Abstract
A-to-I RNA editing can recode mRNAs, giving organisms the option to express diverse, functionally distinct protein isoforms. Here, we propose that RNA editing is inherently geared for temperature adaptation because it tends to recode to smaller, less stabilizing amino acids. Studies on how editing affects protein function support this idea.
The central dogma to biology states that information is stored in genes and realized in proteins. Before being translated, it passes through RNA, a transient molecule that is relatively easy to manipulate. By altering RNA, an organism can regulate the information that it conveys. Organisms edit RNA by a variety of mechanisms, most notably by splicing. In recent years, nucleotide deamination by a variety of mechanisms has been shown to be important as well. In animals, the most common form of editing involves the hydrolytic deamination of adenosines to inosines, a process catalyzed by the ADAR (Adenosine Deaminase Acting on RNA) family of enzymes. Because inosines are read as guanosine by the ribosome (2), editing can result in amino acid recoding. This process is probably ubiquitous in true metazoans, since they all express ADAR homologs (28). By rewriting genetic information, organisms create novel protein isoforms that, in many cases, have modified functions. Furthermore, editing is seldom complete: both edited and unedited isoforms are expressed, often in different proportions depending on the tissue. Editing, then, gives an organism options. If we assume that it can be controlled, then the use of these options provides a plausible mechanism for responding to a variable environment. In this review, we highlight why A-to-I RNA editing is intrinsically well positioned to act as a mechanism for cold temperature adaptation or acclimation. We discuss evidence that it is in fact a plastic process, a prerequisite for its use in this capacity. Finally, we also highlight the evidence, both direct and indirect, that it is being used to respond to the cold.
RNA Editing Tends to Recode for Smaller Amino Acids
In 2002, our laboratory (52) proposed that A-to-I editing is well suited for cold adaptation because of the program of the universal genetic code. For those codons that contain an A, converting it to a G almost always recodes to a smaller side chain, in theory a result that should enhance protein entropy to compensate for the cold. With the aid of high-throughput sequencing technology, over the past decade an abundance of new editing sites has been discovered in RNAs from humans and Drosophila. With this data in hand, we can now more thoroughly address our original idea. Solely based on the genetic code, which amino acid conversions do we expect editing to make most frequently and then, based on datasets of real editing sites, which conversions are actually observed? It should be pointed out that cephalopods and Drosophila inhabit a broad range of thermal environments that can change on a rapid time scale. At great metabolic cost, humans, as all mammals, maintain a constant body temperature. Drosophila and cephalopods use editing to recode amino acids extensively (11, 18, 22, 35, 49, 52). Humans do not (34).
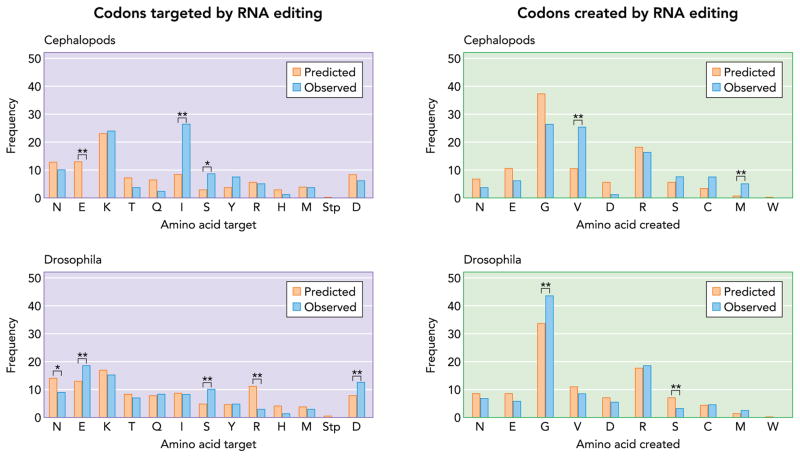
Our first question is whether the frequencies of those codons targeted and created by editing are what we would expect based purely on codon frequencies. According to the genetic code, there is a certain subset of codons that can be targeted by adenosine deamination and likewise a subset that can be created. The potential targets, of course, are those codons that contain an A. We tabulated the usage frequency of these codons in humans, Drosophila, and cephalopods to compensate for any biases toward or against codons with adenosines. If we assume that RNA editing randomly targets As within a codon, we expect lysine codons to be edited ~20% of the time and the remainder, which encode 12 possible amino acids, between 1 and 14% of the time (FIGURE 1). Among known editing sites in cephalopods and Drosophila, lysine is the most frequent target of editing, as expected. However, certain other amino acids were edited at frequencies significantly different than what we would expect for a random selection. In cephalopods, for example, isoleucine and serine were targeted approximately three times more frequently than expected, whereas glutamate codons were never edited, despite the fact that they would be predicted to be with a frequency of 13%. On the other hand, in Drosophila, glutamate codons were edited twice as frequently as expected, as were serine and aspartate codons. Arginine and asparagine codons were edited far less frequently than expected. These cases were the exceptions, however. The majority of codons were edited as expected by chance. The data for humans also suggest that the frequencies of edited codons followed the pattern predicted by random targeting of adenosines; however, the data set was too small [38 sites (34)] to make confident statistical comparisons and accordingly is not shown. A more recent study has confirmed that RNA editing is responsible for only ~40 amino acid recoding events (50). We should point out that, while searching for editing sites in cephalopods, experiments focused on ion channels and transporters. The codon usage patterns for these transmembrane proteins may be slightly different from those of the general pool of proteins.
FIGURE 1. Predicted and observed frequencies of codons targeted and created by A-to-I RNA editing.
All codons that could be edited to produce a different amino acid were identified. The relative frequencies of these codons in gene coding regions were then determined with the codon usage tables available at http://www.kazusa.or.jp (compiled using sequence data from GenBank, NCBI). Predicted editing percentages were calculated assuming that any adenosine within a codon would be randomly targeted for editing. Observed editing frequencies for Drosophila were based on the 611 sites identified in the developmental transcriptome project (22). For Cephalopods, a list of 79 sites was compiled using published data for Loligo and octopus (11, 18, 35, 49, 52) and also unpublished data from the Rosenthal laboratory. To determine whether the differences between predicted and observed editing frequencies were significant, we calculated the expected standard deviation for sample sizes of 611 and 79. If the observed difference was >2 SD (95% confidence interval) from the expected value, we considered the deviation significant. *Confidence interval of ≥95%. **Confidence interval of ≥99.7%.
The other side of the coin is which codons do we expect to be produced? Based solely on codon frequencies, over half of the amino acids produced by editing should be glycine and arginine, and this was in fact observed (FIGURE 1). Almost 50% of the amino acids created by Drosophila are glycine, a frequency even higher than expected. Cephalopods and humans produced about as many glycines as expected. Cephalopods, however, produced more valines, and slightly more methionines, than predicted, a result stemming from the fact that they were editing proportionally more isoleucine codons. In summary, A-to-I RNA editing appears intrinsically suited to create glycines and arginines. However, cephalopods tend to use it to create valine from isoleucine, and this cannot be explained purely by codon frequency. The same can be said for the targeting of glutamates and serines in Drosophila. Globally, editing tends to eliminate lysines, isoleucines, and glutamates (and to a lesser extent serines and aspartates) and to enrich glycines, arginines, and valines. Do these changes make sense for cold-temperature adaptation?
Amino Acid Usage and Temperature Adaptation
According to a well established hypothesis for cold adaptation, organisms lower the cold sensitivity of their enzymes by decreasing the enthalpy of the rate-limiting step of catalysis. This can be achieved by increasing flexibility in the vicinity of the active site (57). If we assume that it is beneficial to destabilize enzyme structure in response to the cold, then the changes made by editing make sense. Glycine, arginine, and valine are the residues most often created by editing, and they normally have a destabilizing effect on proteins. Lysine, isoleucine, and glutamate are selected for conversion most frequently, and they normally have a stabilizing influence. Glycine, with its small side chain, is thought to lend greater flexibility or conformational entropy to hinge and helix regions of proteins (15, 29, 41). Lysine, like arginine, has a large basic side chain; however, it provides for greater entropic stabilization than arginine (3), and the creation of arginine by editing is usually by the conversion of lysines. The isoleucine to valine conversion so common in cephalopods likely has a destabilizing effect within hydrophobic regions. The loss of the methyl group would tend to disrupt packing by producing a small cavity and reducing hydrophobicity (63). The large acidic side chain of glutamate is often involved in salt bridges that stabilize proteins (23, 58, 61). Of course, edited residues only represent a tiny fraction of the total. However, if they lie at key positions, then their destabilizing effects could have disproportionately large influences on function.
Moving beyond the general predicted effects, many studies have focused on the specific DNA-level amino acid changes that various organisms do in fact use for temperature adaptation. These studies provide a useful reference when we speculate on whether the amino acid changes caused by RNA editing fit into the same pattern. There are numerous examples showing that thermal adaptation involves specific changes to enzyme stability (5, 15, 19, 25, 56). In general, cold-adapted enzymes have evolved greater flexibility, presumably to counteract the conformational rigidity induced by the cold. Warm adapted enzymes, on the other hand, have evolved stabilizing features, like well packed hydrophobic cores (10) and surface salt bridges (23, 58, 61) to counteract the melting effects of heat and to pre-organize the enzyme toward the transition state (5). Interestingly, when critical residues have been identified, they are often the same amino acids as those most targeted by editing. For psychrophiles, the relatively small body of data suggests that glycine is an important amino acid for providing added flexibility to cold-adapted enzymes. Dong and Somero (15) found that a serine to glycine mutation in a malate dehydrogenase from a cold-adapted limpet provides greater flexibility and higher activity. Glycines are also generally overrepresented in the proteomes of psychrophilic prokaryotes (40).
The majority of studies on temperature adaptation have focused on thermophilic organisms. In these organisms, glycine is underrepresented. In fact, heat-tolerant enzymes produced by directed evolution have lost multiple glycines (8, 46). Furthermore, a multi-gene comparison between a thermophilic deep-sea vent worm and a mesophilic relative revealed that glycines were lost in the thermophile (26). The same study showed a preference for lysines in the thermophile, a bias previously noted in thermophilic microorganisms (3, 17, 62). Residues with long charged side chains, particularly lysines and glutamates, allow for stabilizing salt bridges on the surface and intersubunit interfaces of warm-adapted enzymes (23, 58, 61). These amino acids become more frequent when going from mesophilic to thermophilic to hyper-thermophilic bacteria (17). A third striking observation from the vent worm study was the high frequency of isoleucine to valine substitutions when going from the “hot” worm to its “cooler” cousin (26), and a preference for isoleucine over valine has been noted in thermophilic prokaryotes (7, 61). Although the structural underpinnings of temperature adaptation are complex, some general strategies and key amino acid players emerge. Glycines decrease stability and are favored in the cold, as are valines over isoleucines. The charged side chains of lysine and glutamate make them advantageous in warm conditions. Since RNA editing disproportionately eliminates lysines, isoleucines, and glutamates and creates glycines and valines, it would appear well suited for cold adaptation.
For a relatively small group of editing sites, the effects of amino acid recoding on protein function have been assessed. A general trend among these studies is that editing sites destabilize key conformations and interactions (Table 1). For example, in the human voltage-gated potassium channel Kv1.1, an isoleucine to valine edit in the channel’s pore dramatically speeds the rate at which the channel recovers from the inactivated state (4). Normally, the inactivation particle binds inside the pore through a hydrophobic interactions (42). Editing partially disrupts this interaction, leading to a faster disassociation of the occluding inactivation particle (4, 21). Potassium channels are also targeted in cephalopods. In squid Kv1.1, there are multiple editing sites in the tetramerization domain, a region that drives the oligomerization of the individual α-subunits into tetramers. One edit that involves an arginine to glycine conversion (R87G) on the interdomain interface strongly disrupts this interaction (52).
Table 1.
RNA editing destabilizes target conformations and interactions
| Target protein | Editing Event | Phenotype | Reference(s) |
|---|---|---|---|
| Human Kv 1.1 | I400V | Destabilized fast inactivated state | 4, 21 |
| Squid Kv 1.1 | R87G | Destabilized tetramerization | 52 |
| Octopus Kv 1.1 | I321V | Destabilized open state | 18 |
| Shab (Kv 2.1) | I681V | Destabilized open state* | 53a |
| Squid Na+/K+ ATPase α1 | I877V | Destabilization of Na+-bound state | 11 |
| Vertebrate 5-HT2CR | I156V+N158S/G+I160V | Reduced agonist affinity | 45 |
| Vertebrate GABAA receptor | I289M | Destabilized open state-reduced GABA affinity* | 44, 53 |
| Vertebrate Cav 1.3 | I1608M+Q1609R | Destabilized Ca2+ inactivated state* | 24 |
For these examples, precise mechanisms for changes in activity are not available.
There are additional examples from proteins other than Kv1 channels where editing destabilizes a specific interaction or state. In the squid Na+/K+ pump, an isoleucine to valine change (I877V) increases the turnover rate at negative voltages by destabilizing the deeply occluded, three Na ions bound state (11). For the human 5-HT2C serotonin receptor, a classic G-protein coupled receptor, there are five closely spaced editing sites in sequence that encode the intracellular loop that couples the receptor with its G protein (9). The fully edited form reduces the stability of the interaction between the receptor and G protein (45). In vertebrate GABAA receptors, one of the primary mediators of inhibition in the brain, an isoleucine to methionine editing event in the α3-subunit decreases activity, presumably by decreasing the stability of the open, ion conducting state (44, 53). Editing in the mouse voltage-gated calcium channel CaV1.3 also appears to destabilize a specific binding interaction between the channel and calmodulin (24). As expected for a large-scale generalization of functional effects, based solely on the identity of amino acid substitutions, there are exceptions to the rule of destabilization. However, the majority of sites appear to conform. Accordingly, we hypothesize that editing provides a means for targeted destabilization of active states. These studies show that editing can change the properties of target proteins, suggesting that there would be functional roles for edited isoforms. However, there is relatively little direct evidence to show that edited mRNAs are actually translated. In their recent paper, Huang et al. (22) use mass spectrometry to show that edited protein isoforms of Cav1.3 channels are indeed found in mouse brain, along with the unedited isoform, and that these edited isoforms are expressed on the surface and can change physiology. This supports the hypothesis that cells do translate and make use of edited isoforms.
For humans and many other endothermic vertebrates, body temperature does not vary. They maintain their core temperature between 35 and 40°C, depending on the species, which puts them 10–15°C above the “average” ectothermic vertebrate and on the warm end of the mesophilic spectrum. Consistent with the overall trend observed for mesophiles, thermophiles, and hyperthermophiles, mammals and birds exhibit amino acid usage frequencies that are more like thermophiles than those found in their ectothermic relatives, reptiles, amphibians, and fish (60). If RNA editing is used for thermal plasticity, particularly in response to the cold, then endotherms would have inherited a tool for which they had little use. It may be telling, then, that although there are many editing sites in humans, they are rarely used to change codons, most being in repetitive elements, untranslated regions, and introns (1, 6, 30, 34, 50). Drosophila or cephalopods, bonafide ectotherms, use it extensively to change codons. For humans, even in the absence of fluctuating temperatures, destabilization by editing may have proven useful in specific circumstances. As more data is made available, it will be interesting to see whether ectothermic vertebrates use editing more extensively than their endothermic relatives.
Regulating RNA Editing: A Mechanism for Temperature Dependence?
If RNA editing is indeed used to respond to cold temperatures, what mechanism might underlie this response? The distinct spatial and temporal patterns of editing make it clear that it is a closely regulated process, although the details of its regulation are still emerging. Thus far there are two major components of the RNA editing process that are known to modulate the final output: ADAR, the editing enzyme, and the RNA substrates that it recognizes. Temperature sensitivity would probably involve both components. If the process is used as a slow, adaptive mechanism, then new editing sites could arise from the evolution of either element. For example, if a particular site were cold adaptive, then there would be selective pressure to edit it at higher levels in organisms inhabiting colder environments. Higher editing could be achieved by evolving a new version of ADAR with greater catalytic activity or one that better recognizes the specific site. Loligo, which edits extensively, evolved a unique splice variant of ADAR, which adds an extra RNA binding domain that allows it to edit more sites at higher levels (47). Other cold-adapted organisms might use a similar strategy. An alternative would be to evolve more editable RNA structures. Normally ADAR requires complex tertiary structures within an mRNA to properly recognize and edit a target adenosine (12, 33, 37). It has been shown that these structures do indeed evolve to produce new editing sites. For example, Robert Reenan tracked the evolution of specific RNA structures that drive editing of the synaptotagmin mRNA across the order diptera (51). Clearly, this is an important mechanism for evolving editing sites in general.
An intriguing possibility is that RNA editing can respond rapidly to acute temperature changes, thus acting as a system for acclimation. For this to occur, the regulatory components that modulate the response of editing would also need to change rapidly. There is a growing body of work showing that ADAR’s activity can be regulated on a short time scale through alternative splicing, cellular compartmentalization, and posttranslational modifications (13, 14, 39, 48, 54). It is conceivable, then, that changing ADAR’s activity by these mechanisms could allow editing to respond to diurnal, meteorological, or seasonal cooling. How then is ADAR activity regulated?
Available evidence from several species suggests that the expression level of ADAR is not the primary mechanism for regulating its activity. In mice, overall ADAR expression, measured at both the mRNA and protein levels, is stable over development. In contrast, the degree of editing at most sites increases with development (59). In Drosophila, ADAR expression is high in embryos and pupae, low in larva, and intermediate in adults (22, 48). Editing levels at most sites, on the other hand, generally increase over development (22). ADAR, however, can be modified in other ways to change its activity. In flies, mammals, and squid, alternative splicing produces multiple ADAR isoforms, many of which enhance, reduce, or abolish activity (20, 27, 31, 47, 48). For Drosophila, there are two ADAR splice variants, the 3/4 form being more active than the 3a form (27). Unlike with 3a, 3/4 expression turns on late, starting with pupae and is greatest in adults, when editing levels are at their highest (48). This suggests that alternative splicing of ADAR plays a role in regulating editing. In addition, ADAR can recode its own message in Drosophila and Loligo (47, 48). In the case of Drosophila, a single edit in the enzyme’s catalytic domain produces a novel form that edits at lower efficiencies than the genomically encoded protein (27). In Loligo, there are multiple editing sites; however, their effects on function have not yet been described. Autoediting, then, provides another means of regulation. Thus a possible mechanism for temperature-sensitive editing would involve temperature-sensitive regulation of splicing or autoediting. In Drosophila, we would hypothesize that cold might favor the production of the unedited or the 3/4 splice variants.
Other mechanisms of regulation affect ADAR posttranslationally. First, its localization within the cell is important since most of A-to-I editing is thought to occur co-transcriptionally within the nucleus (32). Accordingly, ADAR needs to be imported into the nucleoplasm and maintained at sufficient levels to edit its substrates. Both ADAR2 and ADAR1 have nuclear import signals; ADAR1 also has a nuclear export signal, so it is able to move between the nucleus and cytoplasm (14, 16). Within the nucleus, ADARs concentrate in the nucleolus (14, 54). Chemically inducing the release of ADAR from the nucleolus increases editing, and transcription of ADAR target mRNAs appears to stimulate the migration of ADAR from the nucleolus to the nucleoplasm (14, 54). Second, covalent modifications to ADAR change its activity. ADAR1, for example, can be sumoylated, wherein the small ubiquitin-like modifier molecule, SUMO, is covalently attached to a lysine. This modification suppresses editing activity (13). Marcucci et al. described a two-part regulatory system targeting ADAR2 (39). They found that the enzyme Pin1 (peptidyl-prolyl isomerase NIMA interacting protein 1), which isomerizes the peptide bond of prolines between the cis and trans configurations, acts on ADAR2, enhancing its activity, stability, and nuclear localization. On the other hand, they found that ADAR2 is also a target for WWP2 (WW domain-containing protein 2) ubiquitin ligase, which promotes its degradation. So sumoylation, WWP2 ubiquitination, and Pin1 isomerization give an organism options for ADAR regulation.
Cold acclimation through RNA editing could also work through rapid structural rearrangements in the target mRNAs if the key secondary structures that promote editing are themselves temperature sensitive. For a cold-adaptive editing site, the necessary secondary structure would be stabilized at lower temperatures allowing ADAR to edit it more efficiently. Thus the RNA itself would both sense temperature and then respond in a way that modulates editing. The concept of an RNA “thermometer” has been described before in bacteria, where the mechanism controls translation efficiency. In various bacteria, heat shock protein mRNAs have specialized hairpin structures in their 5′ untranslated regions. At low temperatures, these hairpins obstruct the Shine-Delgarno sequence, blocking assembly of the ribosomal complex and inhibiting translation. At higher temperatures, the structure destabilizes and translation proceeds. A similar mechanism, albeit less well studied, regulates translation of some cold shock proteins in bacteria (43). In addition, alternative splicing can drive temperature acclimation. In Drosophila melanogaster, the period gene, which is part of the circadian clock, has a weak splice site. At cold temperatures, splicing is enhanced, and the optimal splice variant is more abundant, speeding the clock so that the flies are more active in the middle of the day. When it is warmer, fewer optimally spliced mRNAs are made, shifting the clock so that there is a longer “siesta” during midday and more evening activity (36, 38). Since RNA editing is coupled to splicing in some cases (32), it may be possible for temperature-dependent splicing to make editing temperature dependent as well.
Evidence for Temperature-Dependent RNA Editing
Thus far, we have argued that editing would be expected to make proteins more cold tolerant. In addition, we show that it could, in principle, be used to respond to temperature simply because it can be regulated. The first direct evidence that RNA editing does serve in this capacity comes from a recent study from our laboratory on octopuses (18). Octopuses inhabit a wide range of thermal environments from shallow tropical reefs to deep polar waters. We compared Kv1.1 channels isolated from two species that inhabit these temperature extremes. Pareledone sp. were collected near McMurdo Station in Antarctica, where water temperatures are below 0°C. Octopus vulgaris were collected from a shallow reef on the north coast of Puerto Rico, where temperatures are ~30°C. The sequences of the two genes were nearly identical, as were the electrophysiological properties and temperature sensitivities of the channels they encoded. However, a comparison of the channels’ mRNAs revealed multiple editing sites, some of which were differentially edited between the two species. The level of editing at one site in particular (I321V) was much higher in the Antarctic octopus than in the tropical octopus. This isoleucine to valine change was found to destabilize the channel’s open state, effectively speeding up closing kinetics, a highly temperature-sensitive process. This made sense. Potassium channels must close before the membrane potential can return to rest, allowing the axon to initiate the next action potential. We expected that by speeding up the closing rate the I321V edit would help the Antarctic octopus maintain rapid firing rates in the cold. If this were true, then other cold-water species might make the same change. We went on to collect six more species and found that the editing levels at I321V correlated closely with the water temperatures where the octopuses were collected. I321V was highly edited in arctic and Antarctic species, moderately edited in temperate species, and minimally edited in tropical species. Thus RNA editing appears to play a role in cold adaptation.
Our findings on octopuses raise many questions. Is this phenomena unique to octopus channels or is temperature-sensitive RNA editing widespread? Were the editing differences among the species generated by long-term adaptation, or can editing respond to acute temperature changes, allowing for acclimation? A recent publication has begun to provide answers. Savva et al. (55) found that self-editing levels in the Drosophila ADAR mRNA differed when flies were held at 15, 25, or 35°C for 3 days, with the highest level observed at 15°C and the lowest at 35°C. This demonstrates that temperature-dependent editing does occur outside of cephalopods, and, in Drosophila at least, it can allow for rapid acclimation on the time scale of days. We expect that many new examples of temperature-dependent RNA editing will be forthcoming. How these events tune excitability will lead to exciting new avenues of investigation. It will also be very interesting to see whether other environmental factors can influence this process as well.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Predoctoral Fellow Award FNS-064774A, NIH 2 U54 NS-039405-06, NIH R01 NS-064259, and the National Science Foundation (NSF) IBN-0344070 and HRD-1137725. This work was partially supported by the NSF award HRD-1137725 entitled Puerto Rico Center for Environmental Neuroscience and by the Laura and Arthur Colwin Research Award from the Marine Biological Laboratory in Woods Hole, MA, and was partially conducted at the MBL under the support of this award. Infrastructure support was provided in part by grants from the National Center for Research Resources (2G12 RR-003051) and the National Institute on Minority Health and Health Disparities (8G12 MD-007600).
Footnotes
No conflicts of interest, financial or otherwise, are declared by the author(s).
Author contributions: J.R. and S.C.G. conception and design of research; J.R. edited and revised manuscript; J.R. and S.C.G. approved final version of manuscript; S.C.G. analyzed data; S.C.G. prepared figures; S.C.G. drafted manuscript.
Contributor Information
Sandra C. Garrett, Email: sandracoralgarrett@gmail.com.
Joshua J. C. Rosenthal, Email: joshua.rosenthal@upr.edu.
References
- 1.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basilio C, Wahba AJ, Lengyel P, Speyer JF, Ochoa S. Synthetic polynucleotides and the amino acid code. Proc Natl Acad Sci USA. 1962;48:613–616. doi: 10.1073/pnas.48.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berezovsky IN, Chen WW, Choi PJ, Shakhnovich EI. Entropic stabilization of proteins and its proteomic consequences. PLoS Comput Biol. 2005;1:e47. doi: 10.1371/journal.pcbi.0010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 5.Bjelic S, Brandsdal BO, Aqvist J. Cold adaptation of enzyme reaction rates. Biochemistry. 2008;47:10049–10057. doi: 10.1021/bi801177k. [DOI] [PubMed] [Google Scholar]
- 6.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton KL, Baker PJ, Borges KM, Engel PC, Pasquo A, Rice DW, Robb FT, Scandurra R, Stillman TJ, Yip KS. Insights into thermal stability from a comparison of the glutamate dehydrogenases from Pyrococcus furiosus and Thermococcus litoralis. Eur J Biochem. 1995;229:688–695. doi: 10.1111/j.1432-1033.1995.tb20515.x. [DOI] [PubMed] [Google Scholar]
- 8.Brouns SJ, Wu H, Akerboom J, Turnbull AP, de Vos WM, van der Oost J. Engineering a selectable marker for hyperthermophiles. J Biol Chem. 2005;280:11422–11431. doi: 10.1074/jbc.M413623200. [DOI] [PubMed] [Google Scholar]
- 9.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Stites WE. Replacement of staphylococcal nuclease hydrophobic core residues with those from thermophilic homologues indicates packing is improved in some thermostable proteins. J Mol Biol. 2004;344:271–280. doi: 10.1016/j.jmb.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Colina C, Palavicini JP, Srikumar D, Holmgren M, Rosenthal JJ. Regulation of Na+/K+ ATPase transport velocity by RNA editing. PLoS Biol. 2010;8:e1000540. doi: 10.1371/journal.pbio.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 13.Desterro JM, Keegan LP, Jaffray E, Hay RT, O’Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desterro JM, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Somero GN. Temperature adaptation of cytosolic malate dehydrogenases of limpets (genus Lottia): differences in stability and function due to minor changes in sequence correlate with biogeographic and vertical distributions. J Exp Biol. 2009;212:169–177. doi: 10.1242/jeb.024505. [DOI] [PubMed] [Google Scholar]
- 16.Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farias ST, Bonato MC. Preferred amino acids and thermostability. Genet Mol Res. 2003;2:383–393. [PubMed] [Google Scholar]
- 18.Garrett S, Rosenthal JJ. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science. 2012;335:848–851. doi: 10.1126/science.1212795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georlette D, Blaise V, Collins T, D’Amico S, Gratia E, Hoyoux A, Marx JC, Sonan G, Feller G, Gerday C. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol Rev. 2004;28:25–42. doi: 10.1016/j.femsre.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Gerber A, O’Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez C, Lopez-Rodriguez A, Srikumar D, Rosenthal JJ, Holmgren M. Editing of human KV1.1 channel mRNAs disrupts binding of the N-terminus tip at the intracellular cavity. Nat Commun. 2011;2:436. doi: 10.1038/ncomms1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennig M, Darimont B, Sterner R, Kirschner K, Jansonius JN. 2.0. A structure of indole-3-glycerol phosphate synthase from the hyperthermophile Sulfolobus solfataricus: possible determinants of protein stability. Structure. 1995;3:1295–1306. doi: 10.1016/s0969-2126(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Tan BZ, Shen Y, Tao J, Jiang F, Sung YY, Ng CK, Raida M, Kohr G, Higuchi M, Fatemi-Shariatpanahi H, Harden B, Yue DT, Soong TW. RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca(2)-dependent inactivation. Neuron. 2012;73:304–316. doi: 10.1016/j.neuron.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johns GC, Somero GN. Evolutionary convergence in adaptation of proteins to temperature: A4-lactate dehydrogenases of Pacific damselfishes (Chromis spp.) Mol Biol Evol. 2004;21:314–320. doi: 10.1093/molbev/msh021. [DOI] [PubMed] [Google Scholar]
- 26.Jollivet D, Mary J, Gagniere N, Tanguy A, Fontanillas E, Boutet I, Hourdez S, Segurens B, Weissenbach J, Poch O, Lecompte O. Proteome adaptation to high temperatures in the ectothermic hydrothermal vent Pompeii worm. PLos One. 2012;7:e31150. doi: 10.1371/journal.pone.0031150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keegan LP, Brindle J, Gallo A, Leroy A, Reenan RA, O’Connell MA. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 2005;24:2183–2193. doi: 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keegan LP, McGurk L, Palavicini JP, Brindle J, Paro S, Li X, Rosenthal JJ, O’Connell MA. Functional conservation in human and Drosophila of Metazoan ADAR2 involved in RNA editing: loss of ADAR1 in insects. Nucleic Acids Res. 2011;39:7249–7262. doi: 10.1093/nar/gkr423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempf JG, Jung JY, Ragain C, Sampson NS, Loria JP. Dynamic requirements for a functional protein hinge. J Mol Biol. 2007;368:131–149. doi: 10.1016/j.jmb.2007.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 34.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 36.Low KH, Lim C, Ko HW, Edery I. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron. 2008;60:1054–1067. doi: 10.1016/j.neuron.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O’Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 38.Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 39.Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, Bisso A, Keegan LP, Del Sal G, O’Connell MA. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metpally RP, Reddy BV. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genomics. 2009;10:11. doi: 10.1186/1471-2164-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moiseev VM, Rodina EV, Kurilova SA, Vorobyeva NN, Nazarova TI, Avaeva SM. Substitutions of glycine residues Gly100 and Gly147 in conservative loops decrease rates of conformational rearrangements of Escherichia coli inorganic pyrophosphatase. Biochemistry (Mosc) 2005;70:858–866. doi: 10.1007/s10541-005-0195-z. [DOI] [PubMed] [Google Scholar]
- 42.Murrell-Lagnado RD, Aldrich RW. Interactions of amino terminal domains of Shaker K channels with a pore blocking site studied with synthetic peptides. J Gen Physiol. 1993;102:949–975. doi: 10.1085/jgp.102.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narberhaus F. Translational control of bacterial heat shock and virulence genes by temperature-sensing mRNAs. RNA Biol. 2009;7:84–89. doi: 10.4161/rna.7.1.10501. [DOI] [PubMed] [Google Scholar]
- 44.Nimmich ML, Heidelberg LS, Fisher JL. RNA editing of the GABA(A) receptor alpha3 subunit alters the functional properties of recombinant receptors. Neurosci Res. 2009;63:288–293. doi: 10.1016/j.neures.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 46.Palackal N, Brennan Y, Callen WN, Dupree P, Frey G, Goubet F, Hazlewood GP, Healey S, Kang YE, Kretz KA, Lee E, Tan X, Tomlinson GL, Verruto J, Wong VW, Mathur EJ, Short JM, Robertson DE, Steer BA. An evolutionary route to xylanase process fitness. Protein Sci. 2004;13:494–503. doi: 10.1110/ps.03333504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palavicini JP, O’Connell MA, Rosenthal JJ. An extra double-stranded RNA binding domain confers high activity to a squid RNA editing enzyme. RNA. 2009;15:1208–1218. doi: 10.1261/rna.1471209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patton DE, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- 50.Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, Guo J, Dong Z, Liang Y, Bao L, Wang J. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 51.Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal JJ, Bezanilla F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 2002;34:743–757. doi: 10.1016/s0896-6273(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 53.Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Ryan MY, Maloney R, Reenan RA. Characterization of five RNA editing sites in Shab potassium channels. Channels. 2008;2:202–209. doi: 10.4161/chan.2.3.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savva YA, Jepson JE, Sahin A, Sugden AU, Dorsky JS, Alpert L, Lawrence C, Reenan RA. Auto-regulatory RNA editing fine-tunes mRNA recoding and complex behaviour in Drosophila. Nat Commun. 2012;3:790. doi: 10.1038/ncomms1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin DS, Didonato M, Barondeau DP, Hura GL, Hitomi C, Berglund JA, Getzoff ED, Cary SC, Tainer JA. Superoxide dismutase from the eukaryotic thermophile Alvinella pompejana: structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J Mol Biol. 2009;385:1534–1555. doi: 10.1016/j.jmb.2008.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somero GN. Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:321–333. doi: 10.1016/j.cbpc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Spiller B, Gershenson A, Arnold FH, Stevens RC. A structural view of evolutionary divergence. Proc Natl Acad Sci USA. 1999;96:12305–12310. doi: 10.1073/pnas.96.22.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang GZ, Lercher MJ. Amino acid composition in endothermic vertebrates is biased in the same direction as in thermophilic prokaryotes. BMC Evol Biol. 2010;10:263. doi: 10.1186/1471-2148-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yip KS, Stillman TJ, Britton KL, Artymiuk PJ, Baker PJ, Sedelnikova SE, Engel PC, Pasquo A, Chiaraluce R, Consalvi V, et al. The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures. Structure. 1995;3:1147–1158. doi: 10.1016/s0969-2126(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 62.Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu BY, Zhou NE, Kay CM, Hodges RS. Packing and hydrophobicity effects on protein folding and stability: effects of beta-branched amino acids, valine and isoleucine, on the formation and stability of two-stranded alpha-helical coiled coils/leucine zippers. Protein Sci. 1993;2:383–394. doi: 10.1002/pro.5560020310. [DOI] [PMC free article] [PubMed] [Google Scholar]