Abstract
Antimicrobial photodynamic inactivation (APDI) using phenothiazinium dyes is mediated by reactive oxygen species consisting of a combination of singlet oxygen (quenched by azide), hydroxyl radicals and other reactive oxygen species. We recently showed that addition of sodium azide paradoxically potentiated APDI of Gram-positive and Gram-negative bacteria using methylene blue as the photosensitizer, and this was due to electron transfer to the dye triplet state from azide anion, producing azidyl radical. Here we compare this effect using six different homologous phenothiazinium dyes: methylene blue, toluidine blue O, new methylene blue, dimethylmethylene blue, azure A, and azure B. We found both significant potentiation (up to 2 logs) and also significant inhibition (>3 logs) of killing by adding 10 mM azide depending on Gram classification, washing the dye from the cells, and dye structure. Killing of E. coli was potentiated with all 6 dyes after a wash, while S. aureus killing was only potentiated by MB and TBO with a wash and DMMB with no wash. More lipophilic dyes (higher log P value, such as DMMB) were more likely to show potentiation. We conclude that the Type I photochemical mechanism (potentiation with azide) likely depends on the microenvironment, i.e. higher binding of dye to bacteria. Bacterial dye-binding is thought to be higher with Gram-negative compared to Gram-positive bacteria, when unbound dye has been washed away, and with more lipophilic dyes.
Keywords: phenothiazinium dyes, antimicrobial photoinactivation, Escherichia coli, Staphylococcus aureus, sodium azide, singlet oxygen, azidyl radicals, fluorescent probe for ROS
Introduction
Phenothiazinium-based photosensitizers (PS) have been employed in antimicrobial photodynamic inactivation (APDI) research for nearly 80 years, both as known compounds1 and as novel derivatives2, 3. In broader biomedical application, the phenothiazinium dye series has also been commonly applied in cytology and cytopathology, as well as in hematological staining4.
Methylene blue (MB) has been used as a lead compound in conventional (non-light mediated) antimicrobial therapy research for over a hundred years5. Since the first publication of its efficacy as an antimalarial compound in 18916, MB has also been employed as an antibacterial, e.g., in local antisepsis and an agent against tuberculosis5, and its low toxicity in man is reflected in its current clinical use as an injectable therapeutic for methemoglobinemia7 and its investigational use against Alzheimer’s disease8 and distributive shock9. However, it is unlikely that MB is the optimum member of the class of phenothiazinium derivatives for antimicrobial photoinactivation.
Wainwright et al10 compared five different phenothiazinium dyes as antibacterial photosensitizers against methicillin sensitive Staphyolococcus aureus (MRSA). Dimethyl methylene blue and new methylene blue were found to be the most active compounds but concern was raised about their dark toxicity.
We recently reported the paradoxical potentiation of MB-mediated antimicrobial PDT by sodium azide (Na+N3−)11 – paradoxical because N3− is known physically to quench 1O2, thus protecting the bacteria from1O2-mediated killing12. We concluded that oxygen radicals (or the excited PS itself) could directly abstract an electron from N3−, forming azidyl radicals (N3•), which although they are less reactive than •OH or 1O2, may be more selective and effective bactericidal agents, because they are longer lived than •OH. This longer lifetime may be responsible for enhanced PDT bacterial killing as N3• may diffuse deeply into the cells and then wreak havoc while the more reactive •OH or 1O2 is rapidly consumed at the cell wall.
In the present study we attempted to gain deeper understanding of this intriguing observation of the potentiation of antimicrobial PDI by azide. We used a panel of six phenothiazinium dyes most of which were originally studied by Wainwright et al4, 13, 14. These compounds have varying lipophilicities as measured by the logP value (octanol:water partition coefficient). We also compared the Gram-negative bacterial species Escherichia coli with a Gram-positive counterpart Staphylococcus aureus, and studied the effect of washing unbound dye out of the bacterial suspension. It should be noted however, that the toxicity of azide precludes it being used for any clinical application, but nevertheless it can provide useful information about photochemical mechanisms.
Materials and methods
Photosensitizers
Azure A (AA), azure B (AB), methylene blue (3,7-bis(dimethylamino)-phenothiazinium chloride; (MB) toluidine blue O (TBO), new methylene blue N, zinc free form (NMB), 1,9-dimethyl-methylene blue (DMMB) were all obtained from (Sigma-Aldrich, St. Louis, MO). Each photosensitizer was stored in the dark as an aqueous stock solution (1mM in MilliQ water, for DMMB in 50% H2O/acetonitrile) at 4°C for no longer than 24 hours. Sodium azide was from Fisher Scientific, Waltham, MA. Purity was around 80%.
Log P values
We measured the logP values experimentally because calculated values of logP for phenothiazinium salts give different values depending whether the canonical structure is drawn with the cationic charge on the ring sulfur atom or on the peripheral nitrogen atom. A mixture of 2 mL of octanol and 2 mL of phosphate buffered saline (PBS; pH 7.4, 50 mM; 150 mM NaCl) in a 20-mL scintillation vial was stirred at room temperature for 3 h. Then less than 0.5 mg of phenothiazinium salt was introduced. Stirring was continued at room temperature at 100–200 rpm for 24 h. The mixture was allowed to stand for 30 min to allow separation of the phases. A 30-µL aliquot of each phase was placed in 3.0 mL of dimethylsulfoxide (DMSO) and the absorption spectrum of each phase was measured. The ratio of the peak area of the red absorption band, for the two phases (octanol/PBS) was calculated; the log(10) of the measured ratios are hereafter denoted meas logP values.
Light source
A convenient light source consisted of a non-coherent broad-band lamp capable of delivering light into a fiber-optic probe (FOP) fitted with a band-pass filter 635+15-nm (LumaCare, Newport Beach, CA). A power meter (model DMM 199 with 201 standard head, Coherent, Santa Clara, CA) was used to measure the irradiance (power density in mW/cm2).
ROS probes
The Singlet Oxygen Sensor Green (SOSG, Life Technologies, Bedford MA) reagent is highly selective for 1O2; although it does also show some response to hydroxyl radical (•OH) and superoxide (O2•−)15. This new singlet oxygen indicator in its non-activated state exhibits weak blue fluorescence, with excitation peaks at 372 and 393 nm and emission peaks at 395 and 416 nm. In the presence of singlet oxygen, it emits a green fluorescence similar to that of fluorescein (excitation/emission maxima ~504/525 nm). Stock solutions ~5 mM were prepared in methanol immediately before use.
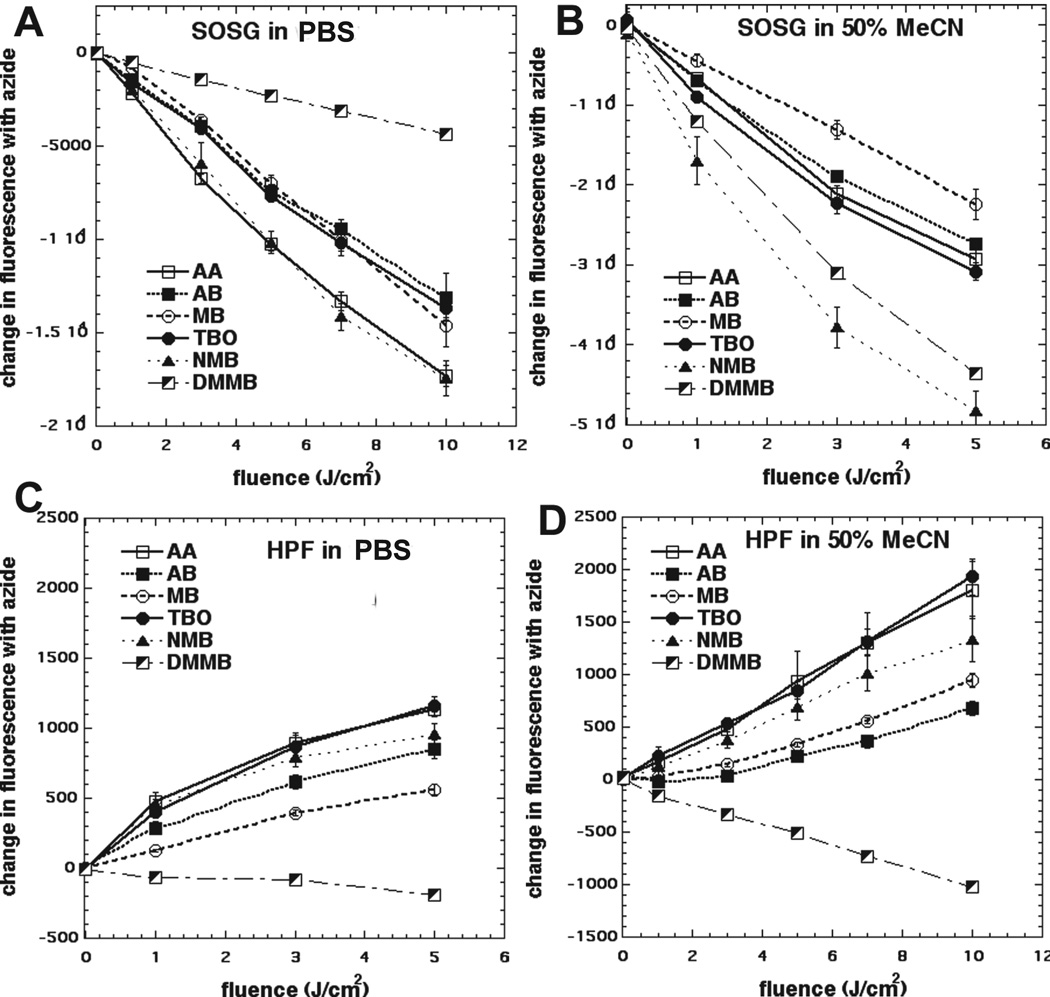
Hydroxyphenyl fluorescein (HPF, Life Technologies) is a fluorescent reagent for the detection of highly reactive hydroxyl radicals. HPF is resistant to light-induced autoxidation. SOSG and HPF were used at 5 µM final concentration and phenothiazinium dyes at 10 µM final concentration in each assay. A microplate spectrophotometer (Spectra Max M5, Molecular Devices) was used for acquisition of fluorescence signals in the “slow kinetic” mode. When HPF was employed, fluorescence emission at 525 nm was measured after excitation at 490 nm using a 2 nm monochromator band pass for both excitation and emission. With SOSG, the corresponding values were emission 525 nm and excitation 505 nm. Increasing fluences (J/cm2) were delivered using 635-nm light at an irradiance of 100 mW/cm2. Each time after an incremental fluence was delivered, the fluorescence was measured. We compared probe activation with and without the addition of sodium azide at 10 mM concentration. Because we were concerned that the different lipophilicities of the dyes might affect their solubility in water and thus their photochemical effectiveness, we carried out these probe activations in two different solutions, (a) PBS and (b) 50:50 mixture of PBS and acetonitrile. The data was plotted by subtracting the fluorescence value from each point in the absence of azide from the fluorescence value of the same point in the presence of azide (see Fig 2).
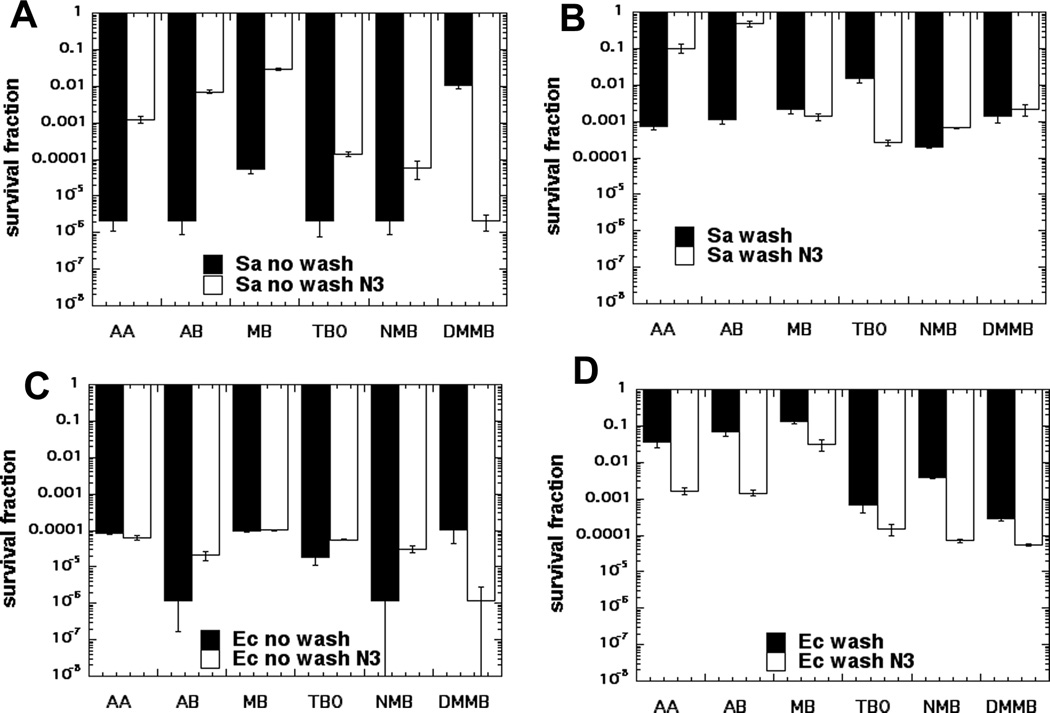
Figure 2. Effect of azide on PDT killing of Gram-positive and Gram-negative bacteria with and without a wash.
S. aureus (A & B) and E. coli (C & D) were incubated with 6 phenothiazinium dyes (10 µM) followed by wash (B & D) or no wash (A & C) and addition of NaN3 (10 mM) and exposure to 10 J/cm2 red light.
Microbial cell culture
Staphylococcus aureus 8325-4 and Escherichia coli K-12 (ATCC, Manassas, VA) were employed. Planktonic bacteria cells were cultured in brain heart infusion broth (BHI) (Fisher Scientific) with shaking at 37°C. PBS Response was used to wash microbial cells and for serial dilution. Liquid growth media was prepared from 200 ml of distilled water and 6 g of BHI powder. All liquid media were autoclaved at 120° C for 15 min before use. Solid growth media was prepared from liquid growth media with the addition of 1,5% microbiological agar (Fisher Scientific). A suspension of microbial cells was prepared by refreshing an overnight stationary phase culture in fresh medium for about 1 h. Cell pellets were isolated by centrifugation (13,690 × g for 5 min) and resuspended the appropriate volume of sterile PBS to give the desired cell-density (OD at 600 nm equivalent to 108 CFU/ml).
In vitro PDI experiments
Bacteria at 10(8) cells/mL were incubated with the dyes at 10 µM concentration using the same incubation time (15 min), and the same light fluence (10 J/cm2 of 635-nm light) for all studies. After 15 min incubation time the bacterial suspensions were centrifuged or not, and the pellets resuspended in the same volume of fresh PBS. Sodium azide at a final concentration of 10 mM was added or not and the cell suspensions were incubated for a further 5 min. We previously showed that 10 mM azide had no bacterial toxicity on its own under these conditions. The light spot was set up to illuminate four wells of a 24 well plate equally by adjusting its diameter to 3.4 cm. At an irradiance of 100 mW/cm2, 10 J/cm2 was delivered in 100 sec. Controls consisted of absolute control (no treatment), photosensitizer without light, and photosensitizer plus azide without light. After delivery of light, suspensions were serially diluted and streaked on square agar plates by the method of Jett et al16. Survival fractions were obtained by dividing the treatment CFU/ml by the CFU/ml in the original cell suspension (absolute control). The experiments were repeated 3 times.
Bacterial binding studies
Since the ability of the dyes to bind to the bacterial cells appeared to be important in determining the degree of potentiation by azide, we measured the absorption spectra of the cell pellet and of the supernatant after incubation by cells (10(8)/mL) with dye (10µM) for 15 min to find out how much of the dye was bound to the cells and how much was not. The cell pellet was dissolved in the same volume (as the supernatant) of 0.1% SDS, while sufficient SDS was added to the supernatant to make the final concentration 0,1% SDS and the volume comparable. Absorption spectra were carried out between 500 and 700 nm. The fraction of dye bound to the cells was then calculated.
Photosensitizer structure and properties
The chemical structures of the 6 dyes are shown in Table 1 together with the absorption maximum wavelength, the redox potential and the experimentally determined logP values. The actual absorption spectra of the dyes both in PBS and in methanol, together with the emission spectrum of the light source are shown in supplementary material Fig S1. Both azure dyes had negative redox potentials (AA = −320 mV; AB = −160 mV) while the redox potentials of the other dyes were close to zero (− 21 mV to +11 mV). The meas logP values are proportional to calc logP values (i.e. the different dyes lie in the same order DMMB > NMB > AB > AA > MB > TBO) although the absolute numbers are different and as previously mentioned the calc logP values depend on which phenothiazinium dye canonical structure is used. Moreover the logP values depend on the ionic strength and pH of the aqueous phase.
Table 1.
Properties of dyes and relative binding by bacterial cells.
| dye | structure | λmax (nm) | Meas logPa | Redox potential | binding S. aureusb | binding E. colib |
|---|---|---|---|---|---|---|
| AA | 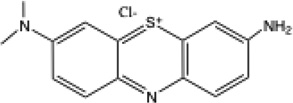 |
622 | 0.08 | −320 mV37 | 0.25 | 0.04 |
| AB | 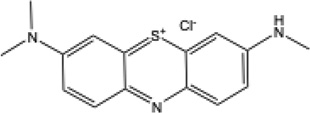 |
647 | 0.26 | −160 mV38 | 0.15 | 0.02 |
| MB | 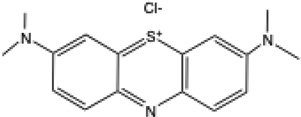 |
667 | 0.00 | +11 mV39 | 0.2 | 0.05 |
| TBO | 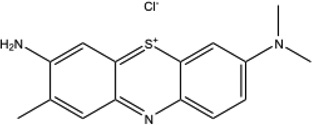 |
617 | −0.30 | −11 mV40 | 0.22 | 0.1 |
| NMB | 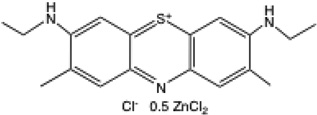 |
628 | 0.78 | −21 mV41 | 0.12 | 0.08 |
| DMMB | 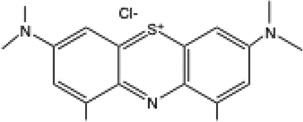 |
649 | 1.53 | c | 0.85 | 0.75 |
log(10) n-octanol:PBS partition co-efficient
fraction of the dye that is bound to cells after 15 min incubation in PBS
not available
The fraction of the dye bound to both S. aureus and E. coli are also shown in Table 1. It can be seen that there is much more dye bound to the Gram-positive S. aureus than to the Gram-negative E. coli for all the dyes.
ROS probes
We had previously found that when a PS was ilIuminated in solution, the balance between activating HPF and activating SOSG gave information on the Type I and Type II photochemical mechanisms11. Furthermore we previously showed that addition of sodium azide was able to increase the activation of HPF by a fullerene excited by white light11. It therefore was reasonable to ask what effect azide had on the activation of these ROS probes by all six of our phenothiazinium dyes. The results are shown in Figure 1 for both PBS (and a 50:50 mixture of PBS and acetonitrile). For SOSG (Fig 1A-B) azide inhibited probe activation mediated by all six photoactivated dyes as expected from its known ability to quench singlet oxygen, with the biggest inhibition seen with NMB and the least inhibition seen with DMMB (PBS) and AB (50 % acetonitrile). There was more pronounced inhibition in 50% acetonitrile than in PBS especially in the case of DMMB which showed much higher inhibition in the organic solvent consistent with its higher lipophilicity. For HPF (Fig 1C) in PBS we saw potentiation of activation for AA, TBO, AB, MB, NMB and in that order with only DMMB showing inhibition. In acetonitrile (Fig 1D) the activation of the probe by AA, TBO, AB, MB, NMB was more pronounced than in PBS, while the inhibition of HPF activation by DMMB was also more pronounced.
Figure 1. Effect of azide on activation of fluorescent ROS probes (SOSG and HPF) by 6 phenothiazinium dyes and light.
The fluorescence value from each point in the absence of azide was subtracted from the fluorescence value of the same point in the presence of azide. (A) SOSG in PBS; (B) SOSG in 50:50 PBS acetonitrile; (C) HPF in PBS; (B) HPF in 50:50 PBS acetonitrile.
In vitro PDI studies
The killing of S. aureus by each of the six phenothiazinium dyes (10 µM with 10 J/cm2 of 635-nm light) with and without a wash and with and without addition of 10 mM sodium azide is shown in Figures 2A and B Under these conditions around 4–5 logs of the Gram-positive bacteria were killed by each photoactivated dye (with the exception of DMMB), with the killing being much more effective with no wash than it was with a wash. This difference (wash vs no wash) has been previously reported in the case of TBO17. The same set of experiments was done with the Gram-negative E. coli as shown in Figures 2C and D. There was about 3.5–5 logs of killing obtained with all six dyes, and the no wash killing was again higher than the killing observed after a wash.
The interesting comparisons are between the killing in the presence of azide and without azide. There was a great variation in the effects of azide on the PDT bacterial killing between dyes and also between species. We found both significant potentiation of killing (> 3 logs), and also significant inhibition of killing (>3 logs) by adding 10 mM azide depending on Gram classification, washing the dye from the cells, and the dye structure. E. coli was more likely to show potentiation of killing by azide after a wash, while S. aureus only showed potentiation of killing by azide with no wash in the case of DMMB. In general more lipophilic dyes (higher log P value such as DMMB) were more likely to show potentiation of killing, while more hydrophilic dyes (AA and AB) were more likely to show inhibition of killing.
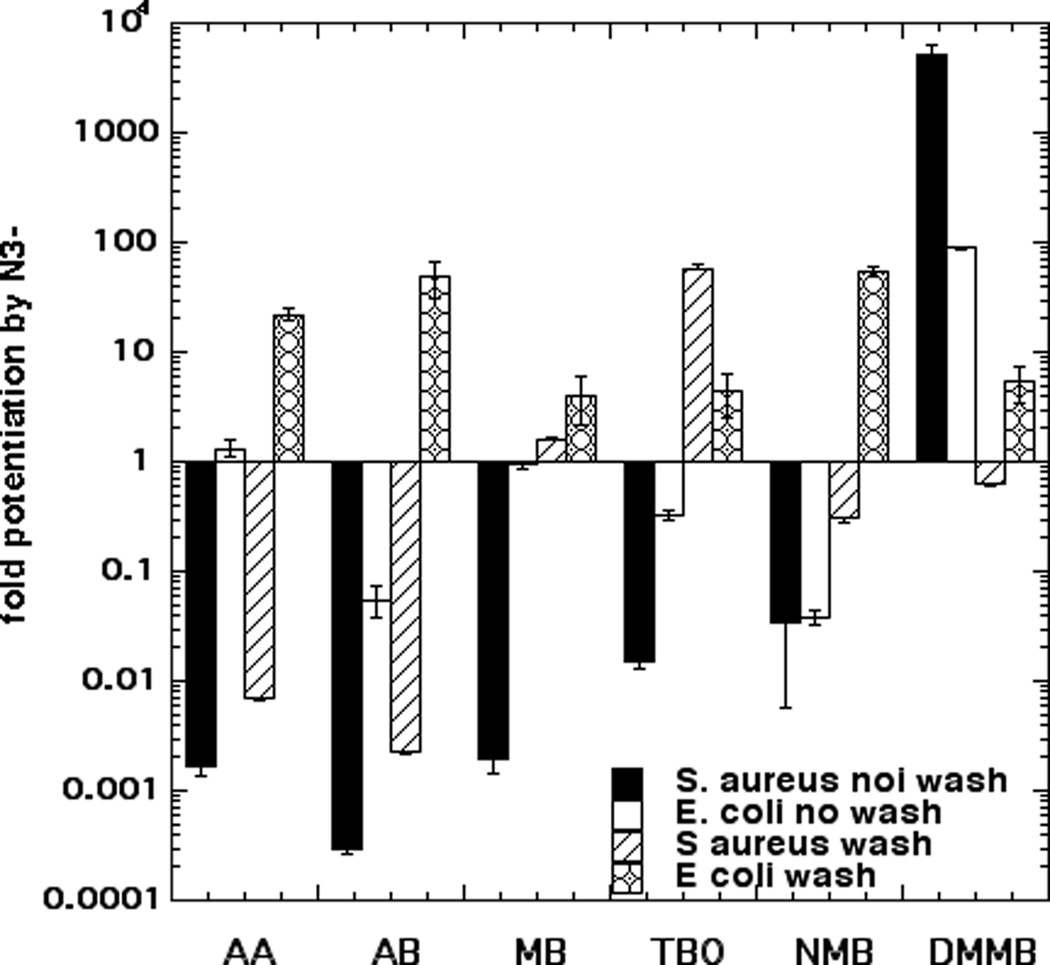
In order to allow a graphical representation of the effects of azide we have plotted the fold-potentiation (or fold-inhibition) of bacterial PDI by addition of azide in all of the experiments in Figure 3. It becomes clear from this figure that E. coli with a wash shows potentiation by azide with every single phenothiazinium dye. Conversely both S. aureus with no wash, and E. coli with no wash, show inhibition of killing by azide by all the dyes except DMMB. AB and NMB gave potentiation of killing by azide in the case of only one condition out of four, AA, MB and TBO gave potentiation of killing by azide in two conditions, while DMMB gave potentiation of killing by azide in three conditions.
Figure 3. Fold-potentiation or inhibition of PDT killing of bacteria by addition of azide to PDT mediated by 6 phenothiazinium dyes shown with and without a wash.
Data extracted from the graphs in Figure 2.
Discussion
Studying and defining the differences between Type I and Type II photochemical mechanisms has occupied workers in the PDT field for decades11, 18–22. Nevertheless the majority of authors maintain that a Type II (singlet oxygen) mechanism is the overwhelmingly more important ROS in PDT and especially in antimicrobial PDI23, 24. Antimicrobial PDI is an attractive biological area to study the different photochemical mechanisms involved in PDT, as the bacterial cells are particularly sensitive to membrane damage from ROS generated in solution. This means that the intracellular uptake of PS is not as strictly necessary to kill bacteria as it would have been in the PDT killing of cancer cells where intracellularly generated ROS (in for instance mitochondria) produce efficient cell death. In other words ROS generated outside the bacterial cells, or actually at the cell wall by PS that are bound to the bacterial cells, may still be able to cause lethal damage to the cell membrane by the process of diffusion.
We previously showed that sodium azide was able to potentiate the bacterial PDI mediated by MB in both Gram-positive and Gram-negative bacteria11. The production of azidyl radicals during the potentiation was demonstrated by ESR spin trapping with DMPO. We initially hypothesized that the mechanism involved the intermediate formation of•OH from MB and light, and then the•OH radical carried out a one-electron oxidation of azide anion to form azidyl radical. However this hypothesis needed to be changed after we observed that both the formation of spin-adduct DMPO-N3 and the potentiation of MB-PDI by addition of azide occurred in the absence of oxygen. This could possibly be used to allow PDT to be used in areas of hypoxic tissue if a less toxic mediator than azide could be found.
It should be noted that azide is toxic to both mammalian cells and to microorganisms including bacteria (all Gram-negative but not many Gram-positive species)34. Azide inhibits the activity of cytochrome c oxidase in a similar manner to cyanide. We checked previously35 that at 10 mM concentration for 30 min there was no azide toxicity to either bacterial species, but toxicity did begin to appear at 50 mM. Therefore it is clear that the potentiation of antibacterial PDI by addition of azide could never be used clinically, but its value is as a mechanistic tool to understand the basic photochemistry.
On discovering the oxygen independence of the bacterial photokilling, we proposed an alternative mechanism whereby the excited triplet state MB could carry out a one electron oxidation of N3− to N3•11. The redox potential of the half reaction: N3• + e− → N3− is +1.3V, while that of the half reaction: HO•+ e− +H+→ H2O is +2.31V, making hydroxyl radicals much more reactive than azidyl radicals25. Nevertheless it is possible that the very high reactivity of HO• is a disadvantage when it comes to killing bacterial cells, because all the reaction happens at the very outermost part of the bacterial cell such as the cell wall structures, while a less reactive radical such as N3• can survive long enough to diffuse closer to sensitive sites such as the plasma membrane. This hypothesis gained additional support when we showed that addition of azide also potentiated bacterial killing by Fenton reagent11. Fenton reagent (a mixture of hydrogen peroxide and ferrous ions) is considered to be a reasonably selective source of hydroxyl radicals26 and azidyl radical is presumably formed from azide by the one electron oxidation produced by hydroxyl radical.
In the present study we compared the potentiation of PDI by addition of azide amongst a series of six different phenothiazinium dyes in order to gain some insight into the structural features governing this interesting photochemical reaction. We initially looked at the effect of azide of the activation of the ROS probes HPF and SOSG. As expected, because SOSG is relatively specific for activation by 1O2, azide quenched activation of SOSG by all six dyes. For HPF the situation was more interesting and surprising. The activation of HPF was potentiated by azide in 5 cases (AA and TBO strongly) while HPF activation was quenched only by DMMB. The relatively lipophilic DMMB was more quenched by azide in MeCN (for both HPF and SOSG) presumably because it was better dissolved in the presence of the organic solvent and its aggregation was less. This disaggregation would increase the yield of 1O2 and allow the physical quenching by azide to be more apparent. We have previously published that HPF activation was potentiated by azide in the case of the tris-cationic fullerene BB6 plus light11. while Kessel et al reported HPF activation by azide in the case of ROS generated by Fenton reagent but gave no mechanism for this observation27. Although it may be tempting to suppose that azidyl radical could attack the phenol ring of HPF releasing fluorescein in the same manner as hydroxyl radical, there is as yet no hard evidence for this hypothesis.
We found that the degree of potentiation (or inhibition) of killing depended strongly on the Gram-classification, whether the dye was washed from the suspension, and on the dye structure. Potentiation was observed for every dye (all six) in only one case, that of E. coli with a wash (between 5 and 150-fold). In the case of S. aureus with no wash Inhibition was observed for 5 dyes with only one dye giving potentiation (DMMB),. Since the wash removes the unbound dye leaving only dye that is bound to bacteria, it may be the case that the more negatively charged Gram-negative bacteria bind cationic dyes more strongly than the less negatively charged Gram-positive cells28–30, although the overall binding is higher for Gram-positive bacteria because their porous cell wall allows much more dye to diffuse inside. The hypothesis that potentiation depends on dye-bacteria binding gains additional support when we consider the lipophilicity of the dyes. The dye with a relatively high meas logP value gave potentiation of killing in three out of four different bacteria/wash combinations (DMMB). Conversely dyes with lower logP values (AA and MB) give modest potentiation of killing only in the case of E. coli with a wash.
Taking all these observations and deductions together we have formed the hypothesis that potentiation of antimicrobial PDT by azide depends on the microenvironment of the photosensitizer. The dye is likely to be more firmly bound to the bacterial cells: (a) for more negatively charged Gram-negative bacteria, (b) after a wash to remove unbound dye, and (c) in the case of more lipophilic dyes. When the dye is firmly bound it may be more likely to receive an electron transfer from azide to its triplet excited state thereby producing a radical pair (PS radical anion and azidyl radical). A complementary hypothesis is that firmly bound dyes can transfer the electron from the PS radical anion into the bacterial cell with its metabolic electron transport chains, thus regenerating the ground state uncharged PS and allowing the process to repeat indefinitely. Non-firmly bound dyes might find it more difficult to carry out this secondary electron transfer because they are not adjacent to the bacterial cell.
Because the radicals produced from bound dyes are by definition produced closer to their target (bacterial cells) than they would have been had the dye not been firmly bound, it may be supposed that the distance the radicals have to travel to encounter their target governs their toxicity. This limitation on effective diffusion length of radicals is in accord with what is known about free radical toxicity31.
We have previously reported11 that the degree of inhibition of bacterial photoinactivation by azide varied according to PS structure according to the proposed photochemical mechanism. We previously found azide inhibited photodynamic killing by PEI-ce6 (polyethylenimine chlorin(e6) conjugate) and red light much more than it did the photokilling of BB6 (cationic fullerene) and white light11. These findings are supported by the relevant redox potentials (C60 fullerene = −169 mV32 and (related) Zn-chlorin(e6) = +540 mV33). The similarity of the redox potentials of phenothiazinium dyes (see table 1) to that of C60 suggests that low redox potentials encourage electron transfer and Type 1 photochemistry. Since high inhibition of killing correlated with more activation of SOSG (PEI-ce6) and low inhibition correlated with more activation of HPF (BB6) we proposed that azide could perhaps potentiate killing by Type I photochemistry (formation of hydroxyl radicals). In fact we proposed11 that a one electron-transfer from azide to hydroxyl radical to form azidyl radical might be responsible. Nevertheless it is important to reiterate that we did not observe any actual azide potentiation of bacterial killing by either PEI-ce6 or BB6 in that study11, although we did observe potentiation of activation of HPF ROS probe by BB6 and white light when azide was added to the solution. So apparently it is necessary to have photosensitizers with the phenothiazinium structure to see actual potentiation of killing, possible because these molecules can carry out the one-electron transfer reaction from the triplet state referred to above11.
More research will be necessary to see if other classes of antimicrobial photosensitizer molecules (that are not phenothiazinium salts) can have their microbial killing potentiated by addition of azide anion. Our laboratory has recently reported that thiocyanate anion can also potentiate antimicrobial PDI mediated by MB36 and has gained some preliminary evidence that other simple inorganic salts (such as iodide anion) are also able to potentiate antimicrobial PDI, and these results will be reported in due course.
Supplementary Material
Acknowledgments
This work was supported by US NIH grant R01AI050875.
Footnotes
Supplementary material.
Figure S1.
Figure S2.
References
- 1.Wainwright M, Phoenix DA, Marland J, Wareing DR, Bolton FJ. A study of photobactericidal activity in the phenothiazinium series. FEMS Immunol Med Microbiol. 1997;19:75. doi: 10.1111/j.1574-695X.1997.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 2.Walker I, Gorman SA, Cox RD, Vernon DI, Griffiths J, Brown SB. A comparative analysis of phenothiazinium salts for the photosensitisation of murine fibrosarcoma (RIF-1) cells in vitro. Photochem Photobiol Sci. 2004;3:653. doi: 10.1039/b400083h. [DOI] [PubMed] [Google Scholar]
- 3.Wainwright M, Smalley H, Scully O, Lotfipour E. Comparative photodynamic evaluation of new phenothiazinium derivatives against Propionibacterium acnes. Photochem Photobiol. 2012;88:523. doi: 10.1111/j.1751-1097.2011.01021.x. [DOI] [PubMed] [Google Scholar]
- 4.Wainwright M, Mohr H, Walker WH. Phenothiazinium derivatives for pathogen inactivation in blood products. J Photochem Photobiol B Biol. 2007;86:45. doi: 10.1016/j.jphotobiol.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Wainwright M, Crossley KB. Methylene Blue--a therapeutic dye for all seasons? J Chemother. 2002;14:431. doi: 10.1179/joc.2002.14.5.431. [DOI] [PubMed] [Google Scholar]
- 6.Wainwright M, Amaral L. The phenothiazinium chromophore and the evolution of antimalarial drugs. Trop Med Int Health. 2005;10:501. doi: 10.1111/j.1365-3156.2005.01417.x. [DOI] [PubMed] [Google Scholar]
- 7.Sikka P, Bindra VK, Kapoor S, Jain V, Saxena KK. Blue cures blue but be cautious. J Pharm Bioallied Sci. 2011;3:543. doi: 10.4103/0975-7406.90112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schirmer RH, Adler H, Pickhardt M, Mandelkow E. Lest we forget you-- methylene blue. Neurobiol Aging. 2011;32:2325 e7. doi: 10.1016/j.neurobiolaging.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Jang DH, Nelson LS, Hoffman RS. Methylene Blue for Distributive Shock: A Potential New Use of an Old Antidote. J Med Toxicol. 2013 doi: 10.1007/s13181-013-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wainwright M, Phoenix DA, Laycock SL, Wareing DR, Wright PA. Photobactericidal activity of phenothiazinium dyes against methicillin-resistant strains of Staphylococcus aureus. FEMS Microbiol Lett. 1998;160:177. doi: 10.1111/j.1574-6968.1998.tb12908.x. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Xuan Y, Koide Y, Zhiyentayev T, Tanaka M, Hamblin MR. Type I and Type II mechanisms of antimicrobial photodynamic therapy: An in vitro study on gram-negative and gram-positive bacteria. Lasers Surg Med. 2012;44:490. doi: 10.1002/lsm.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavares A, Dias SR, Carvalho CM, Faustino MA, Tome JP, Neves MG, Tome AC, Cavaleiro JA, Cunha A, Gomes NC, Alves E, Almeida A. Mechanisms of photodynamic inactivation of a gram-negative recombinant bioluminescent bacterium by cationic porphyrins. Photochem Photobiol Sci. 2011;10:1659. doi: 10.1039/c1pp05097d. [DOI] [PubMed] [Google Scholar]
- 13.Wainwright M, Phoenix DA, Gaskell M, Marshall B. Photobactericidal activity of methylene blue derivatives against vancomycin-resistant Enterococcus spp. J Antimicrob Chemother. 1999;44:823. doi: 10.1093/jac/44.6.823. [DOI] [PubMed] [Google Scholar]
- 14.Rice L, Wainwright M, Phoemix DA. Phenothiazine photosensitizers. III. Activity of methylene blue derivatives against pigmented melanoma cell lines. J Chemother. 2000;12:94. doi: 10.1179/joc.2000.12.1.94. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Ishiyama K, Ikai H, Kanno T, Sasaki K, Niwano Y, Kohno M. Reevaluation of analytical methods for photogenerated singlet oxygen. J Clin Biochem Nutr. 2011;49:87. doi: 10.3164/jcbn.10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 1997;23:648. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 17.Demidova TN, Hamblin MR. Effect of cell-photosensitizer binding and cell density on microbial photoinactivation. Antimicrob. Agents Chemother. 2005;49:2329. doi: 10.1128/AAC.49.6.2329-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Cruz P, Dejesus-Andino F, Alegria AE. Roles of hydrophilicities and hydrophobicities of dye and sacrificial electron donor on the photochemical pathway. J Photochem Photobiol A Chem. 2012;236:54. doi: 10.1016/j.jphotochem.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mroz P, Pawlak A, Satti M, Lee H, Wharton T, Gali H, Sarna T, Hamblin MR. Functionalized fullerenes mediate photodynamic killing of cancer cells: Type I versus Type II photochemical mechanism. Free Radic Biol Med. 2007;43:711. doi: 10.1016/j.freeradbiomed.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote CS. Definition of type I and type II photosensitized oxidation. Photochem Photobiol. 1991;54:659. doi: 10.1111/j.1751-1097.1991.tb02071.x. [DOI] [PubMed] [Google Scholar]
- 21.Foote CS. Mechanisms of photooxygenation. Prog Clin Biol Res. 1984;170:3. [PubMed] [Google Scholar]
- 22.Ochsner M. Photophysical and photobiological processes in the photodynamic therapy of tumours. J Photochem Photobiol B Biol. 1997;39:1. doi: 10.1016/s1011-1344(96)07428-3. [DOI] [PubMed] [Google Scholar]
- 23.Ragas X, He X, Agut M, Roxo-Rosa M, Gonsalves AR, Serra AC, Nonell S. Singlet oxygen in antimicrobial photodynamic therapy: photosensitizer-dependent production and decay in E. coli. Molecules. 2013;18:2712. doi: 10.3390/molecules18032712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maisch T, Baier J, Franz B, Maier M, Landthaler M, Szeimies RM, Baumler W. The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria. Proc Natl Acad Sci U S A. 2007;104:7223. doi: 10.1073/pnas.0611328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.AIfassi ZB, Hamiman A, Huie RE, Mosseri S, Neta P. The Redox Potential of the Azide/Azidyl Couple. J Phys Chem. 1987;91:2120. [Google Scholar]
- 26.Merli C, Petrucci E, Da Pozzo A, Pernetti M. Fenton-type treatment: state of the art. Ann Chim. 2003;93:761. [PubMed] [Google Scholar]
- 27.Price M, Reiners JJ, Santiago AM, Kessel D. Monitoring singlet oxygen and hydroxyl radical formation with fluorescent probes during photodynamic therapy. Photochem Photobiol. 2009;85:1177. doi: 10.1111/j.1751-1097.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minnock A, Vernon DI, Schofield J, Griffiths J, Parish JH, Brown SB. Mechanism of uptake of a cationic water-soluble pyridinium zinc phthalocyanine across the outer membrane of Escherichia coli. Antimicrob Agents Chemother. 2000;44:522. doi: 10.1128/aac.44.3.522-527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock RE, Farmer SW, Li ZS, Poole K. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1309. doi: 10.1128/aac.35.7.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Q, Yue Q, Zhao J, Wang L, Wang H, Wei X, Liu J, Jia J. How far can hydroxyl radicals travel? An electrochemical study based on a DNA mediated electron transfer process. Chem Commun (Camb. 2011;47:11906. doi: 10.1039/c1cc14699h. [DOI] [PubMed] [Google Scholar]
- 32.Diao G, Li L, Zhang Z. The electrochemical reduction of fullerenes, C60 and C70. Talanta. 1996;43:1633. doi: 10.1016/0039-9140(96)01879-6. [DOI] [PubMed] [Google Scholar]
- 33.Mahboob A, Vassiliev S, Poddutoori PK, van der Est A, Bruce D. Factors controlling the redox potential of ZnCe6 in an engineered bacterioferritin photochemical 'reaction centre'. PLoS One. 2013;8:e68421. doi: 10.1371/journal.pone.0068421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichstein HC, Soule MH. Studies of the Effect of Sodium Azide on Microbic Growth and Respiration: I. The Action of Sodium Azide on Microbic Growth. J Bacteriol. 1944;47:221. doi: 10.1128/jb.47.3.221-230.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L, St Denis TG, Xuan Y, Huang YY, Tanaka M, Zadlo A, Sarna T, Hamblin MR. Paradoxical potentiation of methylene blue-mediated antimicrobial photodynamic inactivation by sodium azide: role of ambient oxygen and azide radicals. Free Radic Biol Med. 2012;53:2062. doi: 10.1016/j.freeradbiomed.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St Denis TG, Vecchio D, Zadlo A, Rineh A, Sadasivam M, Avci P, Huang L, Kozinska A, Chandran R, Sarna T, Hamblin MR. Thiocyanate potentiates antimicrobial photodynamic therapy: In situ generation of the sulfur trioxide radical anion by singlet oxygen. Free Radic Biol Med. 2013;65C:800. doi: 10.1016/j.freeradbiomed.2013.08.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Song SH, Yoo YJ. Selection of mediators for bioelectrochemical nitrate reduction. Biotechnology and Bioprocess Engineering. 2005;10:47. [Google Scholar]
- 38.Chakraborty S, Ahamed S, Subrata Pal S, Saha SK. Cyclic Voltammetric Investigations of Thiazine Dyes on Modified Electrodes. ISRN Electrochemistry, 2013. 2013 http://dxdoiorg/101155/2013/959128. [Google Scholar]
- 39.Bespalov VA, Zhulin IB, Taylor BL. Behavioral responses of Escherichia coli to changes in redox potential. Proc Natl Acad Sci U S A. 1996;93:10084. doi: 10.1073/pnas.93.19.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norris JR, Ribbons DW. Methods in Microbiology. Academic Press; 1970. [Google Scholar]
- 41.Roller SD, Bennetto HP, Delaney GM, Mason JR, Stirling SL, Thurston CF. Electron transfer coupling in microbial fuel cells: 1. Comparison of redox mediator reduction rates and respiratory rates of bacteria. J Chem Technol Biotechnol. 1984;34B:3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.