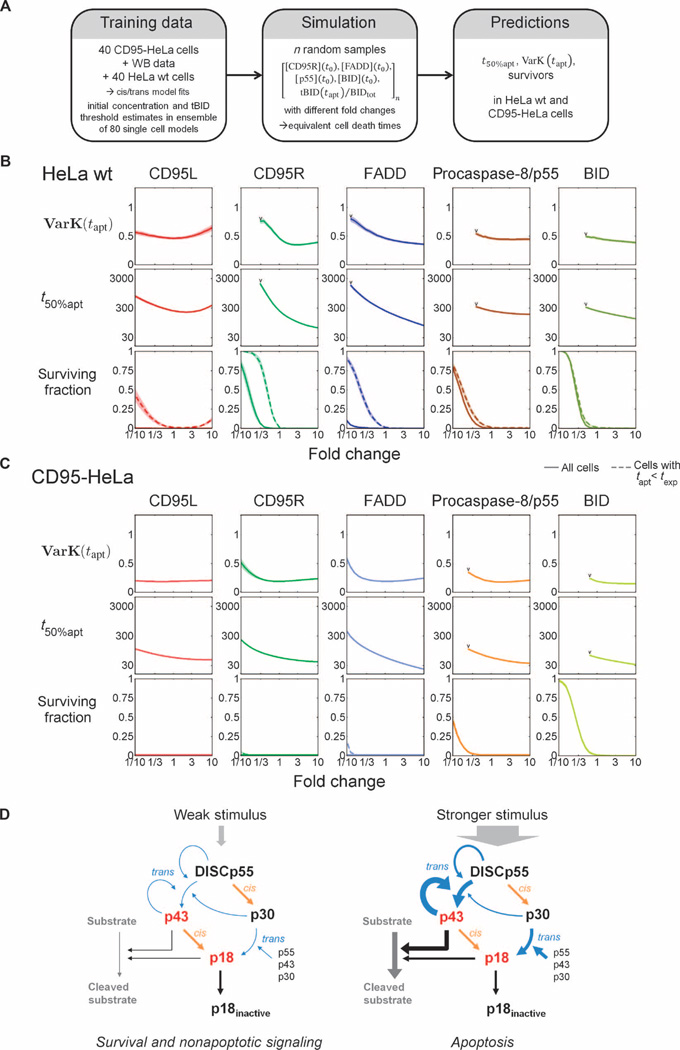
To analyze the impact of individual signaling proteins on apoptosis, we conducted simulations using initial concentration distributions and tBID threshold distributions from cis/trans model fits to the complete set of CD95-HeLa and wild-type HeLa data (Fig. 7A; for details, see text S3: Model analysis and verification, and figs. S19 to S21). Simulations for HeLa wild-type cells showed that cell-to-cell variability and time to cell death strongly depended on amounts of the upstream signaling species, such as the death ligand, death receptors, and FADD (Fig. 7B). In contrast, because of the receptor overexpression, the cell death kinetics and variability of the CD95-HeLa cells were insensitive to changes of initial protein concentrations (Fig. 7C). However, in both wild-type HeLa and CD95-HeLa cells, survival was strongly influenced by the abundance of the downstream signaling proteins p55 and BID (Fig. 7, B and C). In summary, the cis/trans model could be validated using data from CD95-HeLa cells as the training set and then using the resulting model for fitting wild-type HeLa death kinetics and cell death time variability.
Fig. 7. Determinants of cell death kinetics and variability.
(A) To study how concentrations of signaling proteins along the cell death pathway control the dynamics of apoptosis, we used fits of the cis/trans model to the complete set of CD95-HeLa and HeLa wt cell data to generate log-normal multivariate joint distributions of initial protein concentrations and tBID thresholds for the two cell lines. For simulations, random vectors of initial concentrations and tBID threshold fractions were sampled, and simulations were conducted with different fold changes of initial concentrations. (B and C) Predicted coefficients of variation for cell death times [VarK(tapt)], median cell death times (t50%apt), and fractions of surviving cells at different fold changes of initial protein concentrations for CD95L, CD95R, FADD, p55, and BID for HeLa wt (B) and CD95-HeLa cells (C). Predictions at fold change = 1 represent conditions at [CD95L] = 500 ng/ml. In the subplots for surviving fractions, dashed lines represent the surviving fractions within the duration of the single-cell experiments (13 hours), whereas solid lines represent surviving fractions within the total integration time interval. VarK(tapt) and t50%apt are only calculated in cases for which ≥90% of the cells undergo apoptosis (limits marked by “V”). (D) The cis/trans mechanism restricts apoptotic signaling to strong death receptor stimulation. (Left) A weak cell death stimulus generates low caspase-8 cleavage activity: Low numbers of active DISCs generate low amounts of p43 during weak feedback from p43. In this situation, p55 is processed through the inactive intermediate p30 and only non-apoptotic pathways will be activated. (Right) A stronger stimulus enhances production of the active intermediate p43 by causing stronger feedback, therefore increasing the efficiency of the conversion of procaspase-8 into the products with apoptosis-inducing cleavage activity.
DISCUSSION
To study the initiation of extrinsic apoptosis, we developed a model-based method to estimate caspase-8 activity in single cells and a multiscale modeling approach that combined single-cell data with cell population data. Our work extends previous apoptosis models that were trained on cell population measurements only and thereby may not fully capture the detailed kinetics at the single-cell level.
Our caspase-8 activation model complements previous apoptosis models that were more focused on MOMP dynamics (28, 30) or downstream effector caspase activation (31–35). Models that included details of DISC assembly (24, 25, 36) did not discriminate between putative molecular mechanisms of caspase-8 activation and were primarily focused on the role of FLIP proteins in the life or death decision. In our model, we made certain simplifying assumptions concerning the biological activities of individual caspase-8 cleavage fragments. In particular, to keep the complexity of the model low, we did not distinguish between fragments resulting from cleavage at either Asp210 or Asp216 and between fragments resulting from cleavage at either Asp374 or Asp384 but rather collectively described the possible cleavage fragments as the three different procaspase-8 cleavage products p43, p30, and p18.
Using a model selection approach, we excluded intradimeric (cis) processing of procaspase-8 dimers and revealed an inter-dimeric cleavage (trans) mechanism, in which DISCs need to be assembled at least transiently into higher-order clusters at the plasma membrane. Experimental data were best explained by the cis/trans model variant, in which the prodomain site in procaspase-8 is cleaved in cis and the enzymatic domain site is cleaved in trans. The interdimeric trans cleavage mechanism establishes a positive feedback loop with important functional implications for signaling: Autoamplification of p43 allows a distinct transition between low and high activity states (Fig. 7D). As a consequence, the caspase-8 network may enable survival upon exposure to transient, low concentrations of cell death ligand, whereas stronger stimulation efficiently triggers cell death. A delay in caspase-8 activation as a consequence of a positive feedback may help to separate the apoptosis signal from other CD95-dependent pathways involved in tumor cell proliferation, NF-κB (nuclear factor–κB) signaling, or cellular differentiation (24, 26, 37).
Recently, two studies using mass spectrometry of proteins that were extracted together with CD95 or TRAIL receptor DISCs have shown that after stimulation with the synthetic leucine zipper soluble CD95 ligand (LZ-sCD95) or biotin-labeled TRAIL, respectively, the number of procaspase-8 molecules exceeds the number of FADD molecules severalfold, which was interpreted as the formation of a chain of several procaspase-8 at a single FADD molecule (38, 39). Here, we did not explicitly include the formation of such protein conglomerates. This simplification was introduced because our modeling study mainly focused on effective mechanisms of caspase-8 activation while neglecting the stoichiometric details of the complexes. The simplified model of the DISC is justified because DISC formation occurs on a shorter time scale than the subsequent procaspase-8 autoprocessing (1).
By model fitting to a complex data set, we reliably estimated the kinetic parameters describing caspase-8 autoprocessing, revealing that p43 efficiently accumulates at the DISC, whereas p18 is highly unstable. Caspase-8 processing is thus mechanistically similar to procaspase-9 autoactivation at the apoptosome. Apoptosome-bound procaspase-9 cleaves downstream substrates and cleaves itself to caspase-9. After dissociation of caspase-9, the cleavage activity is lost, implying the function as a “molecular timer,” in which the cleavage probabilities control the active time or the extent of cleavage activity (40, 41). Here, procaspase-8 processing at the DISC shows the same phenomenon of the stable intermediate enzyme p43 and the unstable product p18. Accordingly, a previous study showed that the catalytic activity of caspase-8 can be increased by mutating the prodomain cleavage sites (11): This prevents the conversion of the stable p43 intermediate into unstable p18, alleviating the timer effect.
Posttranslational modifications of caspase-8 create another level of complexity in the signaling cascade of apoptosis initiation. Phosphorylation of the tyrosine residue Tyr380 in the linker region between the enzymatic subdomains decreases linker cleavage (42). Polyubiquitination of C-terminal lysine residues in the p10 fragment stabilizes active cytosolic caspase-8 by inducing its recruitment to ubiquitin-rich cytosolic protein complexes (43), whereas K48-linker polyubiquitination of the p18 domain induces its proteasomal degradation (44). In this context, it will be important to quantitatively characterize how posttranslational modifications influence the cleavage efficiency and regulate the stability of caspase-8.
By systematic sensitivity analysis, we found that signaling components upstream of procaspase-8, such as cell death ligand, receptor, and the adaptor protein FADD, primarily shape the kinetics of cell death and produce cell-cell variability in the response. In contrast, downstream species, such as procaspase-8 and BID, influence the fraction of cells surviving the cell death stimulus. The critical role of the procaspase-8 concentration in controlling the fraction of surviving cells can be understood as follows: Because the cleavage activity that can be generated from the available amount of procaspase-8 is limited owing to p18 inactivation, the efficient use of procaspase-8 for downstream substrate cleavage by the active forms p43 and p18 is an important requirement for apoptosis. In summary, the multiscale model presented in this study, calibrating an ensemble of single-cell models to observables of a heterogeneous population of cells, represents a methodological advance that produced detailed insight into the mechanisms involved in the variable response of individual cells to death stimuli.
MATERIALS AND METHODS
Cell lines and cell culture
Stable cell lines were generated from wild-type HeLa cells. CD95-HeLa and the HeLa cell lines stably expressing procaspase-8–mGFP were described in (19). From wild-type HeLa and CD95-HeLa cells, probe- or GFP fusion–expressing cell lines were selected with G418 (0.8 mg/ml; Sigma-Aldrich). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal calf serum (Biochrom AG) and penicillin and streptomycin (100 µg/ml; Invitrogen). Medium for stably transfected cell lines additionally contained G418 (0.2 mg/ml) or puromycin (0.2 µg/ml; Sigma-Aldrich). Cells were transfected with FuGENE 6 (Roche). For microscopy, cells were maintained in eight-well Lab-Tek chambers (Thermo Scientific) with a density of 40,000 per well and, for Western blot experiments, in two-well Lab-Tek chambers at the same density as for single-cell experiments (204,000 per well).
Antibodies and reagents
Caspase-8 was detected with the C-15 antibody (45), which was a gift of P. Krammer. We further used antibodies recognizing FADD (Cell Signaling), BID (Cell Signaling), 5F8 (ChromoTek), and GFP (Roche Diagnostics). Horseradish peroxidase–conjugated secondary antibodies (Southern Biotech) were used for detection. For flow cytometry, we used secondary antibodies coupled to Alexa Fluor 488, goat anti-rabbit or goat anti-mouse (Invitrogen). Cell death was induced with T4-CD95L (Apogenix) in all experiments, except for the combined FRET and NES probe cleavage experiment (fig. S4); the prodomain probe cleavage experiments (Fig. 5, A to D), in which we used LZ-sCD95L (46), expressed in 293T cells as described in (19); and the experiment with mutant caspase-8 (fig. S5), in which we used KillerTRAIL (Enzo Life Sciences). Similar to the T4-Foldon motif in the T4-CD95L, the leucine zipper motif in the LZ-sCD95L and the Killer linker peptide in the KillerTRAIL stabilize the ligands in their trimeric forms.
DNA constructs
We built the ER-anchored probe from the calnexin-LQTDG-YFP described in (19) by replacing the ELQTDG cleavage sequence with GIETDS and by shortening the luminal domain of calnexin by removing residues 25 to 462. For the NES probe used in live-cell experiments to generate single-cell observables for model fitting, the NES sequence MNLVDLQKKLEELELDEQQ was fused to the cleavage sequence GIETDS (caspase-3), followed by a linker and mCherry (NES-GIETDS-mCh) as described in (19). We also created NES probes containing the part Cys164 to Lys231 of the caspase-8 prodomain linker with cleavage sites Asp210, Asp216, and Asp223 as cleavage sequence or containing the noncleavable sequence DEVA. The calnexin construct was cloned in the pEGFP N1 vector (BD Biosciences), and the NES sequence, its promoter region, and its polyadenylation signal were cloned into the calnexin construct on the other side of the enhancer region of the cytomegalovirus (CMV) to generate a bidirectional CMV promoter.
Procaspase-8 was cloned by replacing the enhanced GFP (eGFP) in the pEGFP N1 vector. The double mutants D374A and D384A for the cleavage sites in the catalytic domain were generated using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies).
FADD was fused to mGFP (47) with the linker PRARDPTSGGGGGPVAT and cloned into the pIRES-Neo3 vector (BD Biosciences). BID was fused to mGFP C-terminally and mCherry N-terminally with the linkers GSIAT and RSRG, respectively, and cloned into the pEGFP N1 vector. Myristoylation-palmitoylation (MyrPalm) probes were constructed as described in (19). The MyrPalm domain was fused to a spacer of snap tags, alternative cleavage sequences, and either mCherry or mGFP. Cleavage sequences were either MTISDS (caspase-8 prodomain, Asp210), PREQDS (caspase-8 prodomain, Asp216), PVETDS (caspase-8 enzymatic subdomain linker, Asp374), or ELQTDG (BID).
Analysis of total cellular lysates and protein quantifications
Western blot samples were lysed with lysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich), protease inhibitor cocktail, 1% Triton X-100 (Serva), and 10% glycerol]. Lysates were analyzed with SDS–polyacrylamide gel electrophoresis gels (Invitrogen). The different time points were loaded in a random manner to avoid quantification bias (figs. S7 and S8). Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore) by wet blotting. Detection was performed with the Pico Chemiluminescent Substrate from Thermo Scientific and a charge-coupled device camera (Intas).
In probe-expressing CD95-HeLa and wild-type HeLa cell lines that were used for time series experiments, we quantified endogenous FADD, p55, and BID concentrations using calibrated GFP fusion proteins (figs. S9 to S11). First, GFP fusion protein lysates were calibrated to recombinant eGFP (BioVision). We combined equal amounts of lysates of nontransfected cells with lysates of the fusion protein–expressing cells. This mixed lysate was combined with different amounts (0.1 and 30 ng) of GFP and then loaded onto Western blot gels. Different amounts of these calibrated fusion protein lysates were loaded onto Western blot gels along with lysates from the cleavage probe–expressing cell lines that were used for time series experiments.
Flow cytometry
To estimate the numbers of surface CD95Rs and parameters µ and σ for log-normal distributions by flow cytometry, we used QIFIKIT calibration beads (Dako) with five different antibody epitope numbers per bead, according to the manufacturer’s protocol (fig. S12). To obtain parameters σ for log-normal distributions of initial protein concentrations for FADD, p55, and BID, we used a protocol for intracellular immunostaining of fixed permeabilized cells (fig. S13). For each experiment, 5 × 105 cells were washed in blocking buffer [0.04 M sucrose, 1× phosphate-buffered saline (PBS; pH 7.4), 1% bovine serum albumin] and fixated with 4% para-formaldehyde before primary and secondary antibody staining in 1× PBS with 1% bovine serum albumin. After each step, cells were washed three times with 0.04 M sucrose in 1× PBS. Samples were measured on a modified flow cytometer (FC500/MPL, Beckman Coulter) at 488 nm with a 20-mW solid-state laser (Coherent Inc.). For estimation of distribution parameters, we gated for narrow forward scatter and side scatter intensity intervals to include cells with similar volumes.
Live-cell imaging and simultaneous Western blot sample preparation
Live-cell experiments were performed in a 37°C, 5% CO2 incubation chamber on a Yokogawa spinning disk confocal on a Nikon Ti inverted microscope equipped with 63× Plan Apo (numerical aperture 1.4) objective lens and the Perfect Focus System for continuous maintenance of the focus, using MetaMorph 7 software (Nikon). GFP (Calnsh-GIETDS-mGFP) fluorescence was excited at 491 nm with a 100-mW solid-state laser and collected with a 525/50 emission filter (Chroma Technology Corp.) and an exposure time of 70 ms. Cherry (NES-GIETDS-mCh) fluorescence was excited at 561 nm with a 200-mW solid-state laser and collected with a 620/60 emission filter (Chroma Technology Corp). A binning of 4 × 4 pixels was used. At the beginning of each experiment, a z-stack with 20 slides at 0.7-µm step size was recorded, followed by recording of single slides in the stack center every 2 min (CD95-HeLa cells) or 3 min (wild-type HeLa cells).
Western blot samples were obtained for each time point from 2 × 105 cells in two-well Lab-Tek chambers, which were kept inside a microscope incubation chamber to obtain equivalent experimental conditions as for the imaged single cells. For every time point, Lab-Tek chambers were instantly transferred on an ice-cold metal block, washed in ice-cold 1× PBS before treatment with ice-cold lysis buffer, and harvested with cell scrapers (BD Biosciences).
Image processing
We used custom scripts in MatLab (MathWorks) and ImageJ (National Institutes of Health) for segmentation and analysis of cell shapes and intensities from stack data and of nuclear intensities from slide data for every time point. NES-probe signals were used to segment inner and outer cytoplasm and nucleus borders. Volumes were calculated by voxel numbers and voxel volumes of 0.427 × 0.427 × 0.7 µm3, whereas cell surface areas were calculated by adding face areas of polygon meshes.
Mathematical modeling and simulation
All ODE models were implemented with the MatLab toolbox PottersWheel, which was used for parameter calibrations (http://www.potterswheel.de) (48). Model analysis and simulations were performed with custom MatLab scripts. As a measure for the goodness of fit, we used χ2/N, that is, the sum of weighted model-data differences, divided by the number of data points N. A value of χ2/N smaller than 1 indicates that a model topology fits the experimental data well because the distance between model and data is smaller than the experimental SD. Other common indicators that take into account the numbers of estimated model parameters, such as the corrected Akaike information criterion (AICc), would not improve model selection. Here, differences in parameter numbers Npar of different model variants (Npar, cis/cis = 493, Npar, cis+trans/cis+trans = 497) contribute much less to AICc values than do differences in log-likelihood after model fitting to data (LL1%best fits, cis/cis = −4093…−3868, LL1%best fits, cis+trans/cis+trans = −3419…−3277); that is, all evaluated models are about equally parsimonious with regard to parameter numbers. In this case, AICc values are approximately proportional to χ2 values. Model equations can be found in tables S2 and S3, and kinetic parameter estimates, in table S8.
Supplementary Material
Acknowledgments
We thank C. Cremer, O. Hill, and C. Schmidt for helpful discussions and critical comments, and S. Aschenbrenner for laboratory assistance. We also thank the Nikon Imaging Center at Harvard Medical School for technical advice and support.
Funding: S.M.K. was supported with a short-term fellowship from the German Academic Exchange Service (DAAD). S.L was supported by the BMBF (Virtual Liver Network, e:bio junior group program). This work was supported by the Initiative and Networking Fund of the Helmholtz Association within the Helmholtz Alliance on Systems Biology/SBCancer, the BMBF program SysTec-EpiSys (FKZ 0315502D), and NIH grant GM68762.
Footnotes
Author contributions: S.M.K., S.L, J.B., and R.E. designed the study. S.M.K. and J.B. carried out the experiments. S.M.K., S.L., and J.C. performed the modeling. C.F. provided the T4-CD95 ligand. S.M.K., S.L, J.B., P.K.S., and R.E. discussed and analyzed the results. S.M.K., S.L, J.B., and R.E. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Model files are available at the BioModels database (http:/Mfww.ebi.ac.uk/biomodels/).
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/7/316/ra23/DC1
Text S1. Data analysis.
Text S2. Model implementation and fitting.
Text S3. Model analysis and verification.
Fig. S1. Procaspase-8 and its cleavage products.
Fig. S2. Total cleavage probe concentrations and apoptosis times.
Fig. S3. Single-cell caspase-8 trajectories for CD95-HeLa and wild-type HeLa cells.
Fig. S4. Single-cell trajectories for NES- and FRET-probe cleavage related to caspase-8 activity.
Fig. S5. Western blot of wild-type and prodomain mutant caspase-8.
Fig. S6. Model graphs for oligomerization-driven receptor activation and caspase-8 activation.
Fig. S7. Caspase-8 Western blot time series were used for model fitting of population observables.
Fig. S8. BID and tBID Western blot time series served for model fitting of population observables.
Fig. S9. Estimation of average FADD and p55 amounts by calibrated Western blot.
Fig. S10. Estimation of average BID amounts by calibrated Western blot.
Fig. S11. Estimation of average cleavage probe amounts by calibrated Western blot.
Fig. S12. Flow cytometry of cells immunostained for CD95.
Fig. S13. Flow cytometry of cells immunostained for p55, FADD, and BID.
Fig. S14. Fit qualities of extended cis/cis and cis/trans model variants.
Fig. S15. Reaction fluxes for the cis+trans/cis+trans topology in separate cleavage reactions.
Fig. S16. Reaction fluxes and half-lives of caspase-8 intermediates for the cis/trans topology.
Fig. S17. Reaction fluxes and half-lives of caspase-8 intermediates for the trans/trans topology.
Fig. S18. Monte Carlo modeling supports discrimination between cis/cis and cis/trans topologies.
Fig. S19. Predicted coefficients of variation of apoptosis times at different CD95 ligand concentrations.
Fig. S20. Variability of active receptor fractions for CD95-HeLa and wild-type HeLa cells.
Fig. S21. Model predictions of the effects of twofold increases in initial protein concentrations.
Table S1. Reactions of the complete model cis+trans/cis+trans and smaller variants.
Table S2. Reaction rates in the complete cis+trans/cis+trans model and smaller variants.
Table S3. Model equations of variants cis+trans/cis+trans and cis/trans.
Table S4. Estimates of protein numbers in CD95R-HeLa and wild-type HeLa cells.
Table S5. Receptor number estimates.
Table S6. Initial values and lower and upper limits for estimations of initial concentration.
Table S7. Cellular and cytosolic volumes, cell surface areas, and average fluorescence intensities.
Table S8. Parameters of the fitted cis/trans model.
Caspase-8 model MatLab scripts (zip file of .m files)
Caspase-8 model SBML files (zip file of .xml files)
Caspase-8 single-cell data (Excel)
REFERENCES AND NOTES
- 1.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge BB, Gaudet S, Lauffenburger DA, Sorger PK. Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis. Mol. Syst. Biol. 2011;7:553. doi: 10.1038/msb.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol. Cancer Ther. 2012;11:3–13. doi: 10.1158/1535-7163.MCT-11-0434. [DOI] [PubMed] [Google Scholar]
- 4.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor–induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3–like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 6.Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH, Peter ME. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann JC, Pappa A, Krammer PH, Lavrik IN. A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol. Cell. Biol. 2009;29:4431–4440. doi: 10.1128/MCB.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X. c-FLIPL is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grütter MG. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 11.Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol. Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 13.Donepudi M, Mac Sweeney A, Briand C, Grütter MG. Insights into the regulatory mechanism for caspase-8 activation. Mol. Cell. 2003;11:543–549. doi: 10.1016/s1097-2765(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS, Green DR. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J. Biol. Chem. 2010;285:16632–16642. doi: 10.1074/jbc.M109.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Orozco A, Spencer DM, Wang J. Activation of initiator caspases through a stable dimeric intermediate. J. Biol. Chem. 2002;277:50761–50767. doi: 10.1074/jbc.M210356200. [DOI] [PubMed] [Google Scholar]
- 16.Keller N, Grutter M, Zerbe O. Studies of the molecular mechanism of caspase-8 activation by solution NMR. Cell Death Differ. 2010;17:710–718. doi: 10.1038/cdd.2009.155. [DOI] [PubMed] [Google Scholar]
- 17.Keller N, Mares J, Zerbe O, Grütter M. Structural and biochemical studies on procaspase-8: New insights on initiator caspase activation. Structure. 2009;17:438–448. doi: 10.1016/j.str.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–4407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- 19.Beaudouin J, Liesche C, Aschenbrenner S, Hörner M, Eils R. Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis. Cell Death Differ. 2013;20:599–610. doi: 10.1038/cdd.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watt W, Koeplinger KA, Mildner AM, Heinrikson RL, Tomasselli AG, Watenpaugh KD. The atomic-resolution structure of human caspase-8, a key activator of apoptosis. Structure. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 21.Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS. Activation of caspases-8 and −10 by FLIPL . Biochem. J. 2004;382:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanchard H, Kodandapani L, Mittl PR, Marco SD, Krebs JF, Wu JC, Tomaselli KJ, Grütter MG. The three-dimensional structure of caspase-8: An initiator enzyme in apoptosis. Structure. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 23.Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, Lavrik IN, Eils R. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol. Syst. Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL . J. Cell Biol. 2010;190:377–389. doi: 10.1083/jcb.201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleber S, Sancho-Martinez I, Wiestler B, Beisel A, Gieffers C, Hill O, Thiemann M, Mueller W, Sykora J, Kuhn A, Schreglmann N, Letellier E, Zuliani C, Klussmann S, Teodorczyk M, Gröne HJ, Ganten TM, Sültmann H, Tuttenberg J, von Deimling A, Regnier-Vigouroux A, Herold-Mende C, Martin-Villalba A. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell. 2008;13:235–248. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK, Modeling a snap-action. variable-delay switch controlling extrinsic cell death. PLOS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Düssmann H, Rehm M, Concannon CG, Anguissola S, Würstle M, Kacmar S, Völler P, Huber HJ, Prehn JHM. Single-cell quantification of Bax activation and mathematical modelling suggest pore formation on minimal mitochondrial Bax accumulation. Cell Death Differ. 2010;17:278–290. doi: 10.1038/cdd.2009.123. [DOI] [PubMed] [Google Scholar]
- 31.Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I. Bistability in apoptosis: Roles of Bax Bcl-2 and mitochondrial permeability transition pores. Biophys. J. 2006;90:1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eissing T, Conzelmann H, Gilles ED, Allgöwer F, Bullinger E, Scheurich P. Bistability analyses of a caspase activation model for receptor-induced apoptosis. J. Biol. Chem. 2004;279:36892–36897. doi: 10.1074/jbc.M404893200. [DOI] [PubMed] [Google Scholar]
- 33.Legewie S, Blüthgen N, Herzel H. Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLOS Comput. Biol. 2006;2:e120. doi: 10.1371/journal.pcbi.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehm M, Huber HJ, Dussmann H, Prehn JHM. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 2006;25:4338–4349. doi: 10.1038/sj.emboj.7601295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Würstle ML, Laussmann MA, Rehm M. The caspase-8 dimerization/dissociation balance is a highly potent regulator of caspase-8, −3, −6 signaling. J. Biol. Chem. 2010;285:33209–33218. doi: 10.1074/jbc.M110.113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, Krammer PH, Eils R. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 2004;166:839–851. doi: 10.1083/jcb.200404158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Park SM, Tumanov AV, Hau A, Sawada K, Feig C, Turner JR, Fu YX, Romero IL, Lengyel E, Peter ME. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L, Schwabe JWR, Cain K, Macfarlane M. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schleich K, Warnken U, Fricker N, Oztürk S, Richter P, Kammerer K, Schnölzer M, Krammer PH, Lavrik IN. Stoichiometry of the CD95 death-inducing signaling complex: Experimental and modeling evidence for a death effector domain chain model. Mol. Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malladi S, Challa-Malladi M, Feamhead HO, Bratton SB. The Apaf-1•procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cursi S, Rufini A, Stagni V, Condò I, Matafora V, Bachi A, Bonifazi AP, Coppola L, Superti-Furga G, Testi R, Barilà D. Src kinase phosphorylates caspase-8 on Tyr380: A novel mechanism of apoptosis suppression. EMBO J. 2006;25:1895–1905. doi: 10.1038/sj.emboj.7601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, Pham VC, Stephan JP, Lill J, Ashkenazi A. TRAF2 sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol. Cell. 2012;48:888–899. doi: 10.1016/j.molcel.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Scaffidi C, Medema JP, Krammer PH, Peter ME. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J. Biol. Chem. 1997;272:26953–26958. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]
- 46.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 47.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maiwald T, Timmer J. Dynamical modeling and multi-experiment fitting with PottersWheel. Bioinformatics. 2008;24:2037–2043. doi: 10.1093/bioinformatics/btn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JHM. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J. Biol. Chem. 2002;277:24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- 50.Tyas L, Brophy VA, Pope A, Rivett AJ, Tavaré JM. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 2000;1:266–270. doi: 10.1093/embo-reports/kvd050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 52.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 53.Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 54.Papoff G, Hausler P, Eramo A, Pagano MG, Leve GD, Signore A, Ruberti G. Identification and characterization of a ligand-independent oligomerization domain in the extracellular region of the CD95 death receptor. J. Biol. Chem. 1999;274:38241–38250. doi: 10.1074/jbc.274.53.38241. [DOI] [PubMed] [Google Scholar]
- 55.Siegel RM, Frederiksen JK, Zacharias DA, Chan FKM, Johnson M, Lynch D, Tsien RY, Lenardo MJ. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 56.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 57.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 58.Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 59.Smith CA, Farrah T, Goodwin RG. The TNF receptor superfamily of cellular and viral proteins: Activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 60.Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLOS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schilling M, Maiwald T, Bohl S, Kollmann M, Kreutz C, Timmer J, Klingmüller U. Computational processing and error reduction strategies for standardized quantitative data in biological networks. FEBS J. 2005;272:6400–6411. doi: 10.1111/j.1742-4658.2005.05037.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.