Abstract
Animal genomes contain a code for construction of the body plan from a fertilized egg. Understanding how genome information is deciphered to create the complex multilayered regulatory systems that drive organismal development, and which become altered in disease, is one of the greatest challenges in the biological sciences. The development of methods that effectively represent and communicate the complexity inherent in gene regulatory networks remains a major barrier. This review introduces the philosophy of systems biology and discusses recent progress in understanding the development of the heart at a systems biology level.
Systems biology tools are useful for understanding complex heart developmental networks and for elucidating the impacts of genetic mutations in heart disease.
SYSTEMS BIOLOGY
Systems biology is an emerging multidisciplinary field embedded in large-scale data initiatives that seeks to understand the complex interactions occurring within biological systems. The recent increase in popularity of systems biology has been the consequence of technological advances that have flowed from the sequencing of the human genome (Lander et al. 2001). However, the last decade of research has reinforced the notion that to understand biological networks we need to do more than just describe genome organization. A major finding that has reshaped our view of biology is the realization that transcriptional complexity rather than genome size or the number of protein-coding genes is the evolutionary driver for increased biological diversity (Britten and Davidson 1969; Carninci et al. 2005; Fantom Consortium et al. 2014).
Network theory hypothesizes that specific interaction patterns in biology have logic and functional significance. The discovery that universal properties exist in biological and other networks, including road and social networks, and the World Wide Web, has stimulated the application of network theory to biology (Barabasi and Albert 1999; Barabasi and Bonabeau 2003). The highly interconnected nature of networks means that information flow from any part of a network to another involves a short distance or low number of elements. This is a consequence of highly connected network elements, called hubs. Because of their high connectivity, the disruption of hubs has dramatic effects on the stability of networks (centralitylethality rule) (Jeong et al. 2001), with implications for disease mechanism (Vidal et al. 2011).
GENE REGULATORY NETWORKS AND TRANSCRIPTIONAL REGULATION
The Waddington Landscape and Britten and Davidson Models
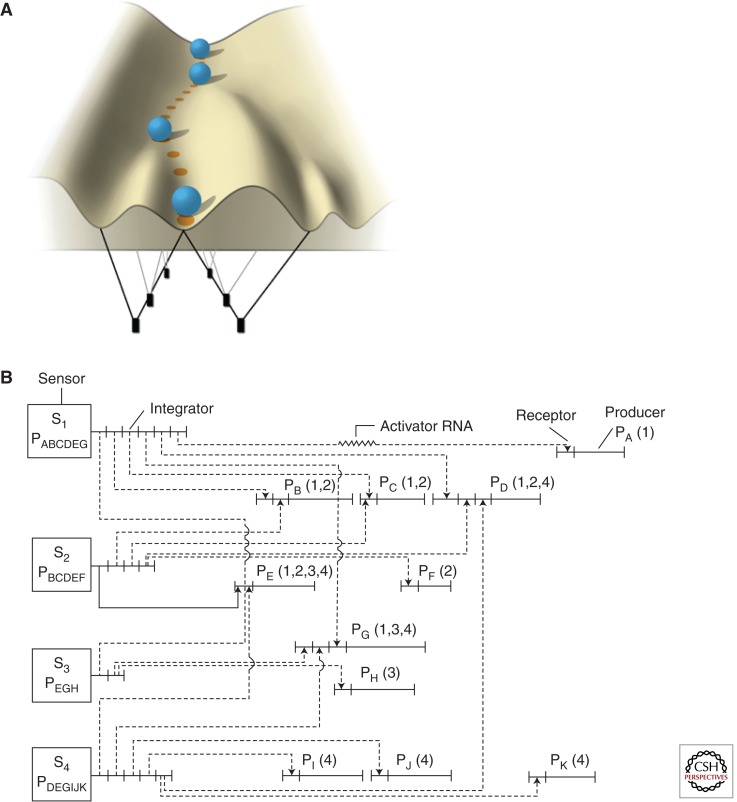
Deciphering the mechanisms that drive development is a long-standing challenge. Emphasis has been placed on revealing the “decision makers” that give rise to cellular identity and to mapping the signaling pathways that enable the reproducible construction of the body plan through successive generations. A key feature of networks is robustness, or resilience to genetic or environmental attack, or random fluctuations in gene expression. Robustness underlying development was captured by Conrad Hal Waddington’s visual metaphor of a ball rolling down a valley system in a terrestrial landscape (Fig. 1A). Waddington envisaged this “epigenetic landscape” as being shaped by genes (pegs in the ground) which were connected to each other and the landscape by guide ropes that mold the valleys that constrain lineage fate (Waddington 1957).
Figure 1.
Two landmark views of gene regulatory network (GRN) architecture. (A) Waddington landscape. A metaphorical view of development representing an organism/cell as a ball rolling down a valley. Bifurcations in the valleys represent decision points/epigenetic influences and are formed through the actions of genes and the environment, represented as pulleys and ropes beneath the landscape. (Adapted from Mitchell 2007.) (B) Britten and Davidson view on the GRN: an integrated/combinatorial view of gene regulation. (From Britten and Davidson 1969; reproduced, with permission.) In this model, developmental inducers termed sensors were linked to multiple and redundant integrators, which regulate batteries of activator RNAs (encoding both transcription factors and noncoding RNAs in our current understanding), which in turn bind in a combinatorial fashion to diverse receptors (cis-regulatory motifs) controlling producer genes that define cellular phenotype.
Britten and Davidson furthered this idea by introducing the concept of gene regulatory networks (GRNs) as hierarchical operating systems regulating mammalian development (Britten and Davidson 1969; Davidson and Erwin 2006). Their early conceptualization of a metazoan GRN (Fig. 1B) was visionary (Britten and Davidson 1969), anticipating many elements of current network theory and evolution, including redundancy, feedback regulation, the relationship between genome size and regulatory complexity, and the role of nuclear RNA in network regulation. The GRN concept appreciated that different elements are spread across the genome, requiring an integrated circuitry with a hierarchical structure, and that a limited number of integrators were required to control many subservient genes.
Emphasis has since been placed on elucidating a general GRN framework that could describe development, and this has given rise to additional concepts: for instance, (1) kernels, (2) subcircuits, (3) switches, and (4) gene batteries (Davidson and Erwin 2006). Kernels are the combination of evolutionarily conserved elements that specify the body plan and perform the executive functions of the network; for example, the combination of transcription factors (TFs) conserved from Drosophila to Homo sapiens that direct heart development (Olson 2006). Kernel TFs show a high degree of cross-regulation (recursive wiring) that provides robustness to kernel function and enables combinatorial coding of downstream target genes—a vehicle for complex decision making (Davidson and Erwin 2006). As such, kernel TFs represent network hubs and tampering with them has catastrophic consequences for development. The kernel then engages subcircuits that establish regional regulatory states controlling, for example, cell division, epithelial state, and/or migration. Analogous to electrical circuits, deployment or detachment of subcircuits can be regulated by GRN “switches.” Switches could be signal or TF dependent, or may involve specific modifications of chromatin that affect accessibility of TFs to DNA elements. These dynamic changes to the structure of networks during development are reflected in the regulation of gene batteries, sets of genes that define the terminal gene expression program and identity of a cell.
Selector Genes, Master Regulators, and Pioneer Factors
Garcia-Bellido formulated the “selector gene” hypothesis to explain the effects of gene mutations that famously convert one body part into another (Garcia-Bellido 1975). Here, gain or loss of single-selector genes invoke a shift in the total program for creation of a morphological structure; for example, the stable transformation of a leg into an antenna through loss of the homeobox (Hox) gene Antennapedia (Struhl 1981). In the contemporary view, selector genes are distinguished from other TFs because they play a deterministic role in “selecting” the overarching developmental program for germ layers, developmental fields, lineage compartments, individual metameric units, or cell types (Mann and Carroll 2002). The notion of combinatorial TF codes underpinning selector protein function was implicit in the hypothesis (see below).
In some circumstances, selector genes may have the properties of master regulators—factors, such as the muscle-specific TF MyoD1, which alone can induce muscle fate when expression is induced in a variety of cell types (Tapscott et al. 1988). However, the qualities of a master regulator are poorly defined and the concept is problematic because it implies autonomous governance of developmental events by a single factor, which is dismissive of broader network functionality (Chan and Kyba 2013). In essence, selector genes are factors that are involved in switching functions within the GRN that impact on cell lineage or morphogenetic fate. Some of these may have the property ascribed to master regulators in being able to cell-autonomously affect a network switch. Others can only do this as part of a TF collective (Ieda et al. 2010; Chen et al. 2014). Certain TFs, termed “pioneer factors,” can find their target sites even in closed chromatin and may “bookmark” these sites during mitosis (Zaret and Carroll 2011). It is likely that selector proteins and kernel TFs, as well as proteins that can reprogram cell fate autonomously, have pioneer qualities.
Restriction of Developmental Potential and Cell Fate
Specification of organ territories or lineages during development requires a progressive restriction of options within cellular fields. This invariably involves a complex reciprocal molecular dialog between neighboring cells (Noseda et al. 2011). A mainstay concept in developmental biology is that the adoption of a lineage fate invokes a global lockdown of possibilities for alternative fates (Davidson et al. 2002). For example, in the early embryo, allocation of blood and cardiac lineage fates within a common mesodermal field is subject to antagonistic principles, with genetic disruption of TFs important for one fate allowing the other to dominate (Schoenebeck et al. 2007; Simoes et al. 2011; Van Handel et al. 2012). At the cellular field level, lineage founder cells may inhibit their direct neighbors that share similar network states from adopting the same fate through a process termed lateral inhibition, often involving the Notch signaling pathway (Carmena et al. 2002). In other contexts, the same fate is reinforced among neighboring cells through so-called community effects (Gurdon 1988). Asymmetric cell division—the division of cells into daughters that have different cell fates—also plays a central role in cell fate determination in stem and progenitor cell populations through the asymmetric partitioning of deterministic components, including ancestral “template” DNA strands, to individual daughter cells (Tajbakhsh et al. 2009). Therefore, in network terms, it is both the internal GRN and the dynamic influences of external signaling that determines regulatory and lineage states.
Epigenetic Memory and Cell Fate Attractors
The general or specific state of a GRN can be “remembered,” to the extent of being inherited by daughter cells across cell divisions, referred to as epigenetic memory. Many mechanisms contribute to epigenetic memory. For example, feed-forward and feedback loops stabilize gene expression programs long after the initial stimulus that initiated them has faded (Alon 2007), and histone and DNA marks that regulate chromatin structure also profoundly contribute to gene expression status and potential (see below) (Orkin and Hochedlinger 2011; Rosa-Garrido et al. 2013).
Stuart Kaufmann considered cell types as stable solutions of genetic networks—or “attractor states” of the GRN (Kauffman 1969; Enver et al. 2009). Lineage choices during development can therefore be visualized as a transition from one attractor state to another, instigated by changes in extracellular signaling, chromatin state, and cross-antagonistic network elements. A former state is overruled or disabled by the changing signaling environment (Herrmann et al. 2012; Mbodj et al. 2013). Developmental progressions in the embryo are “canalized” through a series of major stable network state transitions (valleys in the Waddington epigenetic landscape; Fig. 1A) so as to protect cells from taking inappropriate lineage decisions. Differentiated cells likewise exist in relatively stable network states. Multipotent adult stem cells, in contrast, are likely to maintain metastable states such that when they are stimulated to proliferate and differentiate in response to niche factors they can readily engage lineage-specific attractors, potentially via multiple pathways (Enver et al. 2009).
The experimental conversion of one cell state to another (reprogramming), for example, by cellular fusion, nuclear transfer, or introduction of single or multiple TFs to cells in vitro (Yamanaka and Blau 2010), is at odds with the certainty of developmental progression implied by the Waddington landscape. Therefore, even the most stable network states are not inviolate. Cell division is often required to instigate or stabilize radical changes in epigenetic state (Harrison et al. 2007; Polo et al. 2010), although changes can also be driven purely by histone variants and other nuclear factors in the absence of cell division, as seen after injection of somatic cell nuclei into Xenopus oocytes (Gurdon 1988).
A NETWORK VIEW OF BIOLOGY
Although the GRN model has been widely accepted by developmental biologists, determining the myriad states of a GRN and how it gives rise to biological function remains problematic. This is compounded by the complexity of GRN structure, the many layers of network regulation, and the paucity of tools and computational methods to represent complexity, although this is changing rapidly as a new generation of network scientists enter the field. TFs play a central role in network regulation, as highlighted by recent studies in cellular reprogramming of somatic cells to pluripotency or to differentiated cell fates (Takahashi and Yamanaka 2006; Ieda et al. 2010). The network is in effect hardwired through the global arrangement of TF binding sites within cis-acting elements controlling gene expression. However, TFs do not operate on the genome in isolation and simple occupancy of their binding sites cannot always explain the complex patterns of gene expression occurring in development (Spitz and Furlong 2012; Wilczynski et al. 2012). As alluded to above, other levels of regulation are important—these include combinatorial coding, noncoding RNAs (ncRNAs), long- and short-range epigenetic control through modifications to chromatin and DNA, DNA looping, and the establishment of insulators, boundary elements, and specialized nuclear compartments (Spitz and Furlong 2012). There is still much discussion about which of these events are primary and causal, and which are consequences of TF binding (Spitz and Furlong 2012; Stergachis et al. 2013). This highlights our poor understanding of the structure and function of the noncoding parts of the genome. The concepts described above nonetheless dictate that the reductionist approach (describing individual network connections), while highly successful in generating potent concepts and detailed mechanisms in animal biology and disease, becomes embedded within the broader molecular framework. This framework must consider development as a regulatory system with inherent logic acting within and between cell types, and in the broader view, between individuals and communities, and the physical environment.
HEART DEVELOPMENT
The mammalian heart is a highly modified vessel that develops in the embryo from a simple muscular tube with an endocardial lining, to a dual pump under moment-by-moment electrical and neuroendocrine control. A key feature in mammalian heart development is the septation of a single tubular form to generate four chambers with different roles and identities: two collecting chambers (the atria) and two pumping chambers (the ventricles) guarded by atrioventricular valves (Anderson et al. 2014).
At approximately embryonic day (E) 7.5 in mice, the heart tube begins to develop from a crescent-shaped population of anterior progenitors composed of two distinct fields of cells with different behaviors and lineage fates: the first and second heart fields (FHF and SHF, respectively) (Fig. 2A) (Vincent and Buckingham 2010). The FHF originates from the anterior splanchnic mesoderm and gives rise to the linear heart tube composed mostly of precursors of the left ventricle. The SHF is derived from the pharyngeal mesoderm situated medial to the cardiac crescent and comes to lie dorsal to the forming heart tube. This population is held over from differentiating while expanding by proliferation, and ultimately populates the growing heart at its inflow and outflow poles contributing to the inflow vessels, atria, right ventricle, and outflow tract.
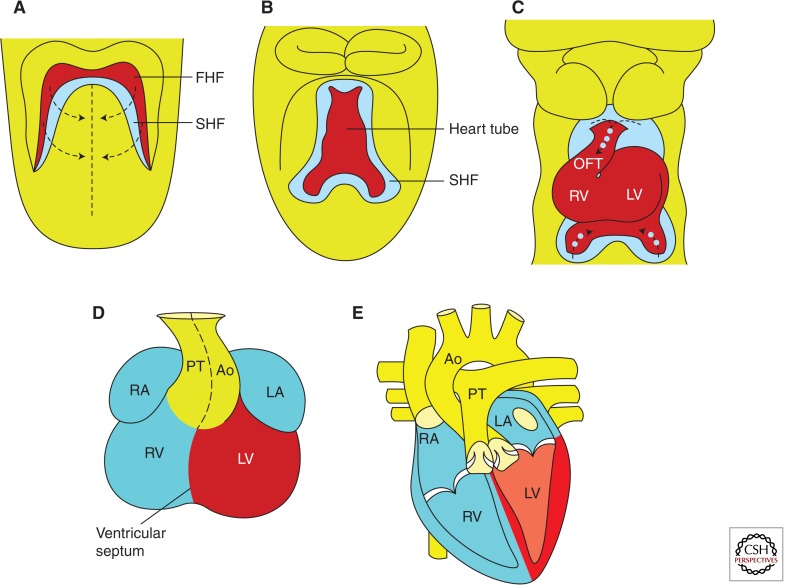
Figure 2.
Graphical representation of murine heart development. (A) Embryonic day (E) 7.75–8.0 showing relationship between progenitors of the first heart field (FHF; red) and second heart field (SHF; blue). Arrows indicate the direction of convergence of progenitors during heart tube formation. (B) E8.0–8.5 showing the linear heart tube derived largely from the FHF with SHF progenitors persisting dorsal to the heart tube. (C) E9.5 and (D) E12.5, showing looped hearts. Arrows and dots in C indicate deployment of SHF cells to the growing poles of the heart. Colors in D indicate approximate origins of chamber cardiomyocytes from FHF and SHF progenitor cells. (E) Late embryonic/postnatal stages, showing final heart structure. Ao, aorta; LA, left atrium; LV, left ventricle; PT, pulmonary tract; RA, right atrium; RV, right ventricle. (Adapted, with permission, from Cohen et al. 2008.)
A more-or-less linear heart tube oriented cranio-caudally is formed by convergence of the FHF progenitors at the midline at around E8.0 (Fig. 2B), and begins to beat owing to the automaticity of its cardiomyocytes (Vincent and Buckingham 2010). This generates blood circulation and hemodynamic forces that also participate in development of chambers, valves, and a more specialized conduction system. Around E8.5, when the linear heart tube begins elongating as a result of deployment of SHF cells, it undergoes a rightward spiral looping under the influence of the left–right asymmetry genetic network (Vandenberg and Levin 2013), and this looped form creates the prepattern for further remodeling (Fig. 2C). Growth, differentiation, and specialization of chamber muscles, valves, and conduction tissue, follow. Precursor cells derived from the neural crest, proepicardial organ, and sinus venosus, also migrate and integrate into the heart, collectively contributing to formation of cardiac ganglia, smooth muscle, and endothelial cells of the coronary vessels, and fibroblasts of the interstitium and annulus fibrosus. By E12.5, the heart contains four distinct muscular chambers ensuring distinct and coordinated patterns of blood flow through the pulmonary and systemic circuits (Fig. 2D,E).
THE CARDIAC KERNEL
A number of deeply conserved cardiac TFs, including GATA4/6, ISL1, NKX2-5, MEF2C, SRF, TBX5, and TBX20, can be considered to be part of a cardiac kernel in that they are expressed regionally in cardiac progenitor cells and differentiating lineages, show extensive cross-regulatory interactions, and their individual knockout phenotypes are catastrophic for early heart development (Davidson and Erwin 2006; Herrmann et al. 2012). However, none of these TFs are absolutely specific to the heart fields and no single knockout in mice eliminates heart formation. Together, however, these factors provide the combinatorial coding for cardiogenesis (discussed further below). It is noteworthy that although knockout of kernel factors generally leads to arrested heart development owing to defective tissue growth and patterning, phenotypes can show distinct features (Watt et al. 2004; Stennard et al. 2005; Prall et al. 2007), indicating overlapping and unique roles within the kernel and in the flow of information from upstream regulators to downstream functions. Limited combinations of cardiac TFs centered on GATA4, TBX5, and MEF2C, with and without the inclusion of specific micro-RNAs (miRNAs) or small molecules modulating signaling and epigenetic states, can reprogram noncardiac embryonic or adult cells to a cardiomyocyte fate (Ieda et al. 2010; Chen et al. 2014). Such combinations presumably represent the minimal cocktail of pioneer and kernel factors, and their modulators, necessary to reconfigure noncardiac networks toward the cardiac attractor state.
The TF MESP1 is expressed in anterior embryonic and extraembryonic mesoderm from gastrulation stages, before the expression of the cardiac-kernel factors, and is capable of enhancing cardiogenesis and repressing alternative cell fates when overexpressed in pluripotent stem cells (Bondue et al. 2008, 2011). This appears to occur through direct regulation of kernel factors, such as NKX2-5, HAND1, and GATA4. MESP1 has therefore been thought of as a cardiac master regulator (Bondue et al. 2008), however, this factor has since been shown to promote the specification of a number of different anterior mesodermal lineages (Chan and Kyba 2013). The network context of MESP1 is not well defined in any one species. In mouse, Mesp1 is regulated directly during gastrulation by the T-box factors Brachyury and Eomesodermin (Costello et al. 2011; David et al. 2011). In ascidians, Mesp1 marks a single pair of blastomeres giving rise to the heart field, and is regulated directly by T-box factor TBX6 in combination with the LIM-homeodomain factor LHX3a (Christiaen et al. 2009). The paired-homeodomain factor, OCT4, known mostly as a gatekeeper of pluripotency, is also involved in driving early lineage states and is coexpressed with MESP1 during mouse pluripotent stem cell differentiation into cardiac lineages where it dose-dependently regulates cardiogenesis (Zeineddine et al. 2006; Blin et al. 2010). MESP1 can thus be regarded as a selector for an anterior mesodermal subdomain acting in divergent networks upstream of lineage-specific kernels.
GENOME-WIDE TECHNOLOGY
The cis-regulatory matrices underpinning development are being deciphered on an unprecedented scale using techniques, such as chromatin immunoprecipitation (ChIP), protein-binding microarrays, and gene expression analysis (Busser et al. 2008). Transcriptomics surveys the total coding and noncoding output of a cell or tissue at a particular moment under particular conditions and is a proxy for TF function and GRN output (Sperling 2011). It has been a powerful discovery tool in biology with the potential for further impact as transcriptomes are accessed at greater depth. As one illustration of its early application to heart development, Rojas et al. used microarrays to survey changes to the transcriptome of murine SHF progenitors induced by loss of the cardiac-kernel factor GATA4 (Rojas et al. 2008). Among dysregulated genes, those involved in the cell cycle were prominent, including direct GATA4 target genes encoding cyclin A2, cyclin D3, and cyclin-dependent kinase 4. Mutations in other cardiac TFs also show pronounced effects on the cell cycle (Yamagishi et al. 2001; Prall et al. 2007) and it seems self-evident that the cardiac kernel should engage cell-cycle networks as subcircuits, although the precise logic underpinning these connections remains unclear as non-cell-autonomous signals to myocardium from epicardium, endocardium, or pharyngeal endoderm (Lavine et al. 2005; Grego-Bessa et al. 2007; Liu et al. 2014) may be involved.
More recently, coupling transcriptomics to the genome-wide analysis of TF targets and chromatin marks has allowed unique insights into network states. ChIP using antibodies specific for native or tagged TFs, or specific histone modifications, followed by microarray analysis (ChIP-chip) or deep sequencing (ChIP-seq), is now commonly used. Mammalian cardiac TFs recently surveyed for target genes or effects on the transcriptome include MESP1, GATA4, MEF2A, NKX2-5, SRF, TBX3, TBX5, and TBX20 (Prall et al. 2007; Holler et al. 2010; Bondue et al. 2011; He et al. 2011; Schlesinger et al. 2011; Shen et al. 2011; Singh et al. 2012). Furthermore, genome-wide binding profiles of the ubiquitously expressed histone acetyltransferases, p300 and CBP, cofactors of many transcriptional activators, have allowed the mapping of active and tissue-specific developmental enhancers without reference to specific TF binding (Blow et al. 2010; May et al. 2012). This approach revealed that the majority of murine heart enhancers occupied by p300 are conserved only among placental mammals, illustrating a feature of network evolution and indicating that conservation-based methods for identifying enhancers undersample regulatory elements and network complexity. It is noteworthy, however, that among putative cardiac enhancers detected in HL-1 cardiomyocytes, only a minor proportion were co-occupied by p300 (He et al. 2011). Chromatin DNase 1 hypersensitivity analysis is also beginning to reveal the fine architecture of TF/cofactor binding and nucleosome assembly in a factor-independent manner and at single nucleotide resolution (Stergachis et al. 2013; Vierstra et al. 2014). These approaches create tissue-specific genome-wide regulatory region maps for comparative network analysis.
Prominent international consortia, including ENCODE (genome.ucsc.edu/ENCODE) and FANTOM (fantom.gsc.riken.jp), are endeavoring to describe all the functional elements of mammalian genomes and provide these data to the research community. Recent findings indicate that ∼80% of the human genome is either transcribed or associated with other chromatin regulatory events, far greater than the ∼3% representing protein-coding exons. Thus, a large proportion of the genome has a regulatory role including specification of long ncRNAs, the functions of which we are only beginning to grasp (Schonrock et al. 2012) (see below). There is also a greater diversity of transcript structure owing to differential splicing than previously appreciated, and many genes utilize alternative transcriptional start sites. Ultimately, it will be vital to incorporate this diversity, as well as posttranslational modifications, microRNA, and long ncRNA networks, and the metabolome, into network models of heart development.
NETWORK REGULATION IN HEART DEVELOPMENT
Many individual genes and pathways important for heart development have been identified, and in some cases collated into a network synthesis (Davidson and Erwin 2006; Vincent and Buckingham 2010; Schlesinger et al. 2011). In the sections below, we illustrate a few examples of how integrated genome-wide and computational approaches have led to further insights into the structure and function of heart developmental networks.
Enhancer Function
The structure of GRNs is hardwired in the genome through the fixed arrangement of numerous regulatory elements, such as core promoters, enhancers, silencers, insulators, and tethering elements (cis-regulatory architecture; Fig. 3; see below) (Spitz and Furlong 2012). Enhancers serve as integrators of complex developmental information. They contain multiple TF binding sites arranged in a specific motif grammar. The overlapping expression of domain-specific TFs provides the combinatorial inputs for enhancer specificity. The autonomy and specificity embedded within individual enhancers is beyond doubt—they can often replicate the complex temporal and spatial patterns of expression of the endogenous genes they normally control when inserted into ectopic positions in the genome. As integrators of information, enhancers have a variety of designs and roles. For example, enhancer output can respond proportionally to TF concentration and act as a rheostat, or higher-order cooperativity between TFs can lead to mass action and an on/off switch-like behavior (Spitz and Furlong 2012; Mbodj et al. 2013). Occupancy of enhancers can lead to bending of DNA, altered nucleosome occupancy and phasing, recruitment of chromatin-remodeling complexes, and protection against inhibitory methylation. Analysis of the cis-regulatory regions of cardiac TF genes including Nkx2-5 and Mef2C highlights the modular nature of tissue-specific enhancers, both positive and negative regulatory regions, partial enhancer redundancy, combinatorial TF coding, and autoregulation (Reecy et al. 1999; Dodou et al. 2004). Enhancer mutations likely underpin many human disease states and the majority of the associations detected between single nucleotide polymorphisms (SNPs) and disease phenotypes in genome-wide association studies lie outside of protein-coding regions. In a recent study, detailed analysis of the architecture of chromosomal looping at the locus encoding cardiac voltage-gated sodium channels, SCN5A and SCN10A, was performed in mouse tissue using high-resolution chromatin conformation capture (4C) technology (van den Boogaard et al. 2014). Heterozygous mutations in the human Scn5a gene are associated with conduction abnormalities including Brugada syndrome, long-QT syndrome, atrial fibrillation, and sudden death. This study revealed that an intronic enhancer within the weakly expressed Scn10a gene contacts the distant promoter region of the more strongly expressed Scn5a gene and regulates its expression. A common haplotype encompassing the human Scn10a intronic enhancer is strongly associated with rhythm disorders including Brugada syndrome, and one of the associated SNPs alters a T-box TF DNA-binding site and leads to reduced Scn5a expression and slow ventricular conduction. This provides a good cardiac example of the relevance of distal enhancers and chromosomal looping to gene expression output.
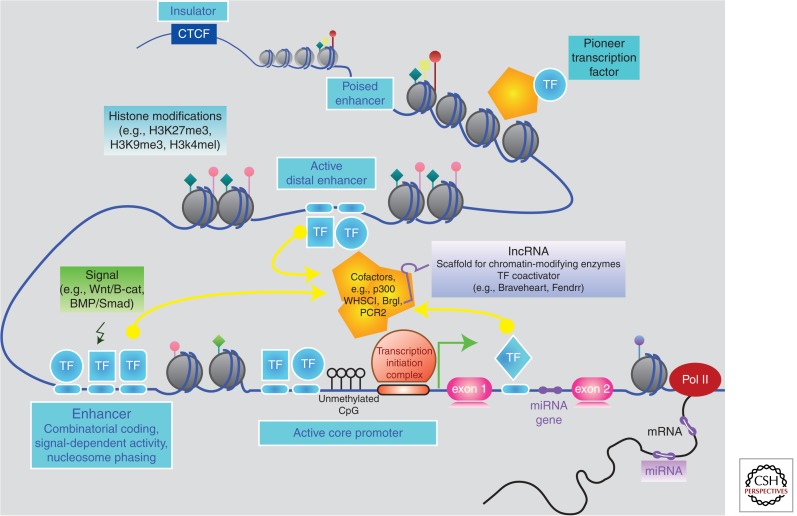
Figure 3.
Regulatory layers of the cardiac GRN. Simplified representation of some of the layers of gene regulation relevant to network biology (epigenetic, transcriptional, and posttranscriptional), as discussed in the text. DNA (blue line); histones (gray circles); transcription factors (TF); RNA polymerase II (Pol II).
Machine Learning for Classification of Cardiac Enhancers
An increasing number of studies have used machine-learning algorithms to classify functional enhancers and to discover new network features. These computational approaches rely mostly on existing statistical learning methods (e.g., support vector machines; lasso) to detect combinations of TF binding that best explain tissue-specific gene expression (Cortes and Vapnik 1995; Tibshirani 1996). Training sets of validated enhancers followed by functional testing of predicted enhancers are critical to the approach. Narlikar et al. considered motifs for known cardiac factors and those enriched in known heart enhancers in a training exercise that then predicted 42,000 cardiac enhancers in the human genome, of which 16/26 (62%) showed reproducible expression in transgenic zebrafish or mouse hearts (Narlikar et al. 2010). This study emphasized the involvement of known cardiogenic TFs MEF2 and SRF, and broadly expressed factors LMO2, ETS, and SP1 in cardiac enhancer function. Clustering of TF motifs is also a feature of predicted murine cardiac enhancers (He et al. 2011; Schlesinger et al. 2011), and additional broadly expressed TFs, such as TEAD, NFAT, STAT, and YY1, were implicated in enhancer function (He et al. 2011) (see below).
The Drosophila Model
The Drosophila model has allowed a number of network features to be analyzed in detail using integrated genomics approaches. The Drosophila heart is a simple linear muscular tube with inflow ports guarded by valve leaflets, and an outflow portion termed the aorta. Its early development relies on a regulatory network of TFs homologous to the kernel factors defined in mammals, including the NKX2-5-like homeodomain factor Tinman, GATA factor Pannier, and T-box factor Dorsocross (Jin et al. 2013). Tinman is expressed first in almost the entire mesoderm, then, under the influence of decapentaplegic (DPP; bone morphogenetic protein [BMP] in vertebrates) signals from dorsal ectoderm, becomes restricted to dorsal mesoderm where it maintains its own expression and induces Pannier. The combination of BMP and wingless signaling (WG in flies/WNT in mammals) independently induces Dorsocross in a segmental pattern, and the three TFs create a cross-regulatory kernel that is necessary for specification of cardioblasts. Like NKX2-5 in mammals, Tinman is expressed in other lineages including pericardial cells, somatic muscle founder cells, and visceral muscles. Through direct activation of downstream regulators, such as bagpipe and even-skipped, in collaboration with fibroblast growth factor (FGF), DPP, and segmental WG signals, Tinman plays a primary role in setting up these distinct lineage territories (Carmena et al. 2002; Jakobsen et al. 2007; Jin et al. 2013). Genome-wide ChIP, transcriptome analysis, and computational approaches have also identified other important regulators of mesodermal lineage specification including MEF2, Biniou, and Ladybird (Sandmann et al. 2006; Jakobsen et al. 2007; Junion et al. 2007, 2012).
Jin et al. (2013) used ChIP to profile the binding sites of Tinman at two developmental stages. High Tinman occupancy was highly predictive of enhancer function in vivo as well as Tinman dependency, and these enhancers showed diverse arrangements of Tinman, Pannier, and Dorsocross motifs. Machine learning identified a new motif classifier, speculated to be bound by an ETS family member. Junion et al. (2012) also explored the occupancy of Tinman, Pannier, and Dorsocross, as well as phospho-MAD, and TCF, the downstream effector TFs of the DPP and WG pathways involved in cardiogenesis, respectively, at three developmental stages. Occupancy of Tinman, Pannier, and Dorsocross was predictive of gene expression in dorsal mesoderm or heart. Clustering analysis revealed six distinct classes of enhancer, with the most prominent class bound by all five TFs analyzed, with relatively few bound by three or four TFs, strongly supporting the combinatorial model for cardiac enhancer function. Out of 26, 24 of the maximally occupied enhancers were active in vivo and 91% of these were expressed in mesoderm, most prominently heart. However, only the Pannier and Dorsocross DNA-binding motifs were preferentially found in such enhancers, whereas Tinman, pMAD, and TCF were preferentially found in enhancers occupied by only two factors. These findings suggest that Pannier and Dorsocross are preferentially recruited to a subset of maximally occupied enhancers through direct DNA binding (perhaps as pioneer factors), and that heterotypic protein–protein interactions account for the occupancy of additional factors detected by ChIP. A further finding from this study was that a rigid motif grammar for TF binding to these enhancers was not evident, supporting the “billboard” or “TF collective” model of enhancer function in which motif spacing and orientation show significant flexibility (Kulkarni and Arnosti 2003; Junion et al. 2012).
Ahman et al. also used similar machine-learning approaches to extract ∼2000 putative heart enhancers from the Drosophila genome (Ahmad et al. 2014). Analysis revealed the motif for the fly homolog of the human proto-oncogene MYB as a positive classifier of cardiac muscle cells and suppressor of Hairless (Su[H]), a downstream TF in the Notch pathway, as a classifier of pericardial cells. Functional studies linked MYB to the control of specific amplifying symmetric cell divisions in cardiac progenitor cells before the divergence of the cardiac and pericardial lineages, which involves the Notch pathway.
An elegant synthetic biology study probed the billboard model further. Erceg et al. generated 63 synthetic cis-regulatory motifs (CRMs) composed of the binding sites representative of mesodermal TFs including Tinman, Pannier, Dorsocross, pMAD, and TCF (Erceg et al. 2014). Different numbers of motifs and different motif architecture were explored and expression was examined in the Drosophila model after insertion into an identical genome site. The study found that motif number as well as arrangement and specific pairing all had significant impacts on expression levels, tissue specificity, and robustness, the latter measured as penetrance and expressivity of protein expression output in different tissues. The heart was much more sensitive to motif grammar than visceral muscle and even the most favorable motif configurations analyzed did not show robust expression in heart, possibly because addition cardiac motifs were missing. Modeling suggested that higher-order functions, for example, protein–protein interactions between TFs that impose defined spatial constraints, were key determinants of robust output. In the context of the growing number of “enhanceropathies” identified in humans (Smith and Shilatifard 2014), this study suggests that even small insertions or deletions in human enhancers that do not specifically affect the binding of TFs, could have significant effects on function, particularly the robustness of gene expression.
Signal Gating of Enhancer Function
An important insight gleaned from the individual and genome-wide analyses of cardiac enhancers in both Drosophila and mammals (Yin et al. 1997; Xu et al. 1998; He et al. 2011; Junion et al. 2012; Jin et al. 2013; Ahmad et al. 2014) is that lineage-restricted TFs, such as Tinman/NKX2-5, collaborate extensively with ubiquitously and broadly expressed TFs acting downstream from signaling pathways. In flies, binding sites for the DPP effector pMAD, WG effector TCF, and FGF-responsive ETS factors, are found in Tinman and Pannier target genes and are essential for enhancer function (Junion et al. 2012). In mice, BMP effector pSMAD1, as well as TCF, ETS, TEAD, NFAT, STAT, YY, SP1, LMO2, and MEIS1/2 have been implicated in cardiogenesis (Brown et al. 2004; Narlikar et al. 2010; He et al. 2011; Paige et al. 2012; Wamstad et al. 2012). It is likely that many ubiquitous or widely expressed signal-gated TFs interact directly with cardiac-kernel TFs (Brown et al. 2004; He et al. 2011). In this way enhancers integrate the combinatorial coding potential of heart-restricted TFs, such as Tinman/NKX2-5, with signal-dependent TFs that gate enhancer activity to dynamic and regional signaling inputs. This collaboration implies a massive coding potential in TF networks. The combinatorial mechanism requires certain checks and balances to protect against noisy gene expression and to ensure rapid activation in response to signaling input. For example, in most cases a single TF motif alone would be insufficient to activate an enhancer (activator insufficiency) (Barolo and Posakony 2002; Mbodj et al. 2013). Furthermore, in the absence of extracellular signals, enhancers occupied by positive TFs may be repressed owing to the presence of negative cofactors that modify the surrounding chromatin (default repression) (Barolo and Posakony 2002; Mbodj et al. 2013). Cardiac-kernel TFs often act as both activators and repressors (Prall et al. 2007; Watanabe et al. 2012), and are themselves subject to signal-dependent posttranscriptional modifications affecting activity and stability (Liang et al. 2001; Elliott et al. 2010).
Boolean Models of Cardiac Development
The dynamics of biological systems can be modeled using quantitative methods, such as ordinary differential equations, although rarely are the quantitative data on the concentrations of key players and the kinetics of interactions available (Bolouri and Davidson 2003). Boolean models seek to overcome this limitation by reducing gene expression network inputs and outputs to a set of binary operators, for example, “on” (value 1) or “off” (value 0). This allows dynamic modeling of complex network states and transitions using a qualitative reduction of genetic, biochemical, and gene expression data. Davidson and colleagues have recently shown that it is possible to model all the key regulatory interactions underpinning skeletogenesis in the sea urchin, including (1) acquisition of fate by maternal cues, (2) activation of regulatory genes using a double-negative gate, (3) stabilization of the regulatory state, and (4) exclusion of alternative states (Oliveri et al. 2008). Mbodj et al. have also used the Boolean modeling to graphically represent the off and on states of major signaling pathways active in Drosophila development, and to compile these subnetworks into a more complete network for early mesoderm development (Mbodj et al. 2013).
Working in the mammalian system, Herrmann et al. (2012) have used the Boolean approach to simulate acquisition of the FHF and SHF attractor states using a set of interactions between kernel TFs and their inducers and effectors described in the literature. They defined the FHF as having on values for genes encoding BMP2, NKX2-5, GATA, and TBX5, and off values for SHF genes WNT, TBX1, FGF8, FOXC1/2, GATA (alternative site), NKX2-5, and ISL1. Sampling all possible state combinations (215 states), two main attractors were identified resembling FHF and SHF values (Fig. 4). Transient states involving MESP1 expression and potentially representing common cardiac progenitors were identified. The model could also predict altered states akin to mutant phenotypes when individual elements were removed, including “no-heart” attractors in which mesoderm does not form (as seen with removal of WNT and MESP1), and a delayed transition to the field attractors, as seen with removal of endodermal DKK1 (a WNT antagonist). These interesting preliminary studies support the view that distinct attractors exist for the FHF and SHF, and that attractors need to be disabled by defined signals to ensure directionality in development.
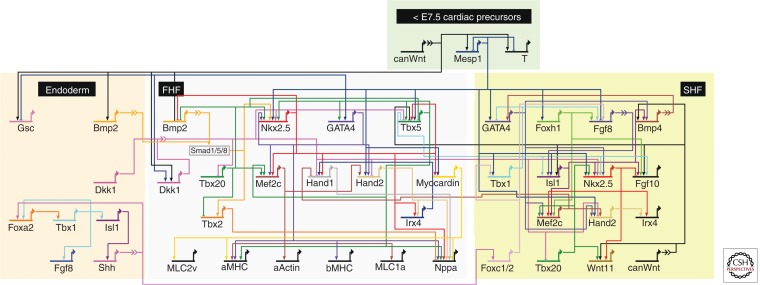
Figure 4.
Early cardiac developmental GRNs. BioTapestry representation of the attractor states discovered for the first and second heart fields (FHF and SHF, respectively) using Boolean inference modeling based on known transcription factor interactions (solid arrows) and signaling inputs (broken arrows) within different territories. (From Herrmann et al. 2012; reprinted, with permission, and under the terms of the Creative Commons Attribution License.)
EPIGENETIC REGULATION OF HEART DEVELOPMENT
GRN output is controlled by both TF occupancy and chromatin structure (Wilczynski et al. 2012), the latter representing a highly malleable vehicle for contributing to cell state specification through modifications to histones and DNA. In the machine-learning study of Junion et al. (2012), which identified mesodermal enhancers in Drosophila, visceral muscle enhancers were found to be co-occupied by both visceral and cardiac TF collectives. This was thought to reflect the common ancestry of these lineages in dorsal mesoderm. It was proposed that Tinman binding to visceral enhancers in dorsal mesoderm before lineage specification might prime them to become active at later stages through induction of an open and accessible chromatin state (a pioneer effect).
Stergachis et al. recently performed a genome-wide analysis of DNase I-hypersensitivity sites (DHS) during embryonic stem cell (ESC) differentiation into cardiovascular lineages and in a range of mature tissues (average 161,000 autosomal DHS/tissue). This enabled them to map the retention, loss, and gain of TF occupancy across developmental time and in both related and distinct tissues (Stergachis et al. 2013). Unsupervised nearest-neighbor clustering showed that DHS signatures were highly predictive of tissue relationships and embryological origins, and in fact these signatures were more discerning for such relationships than transcriptome analysis. Cardiac fibroblasts, for example, clustered with aortic and pulmonary fibroblasts but were more distant from fibroblasts of other tissues. During differentiation, tissue-specific DHSs were activated and those for alternative cell fates deactivated. Although many ESC DHSs were progressively pruned during differentiation, remarkably, a large number of stem cell DHSs (∼37%) were propagated into the chromatin of mature tissues including cardiomyocytes, with each tissue signature being unique. Forward propagation of progenitor cell signatures was also seen. Developmentally stable DHS were occupied chiefly by TFs showing direct autoregulation. This insightful study provides an important glimpse into how lineage history plays a key role in network structure and function, a feature that is lost in the deranged GRNs of malignant cells (Stergachis et al. 2013).
Hundreds of chromatin modification enzymes control chromatin structure and function (Ram et al. 2011) and there is considerable diversity in the grammar of histone modifications with distinct codes associated with different aspects of gene expression (Ernst and Kellis 2010). Table 1 compiles information on different histone modifications and chromatin-modifying enzymes, indicating where they have been analyzed in the cardiac system or associated with cardiac disease.
Table 1.
A compilation of information on different histone modifications and chromatin-modifying enzymes, indicating where they have been analyzed in the cardiac system or associated with cardiac disease
| Marks | cis-Regulatory elements | Genome-wide studies in cardiac cells | Modifiers | Associations with heart defects/disease |
|---|---|---|---|---|
| Histone modifications | ||||
| H3K9ac | Active promoters | Adult hearts (Sayed et al. 2013) HL-1 cells (Schlesinger et al. 2011) |
HATs (p300/CBP; GCN5/PCAF) HDACs |
p300 (Han et al. 2011) HDAC classes I, II, III (Han et al. 2011; Lara-Pezzi et al. 2012) |
| H3K18ac | Active promoters | |||
| H3K27ac | Enhancers or promoters | Differentiating ESCs (Wamstad et al. 2012) | ||
| H4ac (H4K5K8K12K16) | Active promoters and gene bodies | HL-1 cells (Schlesinger et al. 2011) | ||
| H3K4me1 | Promoters and enhancers | Differentiating ESCs (Wamstad et al. 2012) | HMTs | WHSC1 (H3K36me3) (Nimura et al. 2009) |
| H3K4me2 | Active promoters and gene bodies | HL-1 cells (Schlesinger et al. 2011) | ||
| H3K4me3 | Active promoters (at transcription start sites) | Differentiating ESCs (Paige et al. 2012; Wamstad et al. 2012) HL-1 cells (Schlesinger et al. 2011) |
MLL2 (H3K4me) (Ng et al. 2010) EZH2 (H3K27me) (Delgado-Olguin et al. 2012) |
|
| H3K9me2 | Inactive promoters | PTIP (H3K4me) (Stein et al. 2011) | ||
| H3K27me3 | Inactive promoters | Differentiating ESCs (Paige et al. 2012; Wamstad et al. 2012) | HDMs | JARID2, UTX (H3K27), JMJD2 (Lara-Pezzi et al. 2012) |
| H3K36me3 | Active gene bodies | Differentiating ESCs (Paige et al. 2012) | ||
| H3K4me1 + H3K27me3 no H3K27ac | Poised enhancers | Differentiating ESCs (Wamstad et al. 2012) | ||
| Histone variants, H3.3, H2A.Z | Enhancers | |||
| Other chromatin-modifying enzymes | ||||
| Poly(ADP-ribose) polymerases | Bind DNA breaks and regulate transcription | PARP-1 (Han et al. 2011) | ||
| ATP-dependent chromatin remodeler, e.g., SWI/SNF, ISWI, CHD | Alter nucleosomes structure | BAF, BRG1, CHD7, BAF45c/DPF3 (Han et al. 2011) | ||
| RNA polymerase II (Ser5Phos) | Proximal promoters (at transcription start sites) and low at enhancers | Adult hearts (Sayed et al. 2013) Differentiating ESCs (Wamstad et al. 2012) |
||
| DNA modifications | ||||
| 5mC at CpG islands | Inactive promoters | Mothers with CHD pregnancy (Chowdhury et al. 2011) | Chowdhury et al. 2011 | |
| 5hmC | Active promoters, genes, and enhancers | |||
HAT, histone acetyltransferase; HDAC, histone deacetylase; ESC, embryonic stem cell; HMT, histone methyltransferase; HDM, histone demethylase; CHD, congenital heart disease.
RNA polymerase II (Pol II) is known to be recruited to enhancers, which are transcribed into enhancer RNAs (Smith and Shilatifard 2014). Analysis of mesodermal enhancers lying >1 kb from the gene body in Drosophila shows that Pol II occupancy, although transient, is highly predictive of an enhancer’s precise location and temporal activity (Bonn et al. 2012). Trimethylation of histone H3-lysine79 (H3K79me3) and acetylation of H3K27 (H3K27ac) were also enriched in active versus inactive enhancers, and likely create a permissive state over a broad regulatory region encompassing one or more enhancers. However, relatively few active enhancers carried these marks, indicating that other marks can also define active states. H3K27me3 was counterindicative of enhancer activity, consistent with its known association with repressed or poised states (Cheutin and Cavalli 2014).
Bayesian network inference modeling was used to discover dependencies between quantitative histone marks and Pol II occupancy, and gene expression, among mesodermal enhancers with narrow or more general temporal activities. Bayesian models have the ability to integrate diverse data in an iterative probabilistic framework, and are able to cope in a principled way with uncertainty and missing data (Friedman 2004). Modeling identified a link between H3K79me3 and H3K27ac and enhancer activity, whereas H3K27me3 was contraindicative and H3K4me1, H3K4me3, and H3K36me3 had no predictive value. The predictive value of the modeling was tested in vivo considering only Pol II occupancy and H3K79me3 and H3K27ac marks present on mesodermal enhancers active in the 6–8 h window of development, without drawing on information from TF motifs or occupancy. Out of nine, eight (89%) of the predicted enhancers tested were active and directed expression in the defined time window. More extensive Bayesian modeling drawing on TF occupancy showed that neither enhancer occupancy nor chromatin state can accurately predict gene expression patterns, but used together they provide accurate predictions of spatiotemporal activity (Wilczynski et al. 2012). Further validation of 20 genes with unknown expression patterns resulted in an accuracy of 95% for correct temporal expression and 50% for spatial expression.
Two recent studies have explored the dynamics of chromatin changes during directed differentiation of human ESCs into cardiovascular lineages in relation to transcriptome patterns (Paige et al. 2012; Wamstad et al. 2012). Both studies highlighted the dynamic and diverse nature of chromatin changes during differentiation, and the ability to use this data predictively. For example, considering permutations of only three situations (repressive or poised-state mark H3K27me3; active mark H3K4me3; no marks), genes expressed in cardiovascular lineages could be segmented into functional categories (Wamstad et al. 2012). The H3K4me1 mark seemed to indicate a preactivation pattern around promoters that in a minority of genes progressed to H3K4me3, Pol II occupancy, and active expression (Wamstad et al. 2012). The polycomb complex repressive mark H3K27me3 occurred on developmentally regulatory genes but did not participate in regulation of muscle contractile genes (Paige et al. 2012). Putative distal enhancers were scored genome wide as being H3K3me1+ (open; poised) or H3K27ac+/H3K4me1+/− (active). Active enhancers were enriched in Pol II occupancy, and their associated genes were more highly expressed compared with unmarked or poised genes, and corresponded to gene ontology categories appropriate to the stages of cardiogenesis analyzed (Wamstad et al. 2012).
Using their respective prediction platforms, both studies identified the TFs MEIS1 and MEIS2, which interact with HOX and PBX TFs in a variety of developmental settings, as important regulators of heart development (Paige et al. 2012; Wamstad et al. 2012). Studies in zebrafish indicated early expression of meis2b in the heart fields and confirmed its essential role for heart tube fusion and looping (Paige et al. 2012).
Although cellular heterogeneity in these studies is a potential limitation, they form a strong foundation for future work on the dynamics of chromatin structure and how it relates to network output during human cardiogenesis. There is increasing genetic evidence linking epigenetic control to heart disease. For example, mutations in BRG1/BAF (ATP-dependent chromatin-remodeling complex), associated helicase CHD7, Williams syndrome TF WSTF (associates with BAF proteins and is recruited by cardiac TFs NKX2-5, GATA4, and TBX5), and Wolf–Hirschhorn syndrome gene WHSC1 (H3K36me3-methyltransferase), are all associated with cardiac structural defects in humans or animal models (Table 1).
ncRNA REGULATION OF HEART DEVELOPMENT
ncRNAs represent major components of network control and the diversity of their gene structure, processing, and function is only just beginning to be appreciated in the aftermath of the revolution in high-throughput sequencing (Morris and Mattick 2014). Indeed, it is thought that an increase in noncoding DNA and the noncoding transcriptome may have been a major evolutionary driver of organismal complexity (Taft et al. 2007).
miRNAs are ∼22 base single-stranded ncRNAs that inhibit mRNA translation or promote mRNA degradation by annealing to complementary sequences in the 3′ untranslated region of target mRNAs (reviewed in Porrello 2013). Individual miRNAs may have hundreds of targets and individual mRNAs can be targeted by multiple miRNAs. Furthermore, multiple miRNAs may be produced from a single precursor transcript (pri-miRNA) and miRNAs often target functionally related mRNAs. These features suggest the possibility of a vast interactive subnetwork regulating GRN output at each stage of development, homeostasis, adaptation, and regeneration. Whereas germline deletion of the miRNA bisynthesis enzyme DICER leads to early embryonic lethality (Bernstein et al. 2003), cardiac-restricted knockout using the early Nkx2-5-Cre driver leads to midgestation lethality owing to outflow tract, septal, and chamber malformations (Zhao et al. 2007; Saxena and Tabin 2010). Cardiac-kernel TF genes, such as Tbx5 and Hand1, were expressed relatively normally, although Pitx2 and Sema3c involved in outflow tract development were up-regulated. These phenotypes are remarkably mild compared with cardiac knockout phenotypes of kernel or hub genes, and are consistent with studies showing a more prominent role for miRNA networks in the development of cardiomyocyte maturity, growth, function, and physiology (Porrello 2013). Indeed, a number of knockouts in genes for individual miRNA or paralogous clusters have led to mild, partially penetrant, or no detectable phenotypes (Zhao et al. 2007; Porrello 2013; Wystub et al. 2013). Notwithstanding the intricate networks regulated by specific miRNA families in cardiac tissue (van Rooij et al. 2009; Qian et al. 2011), because they act predominantly at a level of the GRN below cis-regulatory architecture, their major function is likely to confer robustness to network output.
Long ncRNAs (lncRNAs) make up the bulk of the human noncoding transcriptome and are central to epigenetic regulation. Indeed many of the protein partners of lncRNAs are chromatin modifiers, and 30% of long intergenic noncoding RNAs (lincRNA) expressed in mouse ESCs were found to associate with at least one of the 12 chromatin-modifying complexes involved in reading, writing, and erasing histone modifications (see Table 1) (Guttman et al. 2011). Many bind to the Trithorax chromatin-activation or polycomb group chromatin-repressing families of histone-modifying enzymes, and guide them to their sites of action. Others may act as scaffolds for chromosomal organization or as miRNA sponges, and many other functions have been described (Morris and Mattick 2014). The analysis of lncRNA function in the heart is only beginning, and this area has been recently reviewed (Schonrock et al. 2012; Peters and Schroen 2014) along with genome tools that facilitate research into the area (Schonrock et al. 2012). New tools, such as ChIRP (chromatin isolation by RNA purification), in which individual ncRNAs are captured using biotinylated oligonucleotides, are enabling genomic occupancy and bound protein and RNA components to be identified at high resolution (Chu et al. 2011).
Here we select two recent examples that illustrate the role of lncRNAs in epigenetic control of heart developmental networks. Klattenhoff et al. identified the lncRNA gene Braveheart (Bvht) from among those expressed in mouse ESCs and heart (Klattenhoff et al. 2013). The Bvht gene produces a 590-nucleotide transcript spliced from three exons. No homologs have been identified in human or rat, consistent with the higher divergence of ncRNAs that are unconstrained by the requirement for codon structure and function (Schonrock et al. 2012). shRNA knockdown of Bvht in ESCs appeared to specifically affect the cardiovascular lineages, and lead to down-regulation of kernel TF genes, such as Hand1, Hand2, Nkx2-5, and Tbx20. Mesp1, which acts upstream of the kernel genes, was also affected and genes for its regulators Brachyury and Eomesodermin were normally expressed or up-regulated. Among several chromatin-modifying enzyme subunits tested, Bvht associated with SUZ12, a component of the polycomb repressive complex, PCR2. In Bvht knockdown ESCs, SUZ12 occupancy at the promoters of Mesp1, Gata6, Hand1, Hand2, and Nkx2-5 remained abnormally high and was lower over the Brachyury and Eomesodermin genes, with the polycomb repressive mark H3K27me3 mirroring these patterns. Although the precise mechanism is unknown, these studies suggest that Bvht has a role in epigenetic control of cardiovascular networks from gastrulation stages.
Grote et al. characterized a mouse 2.4-kb-nuclear-long intergenic ncRNA (lincRNA), Fendrr, which is expressed transiently in lateral mesoderm of the early embryo (Grote et al. 2013). This lincRNA does have a homolog in humans. Homozygous genetic knockout of Fendrr showed omphalocele and malformed hearts with reduced proliferation, leading to midgestation lethality. In the forming heart, kernel TFs Gata6 and Nkx2-5 were significantly up-regulated concomitant with an increase in the Trithorax-activating mark H3K4me3. This held true for other genes (Foxf1 [linked to Fendrr], Gata6, Irx3, and Pitx2) in caudal lateral mesoderm, with all, bar Gata6, also showing a decrease in the repressive mark H3K27me3 and drastically reduced occupancy of polycomb subunits EZH2 and SUZ12. Fendrr was found to bind these polycomb components, as well as the Trithorax subunit WDB5, with further in silico and in vitro assays inferring that Fendrr RNA might bind directly to regions of the Foxf1 and Pitx2 genes through base-pairing to double-stranded DNA.
FUTURE PERSPECTIVES
GRNs, as described by Britten and Davidson (1969), have provided a framework for analysis of development and the acquisition of lineage, organ, and organismal function. The application of this framework to cardiac and broader mesodermal networks in Drosophila and other species using machine-learning approaches has confirmed the combinatorial nature of TF function on CRMs and provided a mechanism for how regulatory regions act as integrators of signaling inputs. It has also provided insights into scalability, plasticity, redundancy, and higher-order interactions in motif grammar. Further, it has revealed how early network states are critical for defining latter networks. The further application of machine-learning and Boolean dynamic models to heart development will certainly advance our understanding of network logic. However, currently, cardiac Boolean models do not take into account the continuous and quantitative nature of gene regulation and grossly oversimplify the spatiotemporal events underpinning developmental progressions (Mbodj et al. 2013). It is self-evident that a regulatory model is only as good as its ability to predict all aspects of development, and progress in this field will depend on renewed efforts to generate quantitative and dynamic data, eventually kinetic data of molecular interactions, and new tools for data analysis and representation. The challenges of describing changing biological context and coping with tissue heterogeneity cannot be underestimated. For example, the SHF consists of subdomains that cannot be reduced to a generic tissue (Vincent and Buckingham 2010). Techniques for obtaining transcriptomes, proteomes, and metabolomes from single cells are advancing rapidly, which will help overcome the averaging effects implicit in working with heterogeneous cell populations. Such technologies are capable of identifying a range of cellular states not previously observed. The ability to remove assumptions about the identity of a cell and/or derived population is attractive and may overcome many hidden biases in our perceptions of development (Jaitin et al. 2014). The elegant work describing skeletogenesis in sea urchins and Drosophila at approaching single-cell resolution (Oliveri et al. 2008; Jin et al. 2013) should be achievable in mammals. The acquisition of data on a scale that nurtures systems biology will require the development of informative high-throughput assays and synthetic biology approaches for testing predictions.
Congenital and adult-onset heart disease is common and debilitating, and there is an urgent need to contextualize disease states in terms of systems biology. The impact of single or multiple gene variants, and genome instability, needs to be understood in network terms. Furthermore, how a GRN interfaces with environmental conditions that influence cardiac disease risk—for example, fetal hypoxia, maternal diabetes, hypertension, and diet (Sparrow et al. 2012)—and how induced epigenetic states and epimutations show transgenerational inheritance (Martin et al. 2011), also warrant systems biology approaches.
Finally, therapies that stimulate cardiac regeneration and rejuvenation have the potential to help control the epidemic of ischemic heart disease and heart failure complicated by age and frailty. Network solutions that allow terminally differentiated cardiomyocytes to reenter the cell cycle (Naqvi et al. 2014), or to rejuvenate the regenerative capacity of endogenous stem cells (Ellison et al. 2013), may ultimately be found. Many tissues reactivate fetal programs of gene expression when injured, highlighting the inherent plasticity in networks, even those defining terminally differentiated states, and this can be explored further at a network level and potentially exploited in regenerative medicine approaches (Bollini et al. 2014). In summary, systems biology provides a philosophical and technological framework for understanding biology and disease that has enormous relevance in contemporary science, and this relevance is set to increase as the field matures.
Footnotes
Editors: Margaret Buckingham, Christine L. Mummery, and Kenneth R. Chien
Additional Perspectives on The Biology of Heart Disease available at www.perspectivesinmedicine.org
REFERENCES
- Ahmad SM, Busser BW, Huang D, Cozart EJ, Michaud S, Zhu X, Jeffries N, Aboukhalil A, Bulyk ML, Ovcharenko I, et al. 2014. Machine learning classification of cell-specific cardiac enhancers uncovers developmental subnetworks regulating progenitor cell division and cell fate specification. Development 141: 878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. 2007. Network motifs: Theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Anderson RH, Spicer DE, Brown NA, Mohun TJ. 2014. The development of septation in the four-chambered heart. Anat Rec (Hoboken) 10.1002/ar.22949 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. 1999. Emergence of scaling in random networks. Science 286: 509–512 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Bonabeau E. 2003. Scale-free networks. Sci Am 288: 60–69 [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. 2002. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev 16: 1167–1181 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, Bellamy V, Rucker-Martin C, Barbry P, Bel A, et al. 2010. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest 120: 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42: 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini S, Vieira JM, Howard S, Dube KN, Balmer GM, Smart N, Riley PR. 2014. Re-activated adult epicardial progenitor cells are a heterogeneous population molecularly distinct from their embryonic counterparts. Stem Cells Dev 10.1089/scd.2014.0019 [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. 2003. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci 100: 9371–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. 2008. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3: 69–84 [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. 2011. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol 192: 751–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. 1969. Gene regulation for higher cells: A theory. Science 165: 349–357 [DOI] [PubMed] [Google Scholar]
- Brown CO 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. 2004. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem 279: 10659–10669 [DOI] [PubMed] [Google Scholar]
- Busser BW, Bulyk ML, Michelson AM. 2008. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev 18: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, Michelson AM. 2002. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol 244: 226–242 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chan SS, Kyba M. 2013. What is a master regulator? J Stem Cell Res Ther 3: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JX, Plonowska K, Wu SM. 2014. Somatic cell reprogramming into cardiovascular lineages. J Cardiovasc Pharmacol 10.1177/1074248414527641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G. 2014. Polycomb silencing: From linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev 25C: 30–37 [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Erickson SW, MacLeod SL, Cleves MA, Hu P, Karim MA, Hobbs CA. 2011. Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS ONE 6: e16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Davidson B, Levine M. 2009. Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev Biol 328: 552–560 [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Tian Y, Morrisey EE. 2008. Wnt signaling: An essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development 135: 789–798 [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. In Machine learning, pp. 273–297. Kluwer Academic, Hingham, MA [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ. 2011. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Jarsch VB, Schwarz F, Nathan P, Gegg M, Lickert H, Franz WM. 2011. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res 92: 115–122 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. 2002. A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. 2012. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 44: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. 2004. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131: 3931–3942 [DOI] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Schaft D, Harvey RP. 2010. NK-2 class homeodomain proteins: Conserved regulator of cardiogenesis. In Heart development and regeneration (ed. Rosenthal N, Harvey RP), pp. 569–597. Elsevier, New York [Google Scholar]
- Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD, Henning BJ, Stirparo GG, Papait R, Scarfo M, et al. 2013. Adult c-kitpos cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 154: 827–842 [DOI] [PubMed] [Google Scholar]
- Enver T, Pera M, Peterson C, Andrews PW. 2009. Stem cell states, fates, and the rules of attraction. Cell Stem Cell 4: 387–397 [DOI] [PubMed] [Google Scholar]
- Erceg J, Saunders TE, Girardot C, Devos DP, Hufnagel L, Furlong EE. 2014. Subtle changes in motif positioning cause tissue-specific effects on robustness of an enhancer’s activity. PLoS Genet 10: e1004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantom Consortium. 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. 2004. Inferring cellular networks using probabilistic graphical models. Science 303: 799–805 [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. 1975. Genetic control of wing disc development in Drosophila. In Ciba Foundation Symposium 29—Cell patterning (ed. Porter R, Rivers J), Wiley, Chichester, UK: [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev Cell 12: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. 1988. A community effect in animal development. Nature 336: 772–774 [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Hang CT, Yang J, Chang CP. 2011. Chromatin remodeling in cardiovascular development and physiology. Circ Res 108: 378–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison NJ, Baker D, Andrews PW. 2007. Culture adaptation of embryonic stem cells echoes germ cell malignancy. Int J Androl 30: 275–281; discussion 281 [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci 108: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Gross A, Zhou D, Kestler HA, Kuhl M. 2012. A boolean model of the cardiac gene regulatory network determining first and second heart field identity. PLoS ONE 7: e46798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler KL, Hendershot TJ, Troy SE, Vincentz JW, Firulli AB, Howard MJ. 2010. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev Biol 341: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343: 776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JS, Braun M, Astorga J, Gustafson EH, Sandmann T, Karzynski M, Carlsson P, Furlong EE. 2007. Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev 21: 2448–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. 2001. Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Jin H, Stojnic R, Adryan B, Ozdemir A, Stathopoulos A, Frasch M. 2013. Genome-wide screens for in vivo Tinman binding sites identify cardiac enhancers with diverse functional architectures. PLoS Genet 9: e1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G, Bataille L, Jagla T, Da Ponte JP, Tapin R, Jagla K. 2007. Genome-wide view of cell fate specification: Ladybird acts at multiple levels during diversification of muscle and heart precursors. Genes Dev 21: 3163–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. 2012. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148: 473–486 [DOI] [PubMed] [Google Scholar]
- Kauffman SA. 1969. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol 22: 437–467 [DOI] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN. 2003. Information display by transcriptional enhancers. Development 130: 6569–6575 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E, Dopazo A, Manzanares M. 2012. Understanding cardiovascular disease: A journey through the genome (and what we found there). Dis Model Mech 5: 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. 2005. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell 8: 85–95 [DOI] [PubMed] [Google Scholar]
- Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol 21: 7460–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kaneda R, Leja TW, Subkhankulova T, Tolmachov O, Minchiotti G, Schwartz RJ, Barahona M, Schneider MD. 2014. Hhex and cer1 mediate the sox17 pathway for cardiac mesoderm formation in embryonic stem cells. Stem Cells 32: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. 2002. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 2: 592–600 [DOI] [PubMed] [Google Scholar]
- Martin DI, Cropley JE, Suter CM. 2011. Epigenetics in disease: Leader or follower? Epigenetics 6: 843–848 [DOI] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. 2012. Large-scale discovery of enhancers from human heart tissue. Nat Genet 44: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbodj A, Junion G, Brun C, Furlong EE, Thieffry D. 2013. Logical modelling of Drosophila signalling pathways. Mol Biosyst 9: 2248–2258 [DOI] [PubMed] [Google Scholar]
- Mitchell KJ. 2007. The genetics of brain wiring: From molecule to mind. PLoS Biol 5: e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. 2014. The rise of regulatory RNA. Nat Rev Genet 15: 423–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, et al. 2014. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 157: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar L, Sakabe NJ, Blanski AA, Arimura FE, Westlund JM, Nobrega MA, Ovcharenko I. 2010. Genome-wide discovery of human heart enhancers. Genome Res 20: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42: 790–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. 2009. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature 460: 287–291 [DOI] [PubMed] [Google Scholar]
- Noseda M, Peterkin T, Simoes FC, Patient R, Schneider MD. 2011. Cardiopoietic factors: Extracellular signals for cardiac lineage commitment. Circ Res 108: 129–152 [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. 2008. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci 105: 5955–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. 2006. Gene regulatory networks in the evolution and development of the heart. Science 313: 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. 2012. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T, Schroen B. 2014. Missing links in cardiology: Long non-coding RNAs enter the arena. Pflugers Arch 466: 1177–1187 [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER. 2013. microRNAs in cardiac development and regeneration. Clin Sci 125: 151–166 [DOI] [PubMed] [Google Scholar]
- Prall OWJ, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. 2007. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128: 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wythe JD, Liu J, Cartry J, Vogler G, Mohapatra B, Otway RT, Huang Y, King IN, Maillet M, et al. 2011. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol 193: 1181–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ. 1999. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development 126: 839–849 [DOI] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. 2008. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol 28: 5420–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Garrido M, Karbassi E, Monte E, Vondriska TM. 2013. Regulation of chromatin structure in the cardiovascular system. Circ J 77: 1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. 2006. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell 10: 797–807 [DOI] [PubMed] [Google Scholar]
- Saxena A, Tabin CJ. 2010. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci 107: 87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, He M, Yang Z, Lin L, Abdellatif M. 2013. Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem 288: 2546–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR. 2011. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet 7: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. 2007. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell 13: 254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N, Harvey RP, Mattick JS. 2012. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 111: 1349–1362 [DOI] [PubMed] [Google Scholar]
- Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al. 2011. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest 121: 4640–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes FC, Peterkin T, Patient R. 2011. Fgf differentially controls cross-antagonism between cardiac and haemangioblast regulators. Development 138: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Hoogaars WM, Barnett P, Grieskamp T, Rana MS, Buermans H, Farin HF, Petry M, Heallen T, Martin JF, et al. 2012. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci 69: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2014. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219 [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O’Reilly VC, Saga Y, Zackai EH, Dormans JP, Alman BA, et al. 2012. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 149: 295–306 [DOI] [PubMed] [Google Scholar]
- Sperling SR. 2011. Systems biology approaches to heart development and congenital heart disease. Cardiovasc Res 91: 269–278 [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: From enhancer binding to developmental control. Nat Rev Genetics 13: 613–626 [DOI] [PubMed] [Google Scholar]
- Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, et al. 2011. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest 121: 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, et al. 2005. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 132: 2451–2462 [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. 2013. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. 1981. A homeotic mutation transforming leg to antenna in Drosophila. Nature 292: 635–638 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29: 288–299 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocheteau P, Le Roux I. 2009. Asymmetric cell divisions and asymmetric cell fates. Annu Rev Cell Dev Biol 25: 671–699 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. 1988. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411 [DOI] [PubMed] [Google Scholar]
- Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc 58: 267–288 [Google Scholar]
- Vandenberg LN, Levin M. 2013. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev Biol 379: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJ, Klous P, McKean D, Muehlschlegel JD, Moosmann J, Toka O, et al. 2014. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest 124: 1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, et al. 2012. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 150: 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ Jr, Olson EN. 2009. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. 2011. Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Wang H, John S, Sandstrom R, Stamatoyannopoulos JA. 2014. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat Methods 11: 66–72 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME. 2010. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr Top Dev Biol 90: 1–41 [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1957. The strategy of the genes: A discussion of some aspects of theoretical biology. Allen & Unwin, Crows Nest, New South Wales [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, Harvey RP, Kelly RG, Buckingham M. 2012. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2–5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci 109: 18273–18280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Battle MA, Li J, Duncan SA. 2004. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci 101: 12573–12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B, Liu YH, Yeo ZX, Furlong EE. 2012. Predicting spatial and temporal gene expression using an integrative model of transcription factor occupancy and chromatin state. PLoS Comput Biol 8: e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wystub K, Besser J, Bachmann A, Boettger T, Braun T. 2013. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet 9: e1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. 1998. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev 12: 2354–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. 2001. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol 239: 190–203 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature 465: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M. 1997. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124: 4971–4982 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, Thurig S, Behfar A, Wallace VA, Skerjanc IS, et al. 2006. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell 11: 535–546 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. 2007. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129: 303–317 [DOI] [PubMed] [Google Scholar]