Abstract
Members of the human KIR class I MHC receptor gene family contain multiple promoters that determine the variegated expression of KIR on NK cells. In order to identify novel genetic alterations associated with decreased KIR expression, a group of donors was characterized for KIR gene content, transcripts, and protein expression. An individual with a single copy of the KIR2DL1 gene but a very low level of gene expression was identified. The low expression phenotype was associated with a SNP that created a binding site for the inhibitory ZEB1 transcription factor adjacent to a c-Myc binding site previously implicated in distal promoter activity. Individuals possessing this SNP had a substantial decrease in distal KIR2DL1 transcripts initiating from a novel intermediate promoter located 230 bp upstream of the proximal promoter start site. Surprisingly, there was no decrease in transcription from the KIR2DL1 proximal promoter. Reduced intermediate promoter activity revealed the existence of alternatively spliced KIR2DL1 transcripts containing premature termination codons that initiated from the proximal KIR2DL1 promoter. Altogether, these results indicate that distal transcripts are necessary for KIR2DL1 protein expression and are required for proper processing of sense transcripts from the bidirectional proximal promoter.
Keywords: human, NK cells, KIR, transcription, ZEB1
Introduction
Natural Killer (NK) cells are lymphocytes of the innate immune system that play an important role in the elimination of neoplastic or virus-infected cells1. NK cell cytotoxic activity is controlled in part by cell surface receptors for class I MHC that survey potential target cells for aberrant class I expression2. Human NK cells use the killer cell immunoglobulin-like receptor (KIR) family3 to interrogate class I status, whereas murine NK cells use members of the C-type lectin-related Ly49 family4, 5. KIR and Ly49 genes are expressed in a probabilistic fashion, generating subsets of NK cells with distinct class I MHC recognition properties6, 7. The selectively expressed human KIR gene family provides an interesting model system for the study of stochastic activation of gene expression. The proximal promoter of most KIR genes is kept in a silent state by promoter methylation8, 9. An AML/RUNX transcription factor binding site in the proximal promoter region has been suggested to play a role in the demethylation of the proximal promoter, since loss of this site in the non-transcribed KIR2DL5*002 allele is associated with non-expression, even though loss of the AML/RUNX site does not abrogate proximal promoter activity in luciferase reporter assays10, 11. Furthermore, treatment of NK cells with the demethylating agent 5-azacytidine, leads to KIR2DL5*002 transcription, supporting a role for AML/RUNX in catalyzing promoter demethylation. Studies of a distal KIR promoter have indicated a role for distal transcripts in gene activation, since distal transcription precedes gene expression, and increased distal transcripts are associated with a higher frequency of gene expression12, 13. A c-Myc binding site in the distal promoter was shown to be important for promoter activity, and over-expression of c-Myc in developing NK cells led to increased distal transcript levels and an increased frequency of NK cells expressing KIR. A model of KIR gene activation has been proposed in which the distal promoter generates transcripts that traverse the proximal promoter region, leading to an opening of this region and allowing access of key transcription factors involved in promoter demethylation, such as AML/RUNX14.
The variegated expression of Ly49 genes is controlled by a bidirectional promoter that acts as a probabilistic switch15. The generation of forward transcripts from this upstream element (Pro1) in immature NK cells leads to activation of the downstream proximal promoter. The expression of a Ly49 gene from the proximal promoter is dependent on distal transcription, since Pro1 deletion eliminates Ly49 transcription16. The probabilistic activation of KIR genes has been linked to the bi-directional nature of the KIR proximal promoter17. Variation in the relative strength of transcription factor binding sites that promote either sense or antisense transcription from the proximal promoter in specific KIR genes or alleles results in bi-directional promoters with differing probabilities of generating a forward transcript18. KIR genes or alleles with a high ratio of sense to antisense proximal promoter activity are expressed by a greater percentage of NK cells than KIR with a low sense to antisense ratio. The KIR antisense transcript has been associated with the production of a 28 base piwi-RNA that may lead to re-silencing of KIR loci that produce the antisense transcript19. Although numerous KIR alleles have been identified that are non-expressed due to the presence of stop codons in the coding region, only one gene (KIR2DL5)10 has been identified that contains non-transcribed alleles. In the current study, we sought to determine if there were additional non-transcribed KIR alleles, to obtain further information with regard to the genomic regions controlling gene expression.
Results
Identification of a weakly expressed KIR2DL1 allele
In order to identify novel genetic alterations associated with a lack of KIR transcription, healthy donors were screened for individuals that possessed but did not show significant expression of KIR genes. A group of 182 National Marrow Donor Program (NMDP) donors were characterized for KIR gene transcription by qRT-PCR, KIR gene content, and FACS analysis of KIR surface expression. An individual was discovered that possessed the KIR2DL1 gene but had barely detectable gene expression by either FACS or qRT-PCR (Figure 1).
Figure 1.
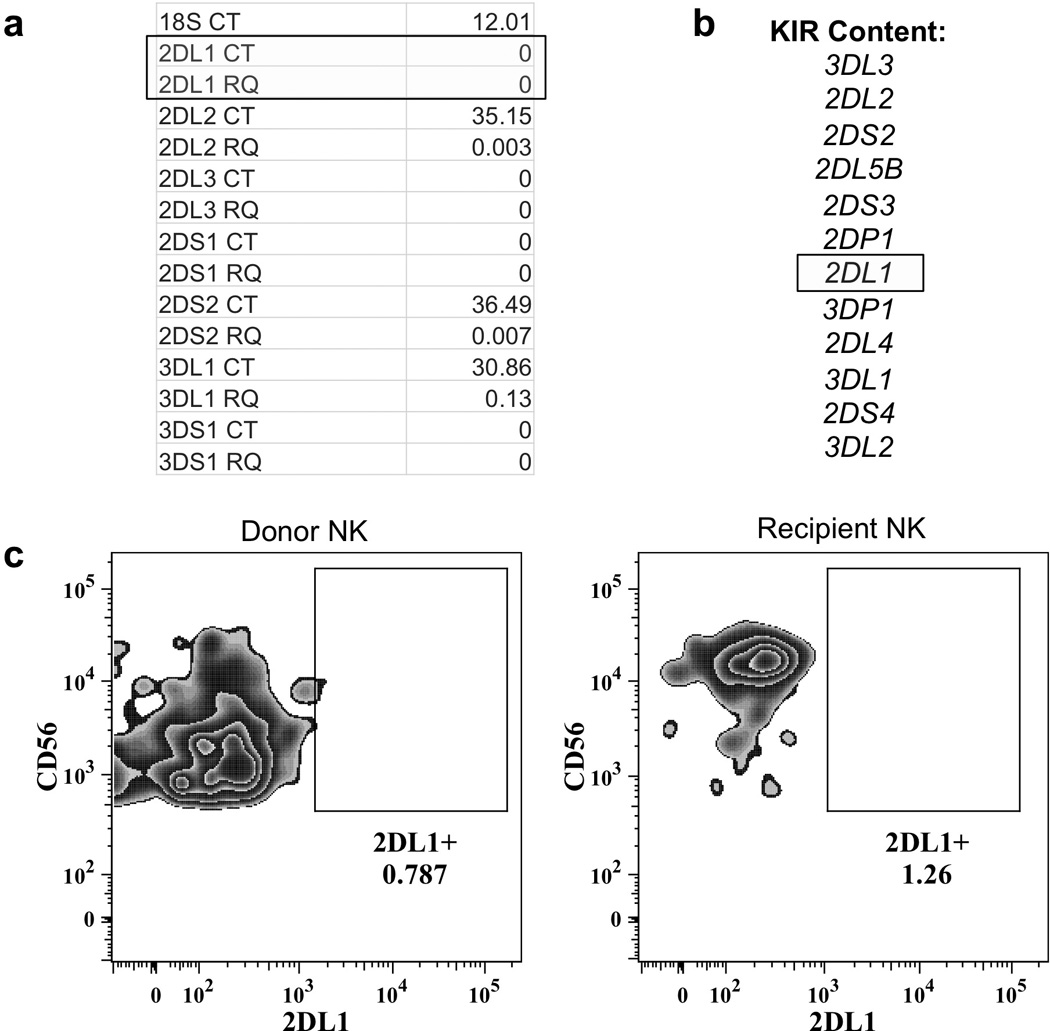
Identification of a poorly expressed KIR2DL1 allele. ( a ) qRT-PCR performed on NK cell RNA from a NMDP donor. ( b ) KIR gene content typing for this individual. ( c ) FACS analysis performed with a KIR2DL1-specific antibody on donor PBMC in the left panel, and PBMC from the corresponding hematopoietic transplant recipient in the right panel, demonstrating an identical pattern of KIR2DL1 expression in reconstituting donor cells.
Identification of SNPs specific to the poorly expressed KIR2DL1 allele
Complete sequencing of the KIR2DL1 gene from the individual with low KIR2DL1 expression revealed the presence of a single allele containing an intact coding region. Analysis of the three known promoter regions as well as the complete 2 kb intergenic region preceding the first exon revealed a cluster of 3 single nucleotide polymorphisms (SNPs) in the distal promoter region approximately 1 kb upstream from the start of KIR2DL1 translation (Figure 2). These 3 SNPs, together with the presence of an exon 4 segment derived from the KIR2DL1*002 allele constituted the only sequence differences between the KIR2DL1 gene in this donor and the previously reported complete sequence of a KIR2DL1*00401 allele (GenBank accession GU182347). The 3 SNPs present in the novel KIR2DL1 allele match the sequence normally found in the distal promoter region of the non-transcribed KIR2DL5*002 allele as well as the KIR3DP1 pseudogene (Figure 2). However, the well-characterized KIR2DL5*002 SNP that disrupts the proximal promoter AML/RUNX site was not present, indicating that the distal promoter variant was able to inhibit KIR expression by a novel mechanism. This also raises the possibility that the KIR2DL1 SNPs identified here could be involved in the silencing of KIR2DL5*002.
Figure 2.

A non-transcribed KIR2DL1 allele is generated by recombination between KIR2DL1*004 and KIR3DP1. The sequence of the region from -1353 to -1093 relative to the translational start of the KIR2DL1*004 gene (2DL1*004) is shown, and only nucleotide differences are shown for the novel KIR2DL1 allele (2DL1-new) and the non-transcribed KIR3DP1 gene (3DP1). The KIR3DP1-derived region of the novel KIR2DL1 allele is shown in bold, and the underlined flanking sequences indicate the regions where recombination may have occurred to generate the novel KIR2DL1 allele. The boxed sequences represent the consensus ZEB1 binding half sites in opposing orientations, followed by a Myc/USF site previously shown to play a role in distal promoter activity. The upstream ZEB1 site is created in the 2DL1-new and 3DP1 sequences due to the A to G SNP.
The KIR2DL1 distal SNP creates a ZEB1 binding site
Examination of the distal SNPs associated with non-expression of KIR2DL1 revealed that the SNP at position -1206 created a binding site for the Zinc finger E-box-binding homeobox 1 (ZEB1) protein, a transcription factor that has been shown to inhibit IL2 gene expression20. ZEB1 requires two E-box motifs (CACCT) for efficient binding to DNA21, and the polymorphism generates a second E-box motif in close proximity to an E-box motif adjacent to the c-Myc/USF binding site (Figure 2) that has been previously implicated in the activation of KIR genes13. EMSA analysis indicated that ZEB1 binds to the E-box pair composed of the novel E-box generated by the polymorphism and the site located next to the c-Myc site (Figure 3). The generation of this novel ZEB1-binding site adjacent to the Myc site shown to enhance distal transcription suggested that the presence of the ZEB1 SNP might decrease activity of the KIR2DL1 distal promoter, reducing the probability of proximal promoter activation. Interestingly, all known KIR2DL5 alleles contain the SNP that generates the ZEB1-binding site, which may account for the low frequency of expression observed for KIR2DL5A*00122. Some KIR2DP1 alleles also contain this SNP, however the complete absence of KIR2DP1 transcripts precludes any observable effect of the ZEB1 SNP on the transcription of this gene.
Figure 3.
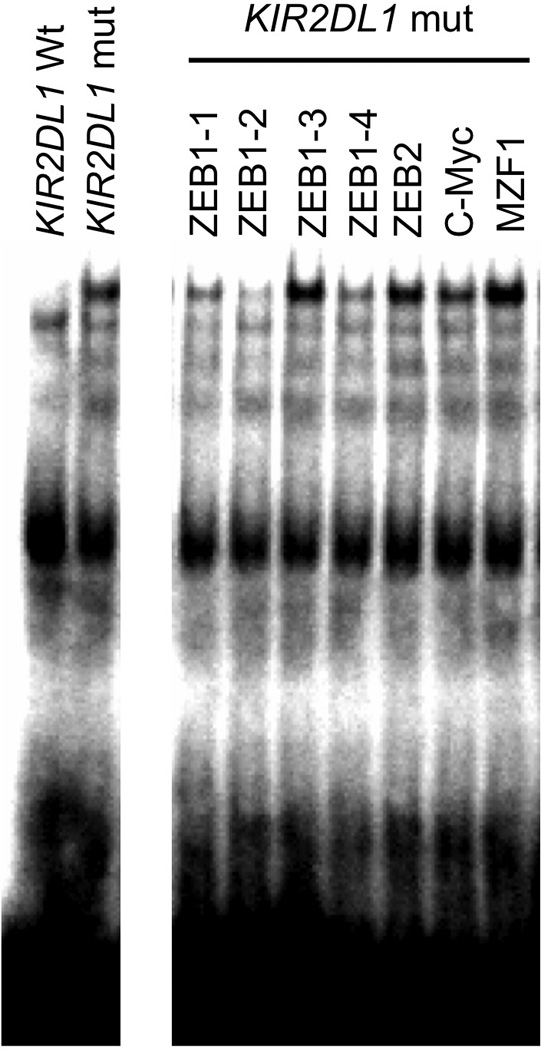
The KIR2DL1 SNP creates a ZEB-1 binding site. An oligonucleotide probe containing the wild-type (wt) or mutant (mut) distal promoter region was combined with nuclear extract from YT cells. The left panel shows the appearance of a novel band associated with the mutant SNP. The right panel shows the ability of several commercial anti-ZEB antibodies (ZEB1-1 to ZEB1-4, ZEB2), together with anti-C-Myc or MZF-1 controls to inhibit formation of the novel complex.
Association of the ZEB1 SNP with decreased KIR2DL1 expression
PCR screening of archived DNA from an additional 376 donors from the NCI Frederick Research Donor Program, resulted in the identification of 6 additional individuals (1.6 %) possessing the ZEB1-associated SNP in KIR2DL1, all of which were associated with a KIR2DL1*004 allele. The complete sequence of the novel KIR2DL1 allele can be found in GenBank under accession number KJ913660. Interestingly, none of these donors had the accompanying substitution of a KIR2DL1*002-like exon 4 found in the original KIR2DL1 low-expression variant. Three individuals with the novel SNP were available for additional characterization, and they were evaluated for cell surface expression of KIR2DL1 as compared to normal donors that possessed either 0, 1, or 2 copies of the wild-type KIR2DL1 gene. FACS analysis of these donors demonstrated low KIR2DL1 surface expression (2.6% of NK cells) in a donor containing a single copy of the mutated KIR2DL1 gene, confirming an association of the ZEB1 SNP with poor expression (Figure 4). The presence of both wild-type and mutant KIR2DL1 alleles produced normal levels of KIR2DL1 expression, indicating the absence of any trans effects of the ZEB1 SNP. The observed higher than expected frequency of KIR2DL1 expression (25.8%) in the individual with only one wild-type allele can be accounted for by the presence of KIR2DS5, as the anti-KIR2DL1 antibody used for detection has been shown to also recognize KIR2DS523. The KIR2DS5 gene was not present in any of the other donors tested. In addition, analysis of an individual possessing a mutant copy of KIR2DL1 together with the KIR2DS3 gene revealed moderate low intensity expression (11.1%), suggesting weak cross-reactivity of the anti-KIR2DL1 antibody with KIR2DS3 in this individual.
Figure 4.
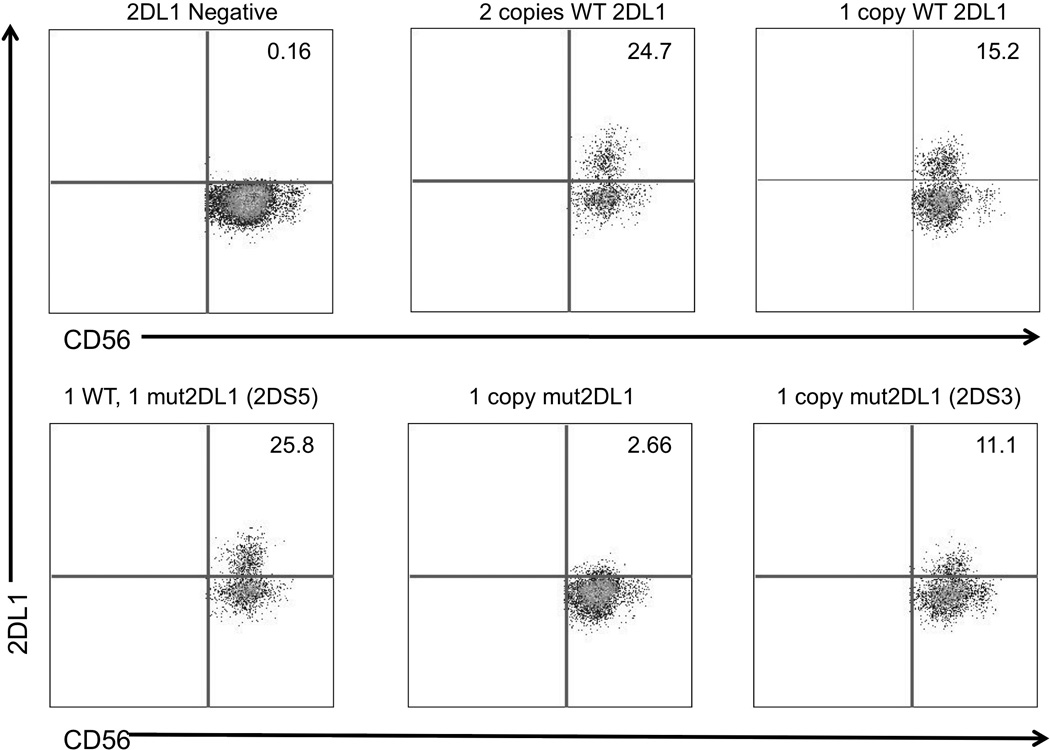
The KIR2DL1 SNP is associated with weak expression. Donors were screened by PCR with primers specific for the ZEB1 SNP. The upper panel shows three donors with 0, 1, or 2 copies of the wild type KIR2DL1 gene (WT 2DL1) stained with an NK marker (CD56) on the horizontal axis, and a KIR2DL1-specific antibody on the vertical axis. The number shown in the upper right quadrant represents the percentage of NK cells reacting with the 143211 anti-KIR2DL1 antibody. The lower panel shows three donors found to possess the ZEB1 SNP. Lack of expression is evident in the individual containing only a mutant KIR2DL1 gene (1 copy mut 2DL1). The presence of either KIR2DS5 or KIR2DS3 genes in two of the donors is also indicated, as these KIR gene products have been shown to react with the 143211 anti-KIR2DL1 antibody23.
The ZEB1 SNP decreases distal transcripts
In order to determine if the ZEB1 SNP affected distal promoter activity, RT-PCR was performed with primers specific for the KIR2DL1 distal transcript. As shown in Figure 5a, there was a substantial reduction in distal transcription in individuals possessing the ZEB1-associated SNP, consistent with an inhibitory effect of ZEB1 binding in the upstream region.
Figure 5.
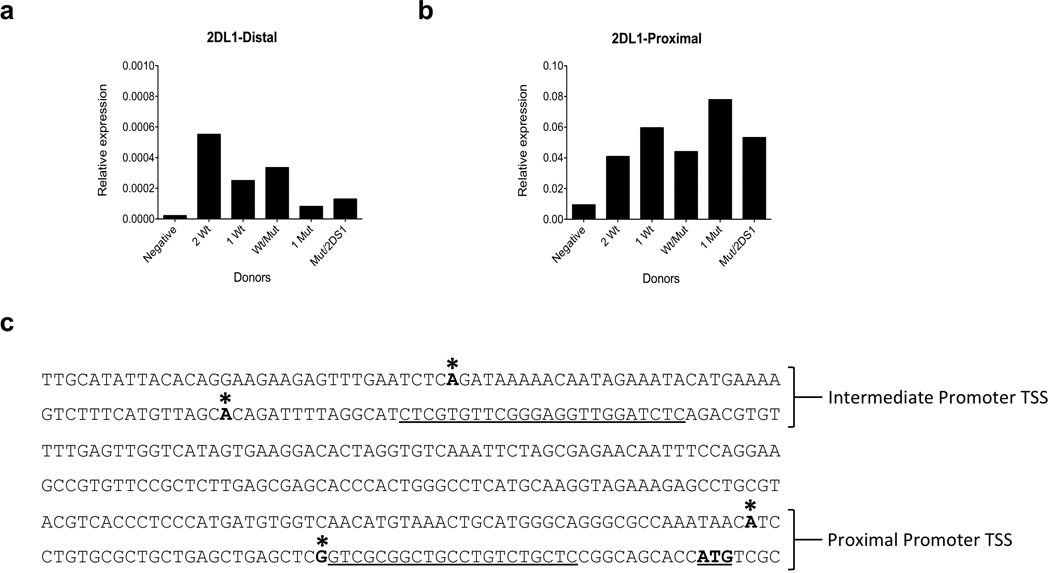
The KIR2DL1 SNP is associated with decreased distal transcript expression. Purified NK cell RNA from purified donor NK cells was subjected to qRT-PCR with KIR2DL1-specific primers detecting distal (a) or proximal (b) transcripts. Expression relative to actin is shown on the Y-axis, and KIR2DL1/KIR2DS1 genotype of individual donors is listed on the x-axis. Donors were: lacking both KIR2DL1/KIR2DS1 (negative); two copies of wild-type KIR2DL1 (2Wt); one copy of Wt KIR2DL1 (1Wt); one Wt, one ZEB1-mutant KIR2DL1 copy (Wt/Mut); one copy of a mutated KIR2DL1 (1 Mut); and one copy of mutated KIR2DL1 and one copy of KIR2DS1 (Mut/2DS1). (c) Transcription start sites (TSS) mapped by 5’ RACE are shown for the 365 bp region immediately 5’ of the KIR2DL1 start codon ( ATG). TSS are indicated by an asterisk. The underlined sequences represent forward PCR primers used for the detection of distal and proximal transcripts in a and b .
Surprisingly, analysis of proximal promoter transcripts showed no decrease in the level of proximal KIR2DL1 promoter activity (Figure 5b), even in an individual that possessed only a single copy of the KIR2DL1 gene containing the ZEB1 SNP. The proximal transcript levels actually increased in the ZEB1 mutant, possibly due to the loss of competition from the upstream promoter. Furthermore, the levels of distal transcript paralleled the level of protein expression observed in Figure 4, whereas proximal transcript did not. Since previous studies had demonstrated distal KIR transcripts initiating approximately 800 bp upstream of the KIR start codon were not translatable, 5’ RACE was performed with primers just upstream of the proximal promoter to look for additional distal transcription start sites. Two novel distal transcript start sites 230 and 273 nucleotides upstream from the major proximal promoter start site were identified in RNA from individuals with wild type KIR2DL1 alleles, indicating the presence of an intermediate promoter (Figure 5c). The lack of correlation between proximal promoter activity and protein expression suggests that transcripts initiated from the novel intermediate promoter may be responsible for KIR2DL1 expression by mature NK cells. To investigate whether or not these novel transcripts are capable of expressing protein, a full-length cDNA originating from the intermediate promoter (GenBank accession number KJ699235) was cloned into an expression vector and transfected into the human 239T cell line. Expression of KIR2DL1 from the intermediate transcript was comparable to expression from the proximal transcript, indicating that the additional 230 base 5’ untranslated region of the intermediate transcript did not inhibit expression, consistent with the association of the novel promoter with KIR expression by mature NK cells. A KIR3DL1 cDNA originating from the intermediate promoter was also tested and found to be capable of producing KIR3DL1 expression (data not shown).
Proximal KIR2DL1 transcripts contain alternative exons
The lack of correlation between KIR2DL1 expression and proximal transcription was perplexing, since the major start site of KIR2DL1 transcription in NK cells has been mapped to the proximal promoter12, 24, 25. However, RT-PCR analysis of individuals with reduced KIR2DL1 distal transcript revealed the presence of two alternative exons containing stop codons (Figure 6), suggesting that proximal promoter transcripts might be alternatively spliced and potentially incapable of producing translatable KIR2DL1 mRNA.
Figure 6.
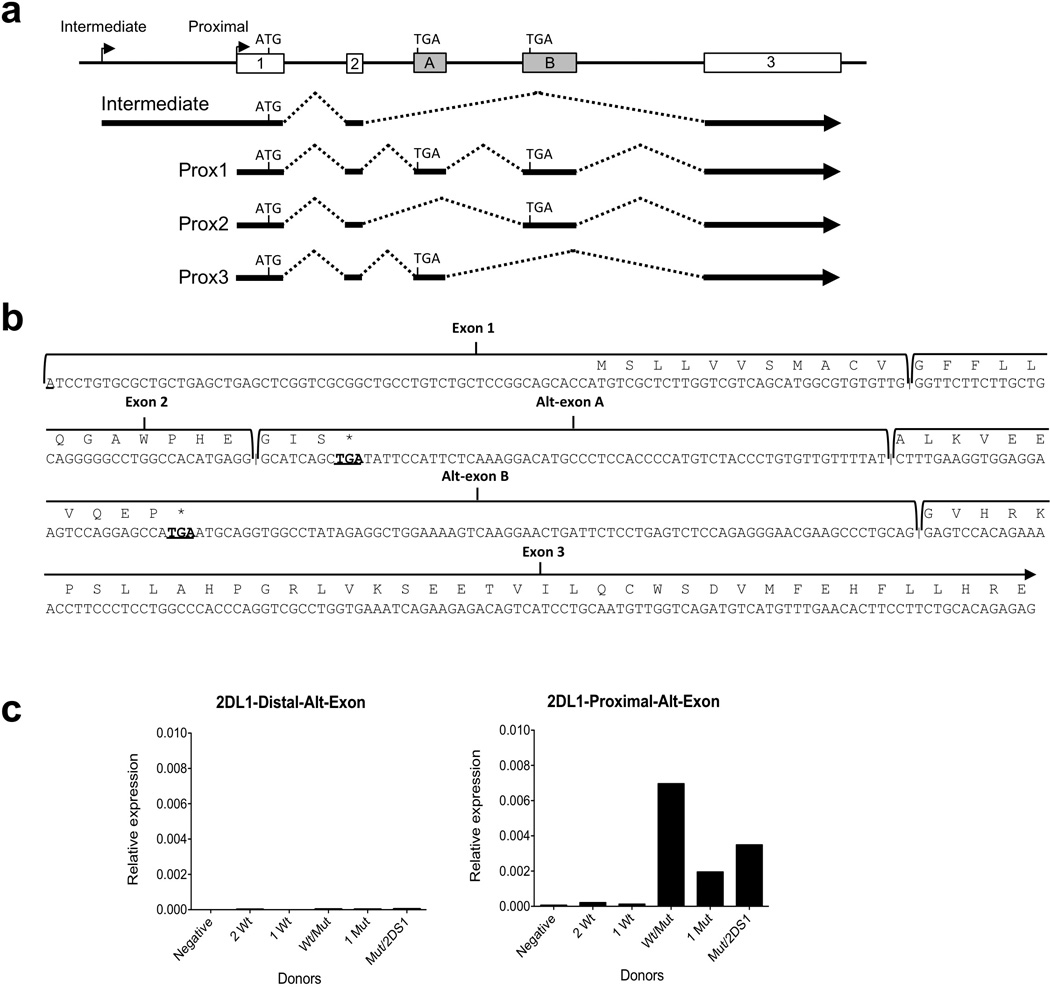
KIR2DL1 proximal promoter transcripts contain additional exons that disrupt the KIR2DL1 open reading frame. (a) A schematic of the KIR2DL1 gene structure is shown. Numbered rectangles represent KIR2DL1-coding exons with the start codon in exon 1 labeled ‘ATG’, whereas the shaded rectangles marked ‘A’ and ‘B’, represent alternative exons containing stop codons indicated by ‘TGA’. Intermediate and proximal transcript initiation sites are marked by vertical lines with arrows attached. Lines below the KIR2DL1 gene diagram represent cDNA clones isolated from purified NK cell RNA obtained from an individual with a single copy of the KIR2DL1 gene containing the ZEB1 SNP. Three distinct isoforms of the KIR2DL1 proximal transcript were identified (Prox1-3), each containing different combinations of the alternative exons. (b) The nucleotide sequence of the Prox1 transcript is shown, with the KIR2DL1 open reading frame indicated by the amino acid sequence listed above. Exon boundaries are marked by vertical lines, and each exon is defined by labeled brackets. The stop codons in alternative exons A and B (Alt-exon A/B) are underlined in bold. (c) qRT-PCR of alternative exon-containing transcripts using either a distal (2DL1-Distal-Alt-Exon) or proximal (2DL1-proximal-Alt-Exon) KIR2DL1 forward primer together with an alternate exon B reverse primer. Expression relative to actin is shown on the Y-axis, and KIR2DL1/KIR2DS1 genotype of individual donors is listed on the x-axis as described in Figure 5b.
To further investigate this possibility, qRT-PCR was performed with antisense primers specific for the alternate exons together with forward primers from either the intermediate or proximal transcripts. Efficient amplification of alternative exon-containing transcripts was only observed with the forward primer from the proximal transcript, suggesting that the alternative exons are specific to the proximal transcript (Figure 6c). However, the alternative exon-containing transcripts were not as abundant in NK cells that contained only wild-type KIR2DL1 genes, suggesting that the presence of distal transcript either inhibits alternative splicing or ZEB1 binding to the distal promoter region affects alternative splicing of the proximal transcript. 5’ RACE performed using primers from the alternative exons generated 5’ ends mapping to the proximal promoter region. No start sites upstream of the proximal promoter were detected with alternative exon primers (data not shown).
Discussion
Studies of the Pro1 distal bidirectional promoter present in the murine Ly49 genes have indicated that forward transcription from the distal promoter is required for activation of the proximal Ly49 promoter (Pro2) that is responsible for Ly49 expression in mature NK cells. This was demonstrated by both naturally occurring deletions of the distal promoter as well as a direct deletion of the Pro1 element in Ly49a transgenic mice26. The distal KIR promoter has been shown to be transcriptionally active in NK precursors that lack KIR expression, suggesting that it may also play a role in the initial opening of the proximal KIR promoter analogous to the murine Ly49 Pro1 promoter13. The weakly expressed KIR2DL1 allele reported herein has revealed the presence of a novel KIR intermediate promoter 230 bp upstream of the proximal promoter, and transcription from this novel promoter is associated with protein expression. For the purposes of this discussion, we will refer to this new intermediate promoter as Pro-I. The bi-directional proximal promoter will be referred to as the probabilistic switch, or Pro-S. The previously described distal promoter will be referred to as Pro-D. The presence of a ZEB1 binding site in Pro-D was correlated with a low level of KIR2DL1 distal transcription in several individuals. The ZEB1 SNP was also associated with decreased in vitro KIR2DL1 Pro-I activity (data not shown) and decreased Pro-I transcripts in NK cells, indicating an enhancer function of the Pro-D element for Pro-I transcription. Pro-S activity was unchanged, indicating an association of Pro-I transcription with KIR2DL1 protein expression. The results obtained in this study necessitate a revision of the model of probabilistic KIR gene activation. The previous observation that CD56dim, KIR-ve NK cells contain antisense transcripts from Pro-S indicated that these cells represented the phase of NK development where the probabilistic Pro-S element was active, however these cells also contain forward Pro-S transcripts, but no KIR protein expression. The discovery of a novel promoter associated with KIR expression indicates that Pro-S transcription alone is not sufficient for protein expression, and it is Pro-I that drives expression in mature NK cells either directly, or by facilitating proper splicing of proximal transcripts. This also reveals another parallel with the murine Ly49 system: forward transcripts from bidirectional promoters active in immature NK cells are non-translatable. Therefore, the process of KIR gene activation should proceed through three stages as defined by the activity of the Pro-D, Pro-S, and Pro-I elements (Figure 7). NK precursors have an active Pro-D element that is associated with the activation of the Pro-S region in developing NK cells. NK cells undergoing the CD56bright to CD56dim transition represent the probabilistic transcription phase marked by Pro-S activity and the presence of either antisense transcripts or forward non-translatable transcript. Mature KIR-positive NK cells should exhibit Pro-I activity driving the expression of KIR protein or preventing alternative splicing of proximal transcripts. The inability of previous studies of KIR transcript initiation sites to detect Pro-I transcripts may be due to a high concentration of proximal Pro-S transcripts masking the presence of upstream start sites. The PCR and subsequent cloning steps performed in 5’-RACE create a bias toward smaller products, making it difficult to identify start sites that are further upstream. There is also a bias toward more abundant transcripts, and Pro-S transcripts are approximately 5-fold higher than the Pro-I transcripts detected in purified peripheral NK cell populations12. As a result, the Pro-I transcription start site could only be mapped by 5’ RACE using primers upstream of the Pro-S start site.
Figure 7.
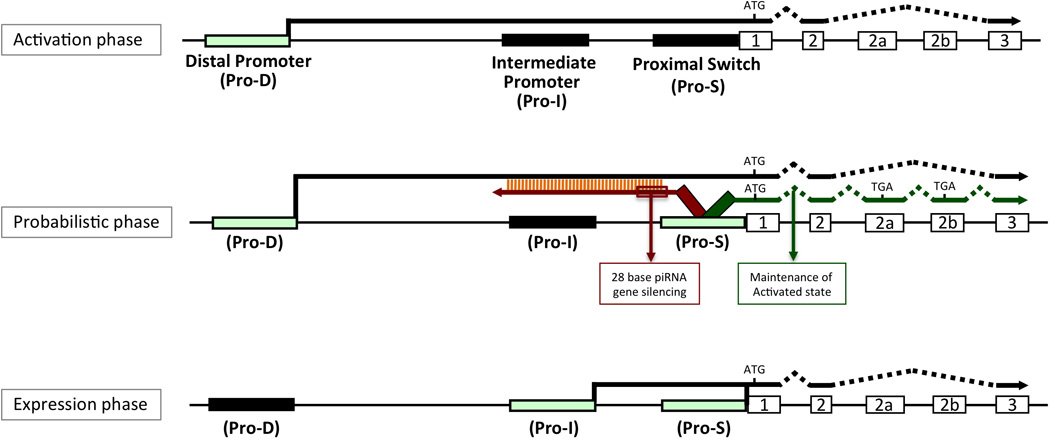
Three promoters define distinct stages of KIR activation. Schematic of the 5’ control region and first 3 KIR-coding exons representing three phases of KIR gene transcription (Activation phase; Probabilistic phase; Expression phase) are shown. Promoters are indicated by filled rectangles, and light green rectangles highlight promoters that define the proposed phases of expression. The site of initiation of KIR protein translation is indicated by a vertical line labeled “ATG”, and the location of premature stop codons in alternative exons found in forward transcripts from Pro-S are indicated by “TGA”. Black lines ending in arrows represent sense coding transcripts, the red and green lines indicate antisense and sense transcripts from Pro-S respectively. The red and green colored boxes indicate the proposed effects of the antisense and sense transcripts on gene activity.
The proposed ability of Pro-I to drive KIR protein expression in mature NK cells and the non-translatable Pro-S transcripts associated with low Pro-I activity suggests that SNPs affecting the level of KIR expression should map to the Pro-I element and the upstream enhancer Pro-D as described in this study, whereas SNPs affecting the percentage of NK cells expressing a given KIR should map to the Pro-S element, as previously demonstrated for the KIR3DL1 gene18. To test the prediction that Pro-I SNPs might affect the intensity of KIR3DL1 expression, the KIR3DL1 Pro-I region was compared amongst the KIR3DL1 alleles represented in GenBank. A single bp deletion was found in the KIR3DL1*005 and KIR3DL1*004 alleles but not in any of the other KIR3DL1 alleles or KIR3DS1. This deletion affects a consensus AP-1 transcription factor binding site, and would be predicted to result in decreased Pro-I promoter activity, consistent with the reported low expression phenotype of KIR3DL1*005.
The analysis of a KIR2DL1 allele associated with low expression revealed a novel KIR variant, but in addition, has resulted in a significant change in our understanding of KIR gene regulation. The promoter previously considered to be responsible for KIR protein expression (Pro-S), may function primarily as a probabilistic switch that governs the frequency of gene activation in developing NK cells. This novel paradigm of KIR gene regulation necessitates a shift away from analysis of the proximal KIR promoter when considering polymorphisms that affect the intensity of KIR expression. Mature NK cells require KIR2DL1 Pro-I transcripts to express the protein, Pro-I transcripts have been identified for the KIR3DL1 gene (data not shown), and the Pro-I element is conserved among all KIR genes except for KIR3DL3, a gene that is not expressed by mature circulating NK cells. This suggests that most KIR genes use Pro-I for the expression of KIR protein in mature NK cells. Of particular interest is the divergence of the KIR2DL3/2DL2/2DS2 Pro-I elements that only share 84% homology with the 3’ half of the KIR2DL1 Pro-I element and possess no significant homology within the 5’ half of the promoter. This may explain the previous observation that NK cells derived from hematopoietic progenitor cells generally acquire the C1-specific inhibitory KIR2DL2/3 receptors at earlier time points than the C2-specific KIR2DL127. Comparison of Pro-S elements of the KIR genes has revealed 5 KIR2DL2/3-specific SNPs, however there is no significant difference in KIR2DL2/3 Pro-S activity as compared to KIR3DL1*002 Pro-S activity18, 28. It will therefore be of interest to compare Pro-I transcripts and Pro-I activity of these genes to see if a correlation exists between divergent Pro-I structures and distinct expression patterns.
In conclusion, the discovery of a novel KIR promoter associated with protein expression by mature NK cells mandates a re-examination of previous data correlating proximal promoter polymorphisms with differences in the timing, tissue specificity, or level of transcription of individual KIR genes or alleles. This paradigm shift promises to provide insight into gene/allele-specific differences that have not been adequately explained by study of the proximal KIR promoter.
Materials and methods
Cell lines
YT cells were cultured in RPMI 1640 media containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin, sodium pyruvate, and L-glutamine. HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin (P/S), sodium pyruvate, and L-glutamine.
Donors and NK Cell Isolation
Collection of DNA and blood samples from 162 donor/recipient pairs from unrelated donor hematopoietic cell transplants performed to treat AML was facilitated by the National Marrow Donor Program (NMDP, Minneapolis, MN) Research Sample Repository. Samples were collected after informed consent and approval from the NMDP and University of Minnesota Institutional Review Boards were obtained in accordance with the Declaration of Helsinki. Healthy volunteers were recruited through the NCI-Frederick Research Donor Program (http://www.ncifcrf.gov/rdp/). The KIR genotype of each donor was determined as previously described29. NK cells were separated from the peripheral blood of healthy donors by Histopaque® (Sigma-Aldrich, Saint Louis, MO, USA) gradient centrifugation using the RosetteSep™ Human NK Cell Enrichment Cocktail (STEMCELL Technologies, Vancouver BC, CA).
Flow Cytometric Analysis of KIR Expression on Peripheral Blood NK Cells
The proportion of NK cells expressing KIR2DL1 was assessed in whole blood by three-color flow cytometry. NK cells were identified using anti-human CD56-APC (Beckman Coulter, Pasadena, CA, USA). T cells were labeled with anti-human CD3-Pacific Blue (eBioscience, San Diego, CA, USA). KIR2DL1 expressing cells were identified with the clone 143211-FITC KIR2DL1-specific antibody (R&D Systems, Minneapolis, MN, USA). Briefly, 200 µl of EDTA-treated blood was incubated with a cocktail of the 3 mAbs for 30 min on ice. Erythrocytes were lysed by the addition of 3 ml of RBC lysis buffer (150 mM NH4Cl, 10 mM NaHCO3, 1 mM EDTA, pH 7.4), and the remaining cells were analyzed with a FACSort flow cytometer (Becton & Dickinson, San Jose, CA). Events (25,000) were collected in the lymphocyte gate and analyzed. The percentage of CD3-CD56+ NK cells expressing KIR2DL1 was determined.
PCR screening and sequencing of KIR2DL1 genes
PCR screening of donors for the ZEB1 SNP was performed with a KIR2DL1-specific forward primer (5’-GGGTTTACAGTTCTTTATTGAACCG-3’) together with a reverse primer recognizing the KIR3DP1/KIR2DL5B sequence found in the low expresser variant (5’-CTCAATAAATAATATTAAACCAATTGGTTAC-3’). To obtain the complete sequence of the KIR2DL1 alleles present in each donor, 50 ng of leukocyte DNA obtained from donor blood was subjected to PCR with a series of KIR2DL1/S1-specific primers to produce overlapping ~ 1 kb amplicons spanning the complete gene from the polyA site of the preceding KIR2DP1 gene to the polyA site of KIR2DL1. PCR fragments were cloned into the pCR2.1-topo vector (Invitrogen, Carlsbad, CA, USA). A minimum of six clones of each fragment was sequenced for each KIR2DL1/S1 allele present.
RT-PCR of KIR transcripts
Total RNA was purified from 1×107 donor NK cells with the RNeasy kit (Qiagen, Valencia, CA, USA) and cDNA synthesis was carried out using Random Hexamer primer, Taqman Reverse Transcription Reagents kit (Applied Biosystems Foster City, CA, USA) according to the manufacturer's instructions. The initial screening of donors for transcripts of all transcribed KIR genes was performed using primer sets previously described30. The primers used in the KIR2DL1-specific qRT-PCR assay were: KIR2DL1-Distal forward primer (5’-CTCGTGTTCGGGAGGTTGGATCTC-3’); KIR2DL1-Proximal forward primer (5’-GTCGCGGCTGCCTGTCTGCTC-3’); KIR2DL1-Exon 3 reverse primer (5’-CTCTCTGTGCAGAAGGAAGTGTT-3’); KIR2DL1-AltExon B reverse primer (5’-CCAGCCTCTATAGGCCACCTGC-3’); Actin-beta forward primer (5’-CCTGGCACCCAGCACAAT-3’); Actin-beta reverse primer (5’- GGGCCGGACTCGTCATACT-3’). Reactions were carried out using FastStart SYBR Green Master kit (Roche Diagnostics, Indianapolis, IN, USA) on the 7300 Real-Time PCR System (Applied Biosystems). The qRT-PCR was performed in duplicate and was repeated in at least three separate experiments using the following conditions. Reaction mixtures contained 12.5 µl of SYBR Green master mix, 2 pmoles each of forward and reverse primers, and 10ng cDNA. Thermocycler conditions included initial denaturation at 50°C and 95°C (10 min each), followed by 40 cycles at 95°C (15 s) and 60°C (1 min). Melting curve analyses were performed to verify the amplification specificity. Relative quantification of gene expression was performed according to the ΔΔ-CT method using StepOne Software 2.0 (Applied Biosystems). All PCR products were cloned into pCR2.1-topo (Invitrogen) and sequenced in order to verify the identity of the transcript and the KIR gene responsible for the product.
RNA Ligase Mediated 5’ RACE Analysis of KIR2DL1 Transcripts
5’ RACE was performed on human donor NK cell RNA using the FirstChoice® RLM-RACE kit (Ambion Inc, Austin, TX, USA). KIR2DL1 gene-specific cDNA was generated using a KIR2DL1 Exon 3 reverse primer: 5’-CAATGAGGCGCAAAGTGTCGTTAAAC-3’. First round PCR amplification was performed using the 5’ outer adapter and the KIR2DL1-Exon 3 reverse primer 5’-CTCTCTGTGCAGAAGGAAGTGTT-3’ at an annealing temperature of 59° C for 35 cycles with Platinum PCR Supermix (Invitrogen). Excess primers from first round PCR reactions were removed with the Charge-Switch PCR Clean-Up Kit (Invitrogen). Nested PCR was subsequently performed with the 5’ inner adapter primer and the KIR2DL1-Alternate exon 2 reverse primer (5’-CCAGCCTCTATAGGCCACCTGC-3’) or a reverse primer upstream of the proximal promoter start sites (5’-TAGTGTCCTTCACTATGACCAAC-3’) at an annealing temperature of 60° C for 30 cycles. PCR products were cloned into pCR2.1 Topo (Invitrogen) and sequences analyzed to map the 5’ start sites.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from YT cells using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich). Protein concentration was measured with a Bio-Rad protein assay (Hercules, CA), and samples were stored at −70°C until use. Double-stranded DNA oligonucleotide probes corresponding to the predicted ZEB1 binding sequence of the mutant KIR2DL1 distal promoter (TTCCTAGGTGTAACCAATTGGTTTAATA-TTATTTATTGAGAAGACATTCTATGCCACCTTAAACCACACGGCAG) or the wild-type sequence were synthesized. Probes were labeled with [α-32P]deoxycytidine triphosphate (3000 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) by fill-in using the Klenow fragment of DNA polymerase I (Invitrogen). 32P-labeled double-stranded oligonucleotides were purified using mini Quick Spin Oligo Columns (Roche Diagnostics, Mannheim, Germany). DNA-protein binding reactions were performed in a 10-µL mixture containing 5 µg nuclear protein and 1 µg poly(dI-dC) (Sigma-Aldrich) in 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5). Nuclear extracts were incubated with 1 µL 32P-labeled oligonucleotide probe (10,000 cpm) at room temperature for 20 minutes and then loaded on a 5% polyacrylamide gel (37:5:1). Electrophoresis was performed in 0.5x TBE for 2 hours at 130 V, and the gel was visualized by autoradiography. For antibody supershift experiments, nuclear extracts were incubated with 2 µL of antibody for 1 h on ice before the addition of 32P- labeled DNA probe. After the addition of labeled DNA probe, the binding reaction was incubated for an additional 20 min at room temperature. The antibodies used were ZEB1-1 (H102, sc-25388x), ZEB1-2 (C-20, sc-10570x), ZEB1-3 (E-20, sc-10572x), ZEB1-4 (R-17, sc-10573x), ZEB2 (SIP1, E-11, sc-271984), and MZF1 (H-45, sc-66991x) from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), and Myc (9E11; Abcam, Cambridge, MA, USA).
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute (NCI), NIH, under contract HHSN261200800001E and NCI grant P01 111412 (JSM, SKA). This research was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Carrington M, Norman P. The KIR Gene Cluster. Bethesda, MD: National Library of Medicine (US), NCBI; 2001. [Google Scholar]
- 4.Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 5.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 6.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 7.Held W, Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur J Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 8.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 9.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J Immunol. 2000;165:6416–6421. doi: 10.4049/jimmunol.165.11.6416. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Lozano N, Trompeter HI, de Pablo R, Estefania E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. 2007;37:1954–1965. doi: 10.1002/eji.200737277. [DOI] [PubMed] [Google Scholar]
- 12.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 13.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, et al. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cichocki F, Miller JS, Anderson SK. Killer immunoglobulin-like receptor transcriptional regulation: a fascinating dance of multiple promoters. J Innate Immun. 2011;3:242–248. doi: 10.1159/000323929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, et al. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Tanamachi DM, Moniot DC, Cado D, Liu SD, Hsia JK, Raulet DH. Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J Immunol. 2004;172:1074–1082. doi: 10.4049/jimmunol.172.2.1074. [DOI] [PubMed] [Google Scholar]
- 17.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, et al. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 21.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1991;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estefanía E, Flores R, Gomez-Lozano N, Aguilar H, Lopez-Botet M, Vilches C. Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J Immunol. 2007;178:4402–4410. doi: 10.4049/jimmunol.178.7.4402. [DOI] [PubMed] [Google Scholar]
- 23.Czaja K, Borer AS, Schmied L, Terszowski G, Stern M, Gonzalez A. A comprehensive analysis of the binding of anti-KIR antibodies to activating KIRs. Genes Immun. 2014;15:33–37. doi: 10.1038/gene.2013.58. [DOI] [PubMed] [Google Scholar]
- 24.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 25.Radeloff B, Nagler L, Zirra M, Ziegler A, Volz A. Specific amplification of cDNA ends (SPACE): a new tool for the analysis of rare transcripts and its application for the promoter analysis of killer cell receptor genes. DNA Seq. 2005;16:44–52. doi: 10.1080/10425170400028202. [DOI] [PubMed] [Google Scholar]
- 26.Pascal V, Stulberg MJ, Anderson SK. Regulation of class I major histocompatibility complex receptor expression in natural killer cells: one promoter is not enough! Immunol Rev. 2006;214:9–21. doi: 10.1111/j.1600-065X.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- 27.Fischer JC, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen DW, et al. Relevance of C1 and C2 epitopes for hemopoietic stem cell transplantation: role for sequential acquisition of HLA-C-specific inhibitory killer Ig-like receptor. J Immunol. 2007;178:3918–3923. doi: 10.4049/jimmunol.178.6.3918. [DOI] [PubMed] [Google Scholar]
- 28.van Bergen J, Stewart CA, van den Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. 2005;35:2191–2199. doi: 10.1002/eji.200526201. [DOI] [PubMed] [Google Scholar]
- 29.Martin MP, Carrington M. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol Biol. 2008;415:49–64. doi: 10.1007/978-1-59745-570-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, et al. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]


