Abstract
Removal of highly abundant proteins in plasma is often carried out using immunoaffinity depletion to extend the dynamic range of measurements to lower abundance species. While commercial depletion columns are available for this purpose, they generally are not applicable to limited sample quantities (<20 μL) due to low yields stemming from losses caused by nonspecific binding to the column matrix and concentration of large eluent volumes. Additionally, the cost of the depletion media can be prohibitive for larger-scale studies. Modern LC-MS instrumentation provides the sensitivity necessary to scale-down depletion methods with minimal sacrifice to proteome coverage, which makes smaller volume depletion columns desirable for maximizing sample recovery when samples are limited, as well as for reducing the expense of large-scale studies. We characterized the performance of a 346 μL column volume microscale depletion system, using four different flow rates to determine the most effective depletion conditions for ~6-μL injections of human plasma proteins and then evaluated depletion reproducibility at the optimum flow rate condition. Depletion of plasma using a commercial 10-mL depletion column served as the control. Results showed depletion efficiency of the microscale column increased as flow rate decreased, and that our microdepletion was reproducible. In an initial application, a 600-μL sample of human cerebrospinal fluid (CSF) pooled from multiple sclerosis patients was depleted and then analyzed using reversed phase liquid chromatography-mass spectrometry to demonstrate the utility of the system for this important biofluid where sample quantities are more commonly limited.
Keywords: Microscale depletion, IgY-14 immunoaffinity resin, Human plasma, Cerebrospinal fluid, MS
Introduction
Mass spectrometry (MS)-based proteomics applications in biomedical studies frequently involve analysis of biofluids such as urine, saliva, blood, and CSF [1–7]. Blood plasma is of particular interest as it can contain signature proteins indicative of disease state; however, the vast dynamic range of protein concentrations in plasma, which spans more than nine orders of magnitude, and the presence of highly abundant proteins inhibit detection and identification of low-abundance proteins by liquid chromatography LC-MS/MS in spite of peak capacities of ~1,000 for a single dimension LC separation [8]. Similar problems have been observed with two-dimensional gel electrophoresis [9].
A number of preanalysis fractionation and depletion methods have been reported as an effective means of reducing sample complexity [10–15]. In particular, immunoaffinity columns have become a preferred tool for isolating low-abundance proteins in plasma prior to comprehensive MS-based proteomic analysis [16–19]. The majority of these columns utilize antibody ligands supported by IgY or IgG [20–22] to bind high-abundance proteins and isolate low-abundance proteins in a flow-through fraction. The immunoaffinity-based column can lose their depletion efficacy over time and a potential removal of nontargeted proteins through nonspecific association to depletion column or a complex with target proteins such as albumin and transferrin have been reported [23, 24]. Nevertheless, the use of immunoaffinity columns can dramatically increase the number of protein identifications; for example, two-dimension LC-MS analysis of nondepleted and immunoaffinity depleted serum identified 262 of unique proteins of which 142 were observed only in the depleted sample and 38 only in the non-depleted sample [25]. Despite the effectiveness of immunoaffinity columns for removing high-abundance proteins, medium-abundance proteins can remain in the flow-through fraction. Removal of these medium-abundance proteins to further increase dynamic range and improve detection of very low-abundance proteins has been achieved via a tandem system that consisted of IgY12 and SuperMix [26] affinity columns [27].
Factors to consider when selecting a depletion column include cost, required depletion efficiency, processing time, reusability, and sample capacity [12]. Another consideration is the potential for nonspecific binding, which causes the potential loss of interesting proteins through the attachment of proteins to the column materials [28, 29]. Both cost and nonspecific binding issues can be relieved by reducing the column volume. Spin column and tip-based methodologies are currently commercially available for this purpose. Spin columns are currently available for 8 μl of plasma; however, this geometry presents significant technical challenges for automation and problems with efficacy and reproducibility have been reported in the literature [30, 31]. Automation is possible with tip-based depletion when using a specialized liquid handler. Nevertheless, integration of tip-based depletion into an online system is not straightforward, the recommended loading volume is more than twice that used in this study, and tips are only rated for 30 uses. From this perspective, a microscale LC column is an attractive solution providing the high reproducibility and reusability of an LC format, and its compatibility with online sample handling provides the basis for full automation and further gains in sensitivity.
Here, we report a depletion methodology that features a microscale immunoaffinity column and offers several advantages, including high depletion efficiency, high reproducibility, reduced sample size requirements, and reduced cost. Furthermore, the microscale column produces elution volumes suitable for incorporating into an online automated system. Results from our evaluation using plasma samples show that the microscale column provides comparable performance in terms of reproducibility and identified peptides/proteins to a commercial column with equivalent sample loading relative to column volume. Following characterization with plasma, we applied the microscale depletion methodology to CSF which shares a similar set of high-abundance proteins with plasma, but has ~102 lower total protein concentration [4]. This result in large volumes required for in-depth proteomic analysis, especially when immunoaffinity depletion is used [32]. CSF is particularly interesting in that it comes in direct contact with the extracellular surface of the brain, and alterations in the biochemical composition of CSF can reflect disorders related to the central nervous system. This makes it an attractive sample for studies related to neurodegenerative disorders such as Alzheimer's, Parkinson's disease, and multiple sclerosis [4, 33].
Materials and methods
Ethics statement
Approval for the conduct of this study was obtained from the institutional review board of Pacific Northwest National Laboratory and the human ethics committee at the Faculty of Medicine of Uppsala University. Written consent was obtained from subjects.
Reagents
All chemicals were HPLC grade. The acetonitrile and isopropanol were purchased from J. T. Baker (Philipsburg, NJ, USA) and formic acid was purchased from Thermo Scientific (Rockford, IL, USA). Dilution buffer, stripping buffer, and neutralization buffer were purchased from Beckman Coulter (Fullerton, CA, USA). DTT, iodoacetamide, and TFA were purchased from Sigma-Aldrich. Formic acid was purchased from Thermo Scientific.
Preparation of depletion column
IgY14 human resin purchased from Sigma-Aldrich (St Louis, MO, USA) was packed into a PEEK column (150×2.1 mm ID, VICI, Houston, TX, USA) by slurry packing method. Briefly, a screen was placed in one end of column and the slurry of IgY14 resin was then pulled into the column using a 1 mL of auto pipette (Gilson. Inc., WI, USA). Once fully packed, a second screen was used to secure the resin in the column. Dilution buffer was kept flowing at 200 μL/min for 1 h to ensure the resin was packed uniformly. The depletion column was stored at 4 °C until use.
Sample preparation and depletion of human plasma and human CSF
Germ-free human plasma was purchased from Sigma-Aldrich (St Louis, MO, USA). The pooled plasma was diluted fivefold with dilution buffer and 30 μL of this sample (equivalent to 432 μg of plasma proteins) was loaded onto a 200-μL sample loop and injected onto IgY14 column. Dilution buffer with flow rates of 20, 80, 140, and 200 μL/min for the flow-through fraction (FF) were collected for 50, 30, 25, and 20 min, respectively, to determine effective depletion conditions. Stripping buffer was then delivered to obtain bound fractions (BF) at a flow rate of 200 μL/min for all test conditions. Column stripping time was determined by UV trace and determined to be 40 min for the 200 μL/min flow rate. Finally, neutralization and dilution buffers were delivered at 350 μL/ min for 15 min for both of them for column regeneration. All buffers were delivered by a quaternary pump (Agilent Technologies, CA, USA). The proteins eluted from the depletion column were monitored using a Spectra 100 Variable UV/Vis CE Detector (Thermo, USA) at a wave length of 580 nm. All experiments were performed in duplicate. Collected fractions were concentrated using Ultra-4 3000 MWCO filter (Millipore, MA, USA) and then Trifluoroethanol (TFE) was added to the solution to be a 50 % of final concentration and incubated at 60 °C for 2 h. Dithiothreitol (DTT) was added to a final concentration of 2 mM and then incubated for 1 h at 37 °C. The sample was diluted 5×with NH4HCO3 followed by addition of Trypsin at a mass ratio of 1 to 50 (trypsin:proteins) and incubated for 3 h at 37 °C [34–36]. The sample was concentrated and stored in at −80 °C until use for MS analysis. The depletion of human plasma with commercial IgY14 column (IgY14 LC-10, Sigma-Aldrich, St Louis, MO, USA) was performed according to the instructions from the company. Briefly, 12.555 mg of sample was loaded using dilution buffer at a flow rate of 0.5 mL/min for 25 min and then for 5 min at a flow rate of 2.0 mL/min. The flow rate remained constant at 2.0 mL/min for all remaining steps: stripping buffer for 18 min, neutralization buffer for 8 min, and finally dilution buffer for 9 min. All results presented in this study were depleted using IgY14 resin, either in micro-scale format or LC-10 format.
A pooled 600 μL CSF sample was derived by combining 200 μL aliquots from three patients with multiple sclerosis. The pooled sample was concentrated using Ultra-4 3000 MWCO filters to a final concentration of 4.49 μg/μL as measured by a BCA protein assay. Depletion was performed on 65 μL of the pooled CSF sample after dilution to a final volume of 200 μL with dilution buffer, equivalent to 292 μg of CSF proteins. The elution flow rate for the FF was 20 μL/ min while the rest of steps were the same as in the flow rate study using plasma. Following depletion, the FF and BF were concentrated using Ultra-4 3000 MWCO filters. The BF was washed with 50 mM NH4HCO3. To each fraction, 8 M urea was added followed by incubation for 1 h at 37 °C, addition of 40 mM DTT, and a second incubation for 1 h at 37 °C in the dark. Following alkylation, samples were diluted 10× with 50 mM NH4HCO3 and then 1 mM CaCl2 was added. Trypsin was added with the enzyme to substrate ratio of 1:50 followed by incubation for 3 h at 37 °C. Desalting was performed using Discovery SPE cartridges with 1 mL of matrix volume (St Louis, MO, USA). The SPE cartridge was conditioned with 3 mL of MeOH followed by 2 mL of 0.1 % TFA. Samples were then slowly loaded, washed with a 4 mL solution of 5 % ACN and 0.1 % TFA, and the peptides eluted with a 1 mL solution of 20 % ACN and 0.1 % TFA. The eluted peptide samples were concentrated in a Speed-Vac sample concentrator, analyzed by BCA protein assay, and then stored at −80 °C until use.
Liquid chromatography–mass spectrometry
The RPLC system used for analysis depleted human plasma samples consisted of an Agilent 1200 nanopump (Agilent Technologies, Santa Clara, CA) for gradient delivery coupled with a PAL autosampler (Leap Technologies, Carrboro, NC) for automated sample injection. An in-house prepared 35 cm× 360 μm o.d. × 75 μm i.d column containing 3-μm Jupiter C18 (Phenomenex, Torrence, CA) was used for reversed phase separations. Trypsin digests from both FF and BF (2.5 μg) were injected at 100 % of mobile phase (MP) A (0.1 % FA in water) and a constant flow rate of 300 nL/min and then the ratio of MP B (0.1 % FA in acetonitrile) was increased to 8, 12, 35, 60, and 75 % over the course of 2, 18, 55, 22, and 3 min, respectively. The reproducibility study (n=5) was performed only on FF samples using these same parameters. Analysis of depleted CSF tryptic digests (1.5 μg) by 1D-RPLC were performed on a nanoAcquity UPLC (Waters, Milford, MA) using the same mobile phases as described separations of human plasma samples. After high-speed loading onto a Symmetry C18 trap column (180 μm i.d. × 20 mm long, Waters) with 99 % MP A, MP B was increased to 8, 12, 35, 45, and 95 % over the course of 6, 54, 165, 66, and 8 min.
A Velos Orbitrap (Thermo Scientific, San Jose, CA) was used for all MS analyses. High-resolution (60 K) precursor scans were acquired from m/z 400 to m/z 2,000 followed by data-dependent ion trap MS/MS acquisition (isolation window=2, collision energy=35) of the top ten most abundant ions. Ions selected for MS/MS were excluded from selection again for 60 s, including any ions falling within −0.6 to 1.6 Da of the selected ion m/z. The ion transfer tube temperature and ESI voltage were 350 °C and 2.2 kV, respectively.
Data analysis
The SEQUEST algorithm (version 27, revision 12) to search MS/MS spectra against reconstructed forward/reverse protein sequence databases, which were generated from two versions of Homo sapiens Uniprot SPROT protein database: H_sapiens_Uniprot_SPROT_2011-04-05 (20,238 protein entries) and H_sapiens_Uniprot_SPROT_2012-04-30 (20,249 protein Entries). The MSGF algorithm [37] was used to rescore peptide-to-spectrum matches (PSMs) generated by SEQUEST. The plasma datasets were searched against the 2011-04-05 version of reconstructed database with no enzyme restraint and oxidation of methionine included as a dynamic modification. The CSF datasets were searched against the 2012-04-30 version of reconstructed database using no enzyme restraint with oxidation of methionine and cysteine alkylation as a dynamic modification. All subsequent data analyses were performed using in-house software, MDART beta, with the SEQUEST first-hit results files. The identified peptides were validated by filtering at 1 % false discovery rate (FDR) using MSGF spectral probability [37], precursor mass error, and peptide length ≥7 as filtering parameters [38]. Protein identifications consisting of ≥2 unique peptide identifications were considered to be confident identifications. Depletion efficiency was defined as PSMs attributed to nontargeted proteins divided by the total PSMs obtained in the dataset times 100. Therefore, a depletion efficiency of 100 % indicates that 0 spectra were matched to IgY14 targeted proteins and a depletion efficiency of 50 % would indicate that 50 % of PSMs were attributed to targeted proteins and 50 % to nontarget proteins.
Results
Microscale online depletion system
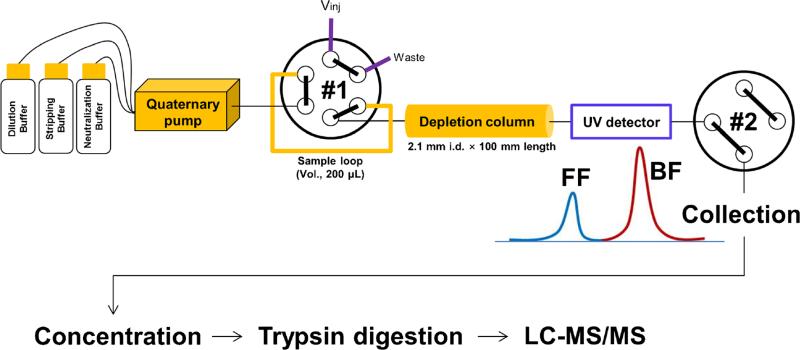
Figure 1 shows a schematic of the microscale depletion system and subsequent sample processing and analysis steps. The system uses capillary tubing to deliver buffers to a 346 μL IgY-14 depletion column via a quaternary pump that is directly connected to the #1 valve equipped with a 200-μL sample loop. Following the vendor recommended pressure limit of 300 psi for the depletion media, the system utilizes 1 mm i.d. Teflon tubing for all plumbing to ensure back pressure remains low. Note that the highest observed back pressure for any experiments at the highest flow rate (200 μL/min) was 174 psi (data not shown). Depletion is monitored by using an in-line UV detector at a wavelength of 280 nm.
Fig. 1.
Schematic of the microscale LC system. Vinj injection valve, Depl. Col. depletion column, UV ultra violet detector, FF flow-through fraction, BF bound fraction
Sample recoveries and depletion efficiencies based on flow rate study
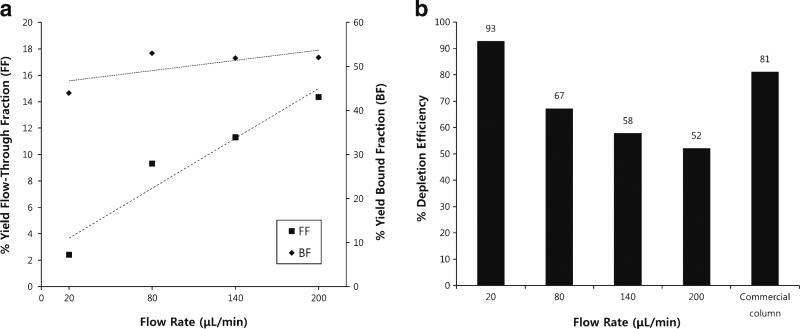
To determine the effect of flow rate on depletion efficiency, we evaluated depletion at four different flow rates: 20, 80, 140, and 200 μL/min. Sample size in each case was 432 μg plasma proteins, which is an equivalent load amount to the commercial IgY14 column (12,555 μg) in proportion to column volume. Following depletion, both the flow-through fraction (FF) and the bound fraction (BF) samples were concentrated using Ultra-4 3000 MWCO spin filters and then protein recoveries were determined using the BCA protein assay (Fig. 2a). Results from BCA protein assay revealed the average protein recoveries for FFs ranged from 2.4 % (10 μg) to 14.3 % (61 μg) for flow rates ranging from 20 to 200 μL/min, respectively. Comparatively, protein recovery in the FF from the commercial IgY14 column operated at 0.5 mL/min (manufacturer suggested flow rate) was 3.3 %. Proteins were eluted from all bound fractions (BF) at 200 μL/min, which resulted in similar sample collection times. Total protein recoveries from the bound fraction ranged from 44.0 % (190 μg) to 53.4 % (230 μg). Protein recovery in the BF from the commercial IgY14 column (elution flow rate of 0.5 mL/min) was 48.7 %.
Fig. 2.
a Sample recoveries after depletion of human plasma with micro-scale online depletion as a function of sample introduction flow rate. FF yield (left axis) showed a linear increase with flow rate (R2=0.93). BF yield did not show significant correlation with flow rate (R2=0.5). The reported values are the mean of two technical replicates. FF flow-through fraction, BF bound fraction. The overall standard deviations of four flow rates were 5.1 % (FFs) and 4.2 % (BFs), respectively. b Measured depletion efficiency of microscale column at different flow rates compared with depletion efficiency of commercial IgY14 column (commercial column) operated at the manufacturer suggested flow rate
Depletion efficiencies of both the microscale and commercial IgY14 columns were evaluated using LC-MS/MS spectral counting, as previously demonstrated for plasma and CSF [39–42]. Figure 2b shows the depletion efficiency of the FF for both the microscale column at different flow rates and the commercial IgY14 column operated at 0.5 mL/min per manufacturer specifications. Note that the depletion efficiency in the FF of the microscale column decreased from 92.8 to 52.2 % as flow rate increased from 20 to 200 μL/min, i.e., depletion efficiency increased as flow rate decreased. The percentage of spectral counts attributed to target proteins in the BFs did not exhibit significant differences, ranging from 80 to 85 %, data not shown.
Identification of nontarget peptides and proteins after depletion
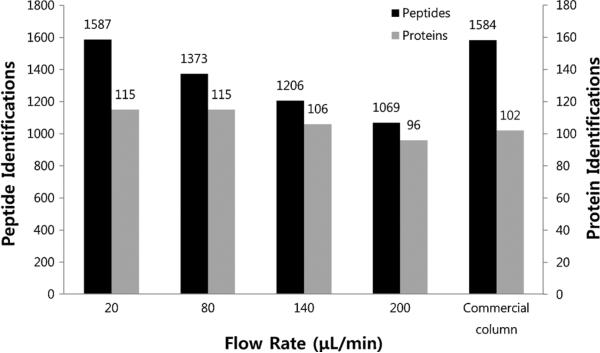
The microscale depletion resulted in increased numbers of non-target (low-abundance) peptide identifications as the flow rate decreased (i.e., as depletion efficiency increased). One thousand five hundred eighty-seven unique peptides were confidently identified in the 20 μL/min depletion test, which is comparable to the 1,584 unique peptides identified following commercial IgY14 column (Fig. 3). In terms of nontarget proteins, the number of identifications following microscale column depletion at 20 and 80 μL/min were both 115, which is slightly higher than the 102 nontarget protein identifications facilitated by the commercial IgY14 column (Fig. 3). Nontarget protein identifications following depletion at 140 and 200 μL/min were comparable to the commercial IgY14 column depletion.
Fig. 3.
The number of a nontarget peptides and b nontarget proteins of human plasma after depletion using the sample obtained from flow rate study with two replicates
Reproducibility of the microscale depletion system
To evaluate the reproducibility of the microscale depletion system, five replicate depletions of high abundance proteins in 432 μg human plasma were carried out at 20 μL/min. The FFs were concentrated, digested with trypsin, and analyzed using LC (100 min)-MS/MS. The depletion efficiency in the FF of the five replicates varied between 91.2 and 90.8 % with a standard deviation of 0.34 (data not shown), which is consistent with the 92.8 % obtained in the flow rate study.
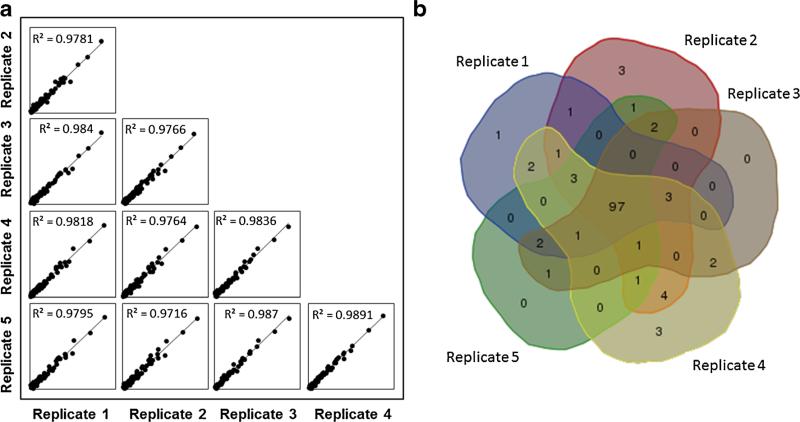
We investigated the linear correlations of spectral counts for 122 protein identifications observed in common among the five replicates using a comparison method that has been reported useful in other studies [36, 29]. In our evaluation, each replicate was plotted against all other replicates (Fig. 4a); only proteins having >2 spectral counts for each replicate were used. All plots showed correlation coefficients (R2) >0.97, demonstrating excellent reproducibility. We further compared the spectral counts for each of the target proteins that were detected in the FF fraction (Table 1), demonstrating that the bleed-through of target proteins is also reproducible. A total of 2,208 unique peptides, mapping to 129 proteins, were identified in these 5 analyses. Of these 129 proteins, only 7 proteins were identified in a single analysis and 97 proteins were identified by 2 or more unique peptides in all 5 datasets (Fig. 4b). Strong overlap of identifications between replicates also suggests reproducible depletions.
Fig. 4.
The reproducibility of the microscale IgY14 column was evaluated using five replicate analyses of human plasma. Flow-through fractions were analyzed by LC-MS/MS and correlation comparisons were carried out using spectral counting data. a Comparisons contain 99 proteins that were present across all 5 replicates. Evaluation of correlation between replicates produced an R2>0.98, indicating strong correlation between replicates. b Symmetric Venn diagram of confident protein identifications by replicate. The Venn diagram was created using web tools hosted by the Bioinformatics and Systems Biology of Gent
Table 1.
Spectral counts of target proteins for five replicates of flow-through fractions (FFs) at 20 μL/min
| Protein names | Replicate 1 | Replicate 2 | Replicate 3 | Replicate 4 | Replicate 5 | Mean±SD |
|---|---|---|---|---|---|---|
| Alpha-2-macroglobulin | 136 | 165 | 146 | 153 | 138 | 148±11.8 |
| Serum albumin | 2 | – | 1 | 1 | 1 | 1±0.5 |
| Apolipoprotein A-I | 4 | 7 | 2 | 4 | 4 | 4±2 |
| Apolipoprotein A-II | 2 | 2 | 2 | 2 | 3 | 2±0.5 |
| Apolipoprotein B-100 | 95 | 112 | 107 | 76 | 95 | 97.0±13.9 |
| Complement C3 | – | 1 | 3 | 3 | 7 | 4±2 |
| Fibrinogen alpha chain | 89 | 80 | 86 | 96 | 90 | 88±5.9 |
| Fibrinogen beta chain | 75 | 66 | 77 | 72 | 74 | 73±4.2 |
| Fibrinogen gamma chain | 53 | 38 | 46 | 49 | 49 | 47±5.6 |
| Ig alpha-1 chain C region | 2 | 4 | 3 | 4 | 4 | 3±0.9 |
| Ig mu chain C region | 10 | 10 | 12 | 10 | 12 | 11±1.1 |
Only target proteins identified in the FFs are listed
Initial demonstration with CSF
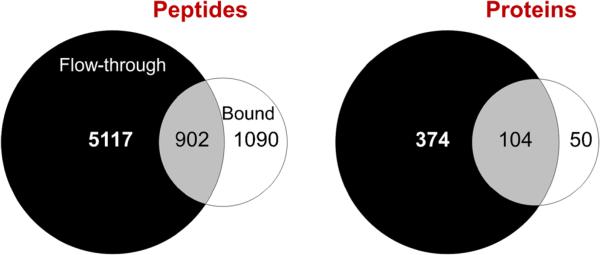
To evaluate performance of the microscale depletion system for another biofluid of interest, 600 μL (292 μg protein) of CSF was depleted at 20 μL/min. CSF is similar in composition to plasma; however, the protein concentration is ~100-fold less [4]. The protein yield determined by BCA protein assay for the CSF depletion was ~10 % (27 μg), which is ~fourfold higher than plasma at this flow rate. This high yield when compared to plasma suggests that our microdepletion column is suitable for even smaller CSF samples than utilized in this study. LC-MS/MS analysis was carried out using 700 ng FF and an extended 300 min LC separation (longer than used for the plasma study, but in line for CSF samples) to increase the number of protein identifications. Seven thousand one hundred nine peptides were identified that covered 528 proteins at 1 % FDR (Fig. 5).
Fig. 5.
Comparison of peptide and protein identifications from CSF after IgY14 microscale depletion followed by 300 min LC-MS/MS
Discussion
This study addresses the need for a microscale depletion system to overcome some drawbacks of using typical immunoaffinity methods in terms of relatively large sample sizes and the cost. Current proteomics technologies based upon nanoESI-MS can achieve sub-attomole protein detection [43]. With this in mind, we fabricated a microscale depletion column to remove high abundance proteins that preclude analysis of lower abundance proteins. Characterization of the column at different flow rates showed that the number of identified peptides and the depletion efficiency of the FF increased as the depletion flow rate decreased (Fig. 2b). We hypothesize that this increase in depletion efficiency is the result of increased binding of the target proteins to IgY antibodies facilitated by longer residence times in the depletion column. More efficient removal of targeted proteins consequently resulted in an increase in peptide and protein identifications. At the 20 μL/min flow rate, our microscale column produced higher depletion efficiencies and delivered comparable peptide and protein identifications to the commercial IgY14 column with much larger sample loadings. This hypothesis is further supported by the increasing protein mass yield as flow rate is increased, shown in Fig. 2a, where additional mass is contributed by increasing contamination of the FF fraction by targeted proteins. Results obtained from the commercial IgY14 column are also consistent with this hypothesis which is evaluated by LC-MS/MS spectral counting (Fig. 2b). The depletion efficiency of the commercial IgY14 column using manufacturer's specifications was 81.2 %, which falls between the efficiency values obtained for 20 and 80 μL/min flow rates. Consistent with our hypothesis, the protein mass yield for the commercial IgY14 column (3.3 %) is intermediate between the yields for the 20 and 80 μL/min flow rates of 2.4 and 9.3 %, respectively. This increase in efficiency is accompanied by an expected decrease in protein yield.
Another interesting observation of these experiments was that the depletion efficiency affected not only the number of nontarget proteins that were identified, but also which proteins were identified. The average overlap between protein identifications at different flow rates was 65 %, which was similar to the average overlap with the commercial IgY14 column, 67 %. This is significantly lower than the average overlap among replicate runs in the reproducibility study of 81 %. This is likely due to the differing presence of proteins co-depleted with target species as well as the impact of undersampling owing to the complexity and dynamic range of plasma [26, 29, 44]. Furthermore, the identification of more unique peptides yielded a limited increase in protein identifications. This effect was most pronounced in the commercial IgY14 column depletion where only 102 proteins were identified from 1,584 peptides as compared to 115 proteins identified from 1,587 peptides at the 20 μL/min flow rate. Closer analysis of results from the commercial IgY14 column depletion revealed higher coverage for large, high-abundance proteins when compared to microdepletion samples. It is likely that this bias towards coverage over higher identifications is introduced by the different sample handling methods used to accommodate large differences in sample size and is not interpreted as inferior performance for the purposes of this study. Comparison of protein identifications in the 20 and 80 μL/min reveals a similar trend where improvements in depletion efficiency result in higher coverage of identified proteins but not necessarily a gain in identifications. These finding suggest that longer separation times and depletion of more high-abundant plasma proteins would enhance the benefits of our microdepletion system.
A major motivator for this work was to decrease the sample losses associated with immunoaffinity depletions allowing for the utilization of smaller sample sizes (<20 μL for plasma). This would allow in-depth proteomic analyses from a single fingerstick specimen, and in the case of CSF, greatly reduce the required amount of this valuable clinical commodity. In our experiments with the commercial IgY14 column, approximately 52 % of the loaded protein was recovered, representing a loss of ≈6 mg of protein. A portion of the observed protein losses is presumed due to small proteins and peptides passing through the MWCO filter, as well as to proteins binding to the MWCO filter, as reported elsewhere [12, 45]. Losses due to small proteins and peptides should contribute the same relative sample loss for both the commercial column and microdepletion columns; however, smaller FF volumes facilitated the use of 1.5 mL spin concentrators instead of the larger 15 mL concentrators used with the commercial IgY14 column reducing losses at this step. Another portion of the losses can be attributed to nonspecific binding of proteins to components of the LC and depletion column. Nonspecific binding can be due to binding between (1) nontarget and target proteins, (2) nontarget proteins and antibodies linked to the column matrix, and/or (3) sample proteins and the solid support of the antibody, column, and tubing walls. In the first case, the relative loss of nontarget proteins would be unaffected by reducing column size. However, in the second two cases, losses can be reduced by decreasing the surface area that the sample comes into contact with. In this study, we were able to recover 49 % of the protein from the 432 μg injection, losing only ≈220 μg of sample proteins.
We chose CSF for the initial application of our microscale depletion column. Because of the greatly decreased protein concentration of this biofluid, the application of immunoaffinity depletion to this sample type has been largely limited to large samples (>1 mL) [32, 41, 46]. In studies with smaller mammals, such as mice where <100 μL of CSF can be obtained [47], immunoaffinity methods become unworkable. The aim in future research is to combine our microscale depletion column with highly sensitive online sample handling approaches [48, 49], permitting reproducible, in-depth proteomic analysis for these limited samples. Thus, CSF analysis is an important driver for our development of micro-scale depletion technologies. In CSF, the depletion efficiency in the FF was found to be 93.4 %, which is similar to the 92.8 % observed for plasma at the same flow rate; however, protein yield by mass increased from 2.4 % in plasma to 10 % in CSF. This observation is primarily attributed to the different relative abundances for targeted proteins in CSF relative to plasma, and suggests that major protein depletion from CSF could benefit from optimizing the antibody composition of the column for this sample type. While we identified 528 proteins from 292 μg (600 μL) protein starting material (LC-MS/MS, 300-min separation), this number will vary depending on the separation methods (i.e., single- or multidimensional LC separation and types of column material), sample loading amounts, and MS instrument type [32, 41]. The results obtained in our study compare well with reports in the literature that utilize larger sample volumes. A summary table of literature results for recent CSF studies for comparison is given in Electronic Supplementary Material Table S1 [32, 41, 46, 47, 50–54]. For instance, Borg et al. identified 156 proteins with 750 μg of CSF sample of a pool from 5 patients using IgY 14 and also identified 535 proteins with the same amount of CSF sample using IgY HAS RP30, in which BEH130TM C18 analytical column and LTQ-FT Ultra mass spectrometer were used for a mass spectrometry [41]. All identified proteins obtained from human plasma and CSF in this study are listed in Electronic Supplementary Material Table S2 including protein identifications consisting of ≥1 unique peptide.
This work has demonstrated the feasibility of microscale immunoaffinity depletion of complex biofluid samples through the successful application of LC-based depletion to biofluid samples of 6 and 600 μL of plasma and CSF, respectively. Our microscale depletion column produces proteomic measurement quality that is comparable to protocols which require much larger sample volumes as well as large volumes of expensive immunoaffinity resins. Additionally, the column format provides ease of automation and high reproducibility, which are critical to successful proteomic investigations. This work represents the first step in building an online, automated biofluid analysis system. We anticipate that the increased sampling handling efficiency obtainable in online systems will allow for further decreases in depletion column size, resulting in further increases in sensitivity.
Supplementary Material
Acknowledgments
This project was supported by grants from the National Center for Research Resources (5 P41 RR018522-10) and the National Institute of General Medical Sciences (8 P41 GM103493-10) from the National Institutes of Health as well as the Department of Energy Office of Biological and Environmental Research Genome Sciences Program under the Pan-omics project. Work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830. We appreciate the favor from Jonas Bergquist at Uppsala University (Department of Chemistry-Biomedical Center, Analytical Chemistry and SciLife Lab, Uppsala, Sweden) for the donation of CSF sample.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00216-014-8058-3) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare no conflicts of interest.
Contributor Information
Seok-Won Hyung, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA; Korea Research Institute of Standards and Science, 267 Gajeong-Ro, Yuseong-Gu, Daejeon 305-340, Republic of Korea.
Paul D. Piehowski, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Ronald J. Moore, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Daniel J. Orton, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Athena A. Schepmoes, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Therese R. Clauss, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Rosalie K. Chu, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Thomas L. Fillmore, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
Heather Brewer, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA.
Tao Liu, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA.
Rui Zhao, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA.
Richard D. Smith, Biological Sciences Division and Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA
References
- 1.Pang JX, Ginanni N, Dongre AR, Hefta SA, Opitek GJ. J Proteome Res. 2002;1:161–169. doi: 10.1021/pr015518w. [DOI] [PubMed] [Google Scholar]
- 2.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 4.Kroksveen AC, Opsahl JA, Aye TT, Ulvik RJ, Berven FS. J Proteome. 2011;74:371–388. doi: 10.1016/j.jprot.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Hu S, Loo JA, Wong DT. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Wang X, Xu LX, Yen Y, Liu P, Jia W. Mol Cell Proteomics. 2011;10(M110):004945. doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Moore RJ, Zhao R, Blonder J, Auberry DL, Masselon C, Pasa-Tolić L, Hixson KK, Auberry KJ, Smith RD. Anal Chem. 2003;75:3596–3605. doi: 10.1021/ac0300690. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed N, Rice GE. J Chromatogr B Anal Technol Biomed Life Sci. 2005;815:39–50. doi: 10.1016/j.jchromb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 10.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Mol Cell Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Shores KS, Knapp DR. J Proteome Res. 2007;6:3739–3751. doi: 10.1021/pr070293w. [DOI] [PubMed] [Google Scholar]
- 12.Polaskova V, Kapur A, Khan A, Molloy MP, Baker MS. Electrophoresis. 2010;31:471–482. doi: 10.1002/elps.200900286. [DOI] [PubMed] [Google Scholar]
- 13.Yuan X, Desiderio DM. Proteomics. 2005;5:541–550. doi: 10.1002/pmic.200400934. [DOI] [PubMed] [Google Scholar]
- 14.Boschetti E, Righetti PG. J Proteome. 2008;71:255–264. doi: 10.1016/j.jprot.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Ly L, Wasinger VC. Proteomics. 2011;11:513–534. doi: 10.1002/pmic.201000394. [DOI] [PubMed] [Google Scholar]
- 16.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. J Proteome Res. 2006;5:1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 19.Pernemalm M, Lewensohn R, Lehtiö J. Proteomics. 2009;9:1420–1427. doi: 10.1002/pmic.200800377. [DOI] [PubMed] [Google Scholar]
- 20.Fang X, Huang L, Feitelson JS, Zhang WW. Drug Discov Today Technol. 2004;I:141–148. doi: 10.1016/j.ddtec.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Proteomics. 2005;5:3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, Liotta LA, Veenstra TD, Conrads TP. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 24.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Proteomics Clin Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yocum AK, Yu K, Oe T, Blair IA. J Proteome Res. 2005;4:1722–1731. doi: 10.1021/pr0501721. [DOI] [PubMed] [Google Scholar]
- 26.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Mol Cell Proteomics. 2008;7:1963–1973. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandow JE. Proteomics. 2010;10:1416–1425. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- 28.Wetterhall M, Zuberovic A, Hanrieder J, Bergquist J. J Chromatogr B Anal Technol Biomed Life Sci. 2010;878:1519–1530. doi: 10.1016/j.jchromb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Angela DN, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corrigan L, Jefferies C, Clive LT, Daly J. Proteomics. 2011;11:3415–3419. doi: 10.1002/pmic.201000587. [DOI] [PubMed] [Google Scholar]
- 31.Seam N, Gonzales DA, Kern SJ, Hortin GL, Hoehn GT, Suffredini AF. Clin Chem. 2007;53:1915–1920. doi: 10.1373/clinchem.2007.091736. [DOI] [PubMed] [Google Scholar]
- 32.Schutzer SE, Liu T, Natelson BH, Angel TE, Schepmoes AA, Purvine SO, Hixson KK, Lipton MS, Camp DG, Coyle PK, Smith RD, Bergquist J. PLoS One. 2010;5:e10980. doi: 10.1371/journal.pone.0010980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujmovic I. Mult Scler Int. 2011;2011:767083. doi: 10.1155/2011/767083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown JN, Ortiz GM, Angel TE, Jacobs JM, Gritsenko M, Chan EY, Purdy DE, Murnane RD, Larsen K, Palermo RE, Shukla AK, Clauss TR, Katze MG, McCune JM, Smith RD. Mol Cell Proteomics. 2012;11:605–618. doi: 10.1074/mcp.M111.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown RN, Romine MF, Schepmoes AA, Smith RD, Lipton MS. J Proteome Res. 2010;9:4454–4463. doi: 10.1021/pr100215h. [DOI] [PubMed] [Google Scholar]
- 36.Cao L, Bryant DA, Schepmoes AA, Vogl K, Smith RD, Lipton MS, Callister SJ. Photosynth Res. 2012;110:153–168. doi: 10.1007/s11120-011-9711-8. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Gupta N, Pevzner PA. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piehowski PD, Petyuk VA, Sandoval JD, Burnum KE, Kiebel GR, Monroe ME, Anderson GA, Camp DG, 2nd, Smith RD. Proteomics. 2013;13:766–770. doi: 10.1002/pmic.201200096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 40.Shuford CM, Hawkridge AM, Burnett JC, Jr, Muddiman DC. Anal Chem. 2010;82:10179–10185. doi: 10.1021/ac102248d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borg J, Campos A, Diema C, Omeñaca N, de Oliveira E, Guinovart J, Vilaseca M. Clin Proteomics. 2011;8:6. doi: 10.1186/1559-0275-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJ, Liebler DC. J Proteome Res. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue G, Luo Q, Zhang J, Wu SL, Karger BL. Anal Chem. 2007;79:938–946. doi: 10.1021/ac061411m. [DOI] [PubMed] [Google Scholar]
- 44.Qian WJ, Liu T, Petyuk VA, Gritsenko MA, Petritis BO, Polpitiya AD, Kaushal A, Xiao W, Finnerty CC, Jeschke MG, Jaitly N, Monroe ME, Moore RJ, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, Smith RD. J Proteome Res. 2009;8:290–299. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liebler DC, Ham AJ. Nat Methods. 2009;6:785. doi: 10.1038/nmeth1109-785a. author reply 785–786. [DOI] [PubMed] [Google Scholar]
- 46.Ogata Y, Charlesworth MC, Higgins L, Keegan BM, Vernino S, Muddiman DC. Proteomics. 2007;7:3726–3734. doi: 10.1002/pmic.200700455. [DOI] [PubMed] [Google Scholar]
- 47.Stoop MP, Rosenling T, Attali A, Meesters RJ, Stingl C, Dekker LJ, van Aken H, Suidgeest E, Hintzen RQ, Tuinstra T, van Gool A, Luider TM, Bischoff RM. J Proteome Res. 2012;11:4315–4325. doi: 10.1021/pr300428e. [DOI] [PubMed] [Google Scholar]
- 48.Slysz GW, Lewis DF, Schriemer DC. J Proteome Res. 2006;5:1959–1966. doi: 10.1021/pr060142d. [DOI] [PubMed] [Google Scholar]
- 49.Massolini G, Calleri E. J Sep Sci. 2005;28:7–21. doi: 10.1002/jssc.200401941. [DOI] [PubMed] [Google Scholar]
- 50.Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, Mann M. J Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 51.Mouton-Barbosa E, Roux-Dalvai F, Bouyssié D, Berger F, Schmidt E, Righetti PG, Guerrier L, Boschetti E, Burlet-Schiltz O, Monsarrat B, Gonzalez de Peredo A. Mol Cell Proteomics. 2010;9:1006–1021. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan S, Zhu D, Quinn JF, Peskind ER, Montine TJ, Lin B, Goodlett DR, Taylor G, Eng J, Zhang J. Proteomics. 2007;7:469–473. doi: 10.1002/pmic.200600756. [DOI] [PubMed] [Google Scholar]
- 53.Bora A, Anderson C, Bachani M, Nath A, Cotter RJ. J Proteome Res. 2012;11:3143–3149. doi: 10.1021/pr300057v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cunningham R, Jany P, Messing A, Li L. J Proteome Res. 2013;12:719–728. doi: 10.1021/pr300785h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.