Abstract
Objectives
The host genetic basis of mixed cryoglobulin vasculitis is not well understood and has not been studied in large cohorts. A genome-wide association study was conducted among 356 HCV RNA positive individuals with cryoglobulin-related vasculitis and 447 ethnically-matched, HCV RNA positive controls.
Methods
All cases had both serum cryoglobulins as well as a vasculitis syndrome. A total of 899,641 markers from the Illumina HumanOmni1-Quad chip were analyzed using logistic regression adjusted for sex, as well as genetically-determined ancestry. Replication of select single nucleotide polymorphisms (SNPs) was conducted using 91 cases and 180 controls, adjusting for sex and country of origin.
Results
The most significant associations were identified on chromosome 6 near the NOTCH4 and MHC class II genes. A genome-wide significant association was detected on chromosome 6 at SNP rs9461776 (OR= 2.16, p=1.16E-07) between HLA-DRB1 and DQA1: this association was further replicated in additional independent samples (meta-analysis p=7.1×10−9).
Conclusions
A genome-wide significant association with cryoglobulin related vasculitis was identified with SNPs near NOTCH4 and MHC Class II genes. The two regions are correlated and it is difficult to disentangle which gene is responsible for the association with MC vasculitis in this extended MHC region.
Keywords: Genome-Wide Association Study, Cryoglobulinemia, Human Leukocyte Antigen, SNP
Introduction
More than 185 million persons have been exposed to hepatitis C virus (HCV) and an estimated 130 million have chronic infection.1 Although the main target of this virus is the hepatocyte, HCV also appears to infect lymphocytes.2 Patients with chronic HCV infection typically produce anti-HCV IgG and can develop not only hepatic disease but also extrahepatic conditions amongst which the most frequent is mixed cryoglobulinemia (MC).3, 4 MC vasculitis is both an autoimmune and B-lymphoproliferative disorder, clinically benign, but evolving in 8–10% of cases into a frank non-Hodgkin’s lymphoma (NHL).5 Cryoglobulins are called “mixed” because they are composed of both IgGs and IgMs, which can be partially monoclonal (type II MC) or totally polyclonal (type III MC) and have rheumatoid factor (RF) activity.6–8 The prefix “cryo” refers to the property that these immune complexes precipitate at temperatures below 37° C and dissolve with rewarming.9
Approximately half of persons with chronic HCV infection have serum cryoglobulins, but only ~5% develop a clinically evident MC vasculitis syndrome.10, 11 This HCV-related, cryoglobulinemic vasculitis most commonly manifests in the skin (palpable purpura, 80%), joints (70%), peripheral nerves (60%), and kidney (immune complex nephritis, 20%).11 Due to the wide range of symptoms, MC vasculitis subjects are often clinically referred to different specialties, thus the actual prevalence may be underestimated.10
The clinical vasculitis syndrome can be improved or resolved by therapy directed at HCV infection and/or the B cell proliferation (e.g., with rituximab), inflammation (e.g., with corticosteroids), or serum cryogolbulins (e.g., with plasmapheresis).12–17 While, the pathogenesis of the MC vasculitis syndrome is still unclear accumulating evidence points to a role for host genetics. There are no consistent differences in the infecting viruses of persons with chronic infection with or without the vasculitis syndrome.4, 18 There are marked ethnic and regional differences in the prevalence of MC vasculitis with the highest prevalence in persons of Mediterranean descent.10, 19–21 Pozzato and colleagues found only Mediterranean descent explained the much higher prevalence of MC vasculitis in Italy compared to Japan.22 In addition, several studies have identified associations between MC vasculitis and particular HLA alleles.23–27. The purpose of this investigation was to perform a genome-wide, multi-center study to investigate the host genetic basis for MC vasculitis in persons with chronic HCV infection.
Results
We identified 356 individuals with cryoglobulin-related vasculitis and chronic hepatitis c virus (HCV) infection. A total of 447 controls with chronic HCV infection but with no cryoglobulin-related vasculitis were selected from a previously published GWAS of spontaneous clearance versus chronic infection of HCV, or were specifically screened for this study.28 After quality control measures, 899,641 single nucleotide polymorphisms (SNPs) were compared between cases and controls.
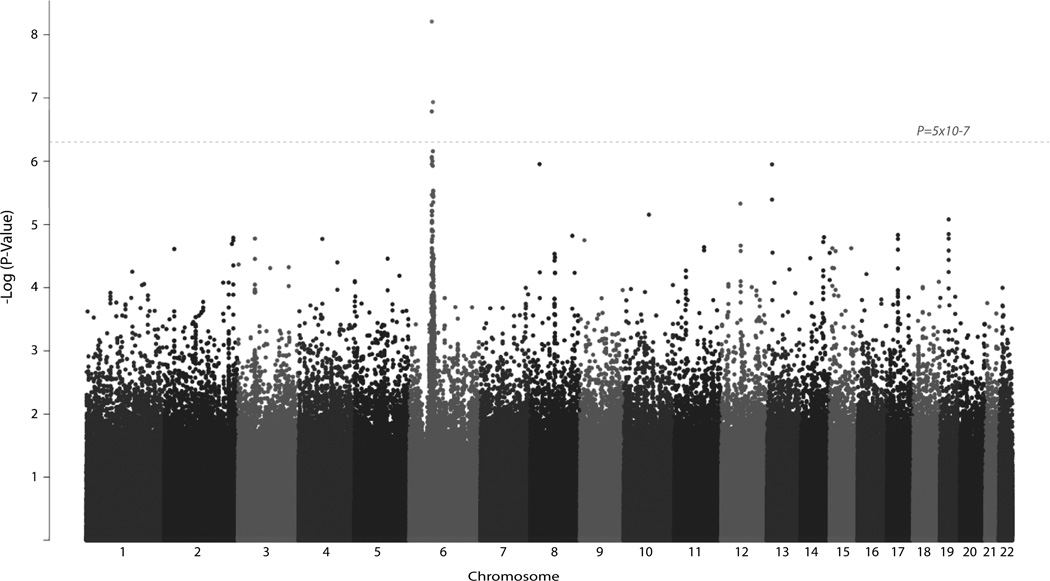
The most significant association was located on chromosome 6p21.32 (Figure 1, Table 1). A SNP, rs2071286, located within an intronic region of NOTCH4 (P=6.2×10−9) conferred 2.15 times the odds of having cryoglobulin-related vasculitis within chronically infected patients for each risk allele. An additional SNP within NOTCH4 at rs2071279 (P=1.6 ×10−7) had a similar effect size with each risk allele (T) resulting in 1.90 times the odds of disease. To confirm these findings, replication was attempted for both of these SNPs, however rs2071286 did not reach significance (P=0.13) and rs2071279 failed in production. Additionally, the next most significant NOTCH4 findings (rs9267820 and rs9267833) also did not meet the replication threshold (P>0.01) (Table 2).
Figure 1.
Manhattan Plot of GWAS Results. Significance is indicated by the −log transformation of the P-value on the y-axis. (e.g. P-value=0.001 denoted as 3) Associations are organized by chromosome on the x-axis. Genome-wide significance is indicated by the dashed line (P<5×10−7).
Table 1.
Study demographics for the discovery and replication populations by case and control status.
| Discovery Samples | Replication Samples | Everyone | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | Total | Control | Case | Total | Control | Case | Total | ||
| % Female (N) |
26.4 (118) |
66.9 (238) |
54.6 (438) |
45.0 (81) |
69.2 (63) |
53.1 (144) |
31.7 (199) |
67.3 (301) |
46.6 (500) |
|
| Study (country) | Toulouse Cohort (FR) | 1 | 0 | 1 | 29 | 0 | 29 | 30 | 0 | 30 |
| BAHSTION (US) | 78 | 0 | 78 | 37 | 0 | 37 | 115 | 0 | 115 | |
| Charles (US) | 0 | 0 | 0 | 0 | 20 | 20 | 0 | 20 | 20 | |
| RomeCryo (IT) | 0 | 74 | 74 | 2 | 16 | 18 | 2 | 90 | 92 | |
| UK Cohort (UK) | 35 | 0 | 35 | 0 | 0 | 0 | 35 | 0 | 35 | |
| Mangia (IT) | 114 | 25 | 139 | 111 | 24 | 135 | 225 | 49 | 274 | |
| REVELL (US) | 127 | 0 | 127 | 0 | 0 | 0 | 127 | 0 | 127 | |
| Cacoub (FR) | 0 | 72 | 72 | 0 | 15 | 15 | 0 | 87 | 87 | |
| MASVE (IT) | 92 | 185 | 277 | 0 | 17 | 17 | 92 | 202 | 294 | |
| Total | 447 | 356 | 803 | 179 | 92 | 271 | 626 | 448 | 1074 | |
Table 2.
Most significant associations from the Discovery GWAS of MC Vasculitis
| Chr | SNP | MAF | OR | P-value | PRep | PMeta | Gene |
|---|---|---|---|---|---|---|---|
| 6 | rs2071286 | 0.29 | 2.15 | 6.2E-09 | 0.18 | 1.7E-08 | NOTCH4 |
| 6 | rs9461776* | 0.21 | 2.16 | 1.2E-07 | 0.01* | 7.1E-09 | HLA-DRB1/DQA1 |
| 6 | rs2071279 | 0.38 | 1.90 | 1.6E-07 | NA | NA | NOTCH4 |
| 6 | rs9267820 | 0.37 | 1.83 | 8.6E-07 | 0.12 | 5.9E-07 | NOTCH4 |
| 6 | rs9267833 | 0.36 | 1.83 | 9.0E-07 | 0.12 | 6.0E-07 | NOTCH4 |
The second most significant association was found nearly 400 kilobases away within the Major Histocompatibility Complex (MHC) between HLA-DRB1 and HLA–DQA1 at SNP rs9461776 (P=1.2×10−7). Each additional copy of the risk allele (G) was associated with 2.16 times the odds of cryoglobulin-related vasculitis. This SNP was significantly replicated in an independent sample of cases and controls (P=0.01). When the discovery and replication stages were combined within a meta-analysis, rs9461776 had a p-value of 7.1×10−9 (OR=2.02, I2=0). Imputation of additional SNPs in both the NOTCH4 and MHC regions areas did not yield more significant signals than the actual genotyped SNPs, which may reflect the strong linkage disequilibrium (LD) in the region.
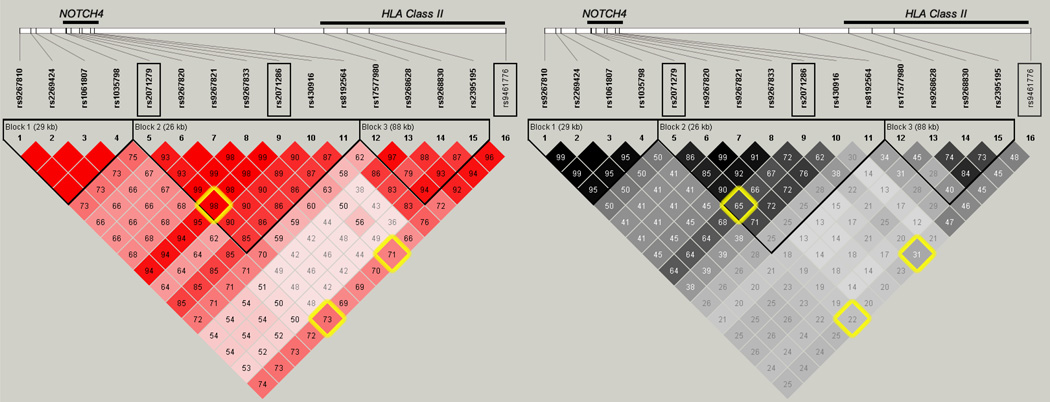
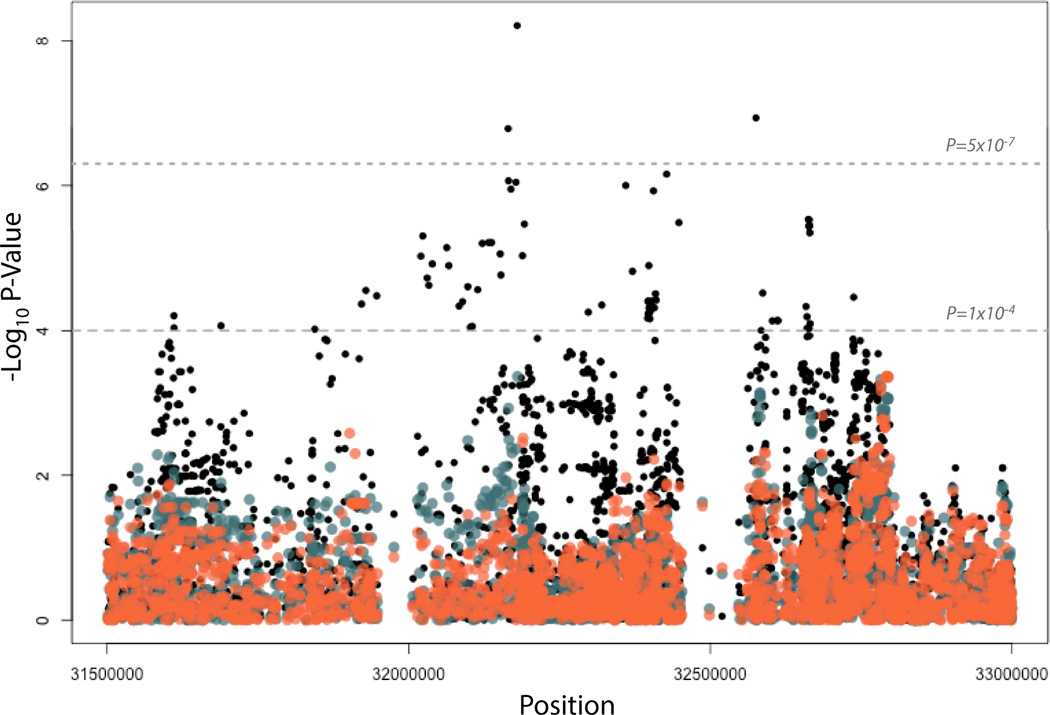
The LD structure around the top NOTCH4 association (Figure 2) suggests that there are two distinct blocks of LD defining NOTCH4 and the MHC region. However, rs2071286 and rs2071279 in NOTCH4 are in strong LD (D’ = 0.98, Figure 2) and both are in LD with the HLA class II SNP rs9461776 (D’ = 0.71 and 0.73, respectively) despite low r2 values likely due to differences in minor allele frequencies (Figure 2). To determine if the two regions might be statistically representing the same underlying association, we performed a conditional analysis. The associations in this region were conditioned on the NOTCH4 SNP, rs2071286, and the other associations in this region were attenuated to P<10−4. (Figure 3) When the associations were conditioned on the top HLA Class II SNP, rs9461776, the other associations in the region were also attenuated to P<10−4. These results do not clarify if the underlying causal allele in this region is attributable to NOTCH4 or to the HLA Class II alleles.
Figure 2.
Linkage disequilibrium for SNPs with P<10−5 in terms of D’ (red) and r2 (grey). The top SNPs’ pairwise linkage disequilibrium measures are highlighted in yellow. Of note is the HLA Class II SNP that is in long-range LD with the NOTCH4 SNPs to the left. Genes are depicted at the top and the corresponding SNPs are below in the LD plot. Black Triangles depict blocks of LD.
Figure 3. Conditional Associations in NOTCH4 and HLA Class II Region.
The original associations are shown in black, with the associations conditioning on the top NOTCH4 SNP shown in orange. The associations conditioning on the top HLA Class II SNP is shown in blue. Genome-wide significance is indicated with the gray dashed line (P=5×10−7). A threshold of 10−4 is shown with the lower dashed line
Discussion
In this investigation we found strong evidence of a host genetic basis for MC vasculitis focused in and around the MHC class II and NOTCH4 genes. Although there have been reported associations of MC vasculitis with the Class II MHC region the findings have been inconsistent. Cacoub and colleagues found an association of the HLA class II allele DRB1*11 with HCV-related cryoglobulinemia, although Amoroso and colleagues failed to find a significant association with HLA DR or DQ loci.26, 27 In a separate study by De Re and colleagues, DR5 and DQ3 alleles were associated with HCV-related cryoglobulinemic vasculitis.29 In a study of 25 HCV positive MC vasculitis patients and 407 controls, Lenzi and coworkers suggested an HLA-B8-DR3 haplotype associated with susceptibility to MC vasculitis, partially confirmed by a Chinese report.23, 24 The complexity of the MHC region, the ethnic differences in populations and the smaller sample sizes in the earlier studies may also contribute to the differences in association results. In this genome wide association study of European individuals we do identify a significant association in the extended HLA Class II region that is independent of the recently associated HLA Class II HCV viral clearance region.28
However, the nearby NOTCH4 gene also shows association with MC vasculitis in the current study. The Notch proteins are a family of transmembrane proteins (Notch 1–4) that serve as receptors for a signal transduction pathway. The Notch genes are involved with promotion and inhibition of cell differentiation and gene expression in a positive feedback loop. Genome-wide associations have shown significant associations of NOTCH4 with systemic sclerosis, myeloperoxidase levels, age related macular degeneration, asthma and schizophrenia.30, 31 Additionally independently of HLA genes, NOTCH4 polymorphisms have been associated with autoimmune disorders.32, 33 The importance of Notch signaling in T cell development makes it a viable candidate gene for MC vasculitis. However, the SNPs that mark NOTCH4 and MHC class II are too strongly correlated to discriminate the degree to which each region contributes to the association, and there is no tractable experimental model of MC vasculitis that can be used to dissect the association further.
In summary, based on our GWAS data based on a large sample of MC vasculitis patients and controls identified two significant regions with implicated polymorphisms: MHC Class II and NOTCH4. However, we do not have evidence to indicate the causal variants for these associations. Each of these genes may play a role but further investigation is needed to determine the biologic link to this syndrome.
Materials and Methods
Subjects
The case participants in both the discovery and replication stages were enrolled in 9 different studies using study-specific case definitions (Supplementary methods and Supplementary Table 1). A total of 1,074 individuals were identified from the following studies: Toulouse Cohort, BAHSTION (Boston Acute HCV Study: Transmission, Immunity and Outcomes Network), Charles, RomeCryo, Mangia, REVELL (Correlates of Resolved Versus Low-Level Viremic Hepatitis C Infection in Blood Donors), Cacoub (Paris VIREP-C Study), United Kingdom Drug Use cohort and MASVE (Center for Systemic Manifestations of Hepatitis Viruses) (see Supplemental Methods). Controls were individuals with chronic HCV infection from a previously published GWAS.28 The research was approved by the relevant institutional review boards for each study.
Genotyping
In the initial analysis, 3,454 individuals were genotyped on the Illumina Human Omni-Quad array at the Center for Inherited Disease Research (CIDR). Samples included both cases and controls that were recruited specifically for MC vasculitis (N=497), as well as a previously published GWAS for spontaneous HCV resolution versus chronic infection (N=2,957). A total of 1,134,514 SNPs were released with genotype and intensity data. Standard quality control measures were performed for both the individual and SNP. Individuals were excluded if the percentage of missing data was >2% and/or individuals were more related than expected (1st degree relatives). Markers were excluded if the percentage of missing genotypes was > 5%, the minor allele frequency (MAF) was < 1% or there were strong deviations from Hardy-Weinberg equilibrium (p > 10−5). Principal components analysis (PCA) was used to determine genetic ancestry using 41,871 autosomal independent SNPs, excluding regions known to be associated with ethnicity, such as the extended HLA region and the lactase gene. Only individuals that clustered with European ancestry were included in further analyses (N=983). Within individuals of European-descent, a gradient was apparent from the United Kingdom to Italy, with the French participants clustering in the middle (Supplemental Figure1A and 1B). This is consistent with prior studies showing a North-South ancestral cline in Europe.34 The association between the principal components (PCs) and outcome was statistically significant for only the first two PCs (P<0.05), therefore both PCs were included in all genome-wide analyses. The inflation factor (λ) after adjusting for the first two PCs and sex was 0.97, suggesting there was no genomic inflation due to confounding. The final genome-wide data set included 983 individuals (356 cases and 627 controls) at 899,641 locations across the genome.
Statistical Analysis
The initial analysis was the discovery phase and included cryoglobulinemia-associated vasculitis cases (n=356) and the HCV GWAS controls (n=447) with similar ancestral clustering (PCA determined) and chronic HCV infection from 8 different studies (Toulouse Cohort, BAHSTION, RomeCryo, Mangia, REVELL, Cacoub (Paris VIREP-C Study), the UK Drug Cohort, and MASVE). The controls consisted of individuals with chronic HCV infection from the HCV GWAS (N=355), as well as a set of controls recruited specifically for this study (N=92). These latter were consecutively recruited patients with chronic HCV infection showing complete absence of features of mixed cryoglobulinemic vasculitis or other autoimmune and/or lymphoproliferative disorders (i.e, purpura, arthralgia, asthenia, polyneuropathy,nephritis, sicca syndrome, cryoglobulinemia, rheumatoid factor activity, complement consumption, autoantibodies, monoclonality) confirmed during at least two years of follow-up in two yearly evaluations.
All tests of association were conducted in the statistical software, Plink.35 GWAS was performed using logistic regression, under an additive genetic model adjusting for the first two PCs and sex. A significance threshold of P <5×10−7 was used to assess genome-wide significance.36 The −log10 P-values from each SNP’s logistic regression was plotted to generate a Manhattan plot (Figure 1). A conditional analysis was conducted using the top-associated SNPs (rs2071286, rs9461776) by adding the SNP as a covariate to the logistic regression and evaluating the association.
Imputation
SNP Imputation was conducted to increase the density of untyped markers around significant and suggestive association signals using the statistical program IMPUTE237 using a multi-ethnic reference panel from four continents (Africa, North and South America, Asia, and Europe) as part of the 1000 Genomes Project.38 The imputed genotypes were analyzed using logistic regression under an additive model adjusting for the first two principal components and sex. An Expectation-Maximization (EM) algorithm was used to incorporate the probabilities of each genotype in the logistic regression.
Replication
To confirm the novel findings from the discovery phase of the GWAS, a replication study was performed using convenience samples for the cases and a subset of controls with chronic HCV from the previously published GWAS. Controls were selected to be matched 2:1 with the cases based on their study’s country-of-origin. (Table 1) SNPs were genotyped that had reached either genome-wide significance (P<5×10−7) or were SNPs in linkage disequilibrium with the genome-wide significant SNPs and had suggestive P-values (P<5×10−6). A total of 4 SNPs were analyzed for replication in 92 cases and 179 controls from 7 study sites including one new site not represented in the discovery stage (Toulouse Cohort, BAHSTION, Charles, RomeCryo, Mangia, Cacoub (Paris VIREP-C Study), and MASVE). Using the standard Qiagen spin protocol, DNA for genotyping was extracted from stored cells using the QIAamp DNA Blood Mini Kit (Qiagen, Germantown, MD). Genotyping for single nucleotide polymorphisms (SNPs) was performed with TaqMan SNP genotyping assays (Life Technologies, Carlsbad, CA), which uses unique primers and probes sets to target each polymorphism. Taqman’s standard protocol for 5.0ul reactions was utilized. The Roche LightCycler 480 Real-Time System (Roche Applied Science, Indianapolis, IN) was used to amplify the DNA and measure the fluorescence from each reaction to determine the genotype using Roche’s Endpoint Genotyping Software.
Tests of association were conducted in Plink using logistic regression, adjusted for study country-of-origin and sex. A conventional P-value threshold was used for replication (P≤0.01). A meta-analysis was conducted between the discovery and replication associations using a fixed-effects model in META39, 40 and evaluated for heterogeneity (I2). Linkage disequilibrium was estimated in Haploview.
Supplementary Material
Acknowledgments
This study was supported in part by research funding from Merck (DT, PD), through the National Institutes of Health, R01DA013324 (DT) and the Office of AIDS Research. The REVELL study was funded by a grant from the National Heart, Lung and Blood Institute (R01HL076902). This work was supported in part by the National Institutes of Health (R01 AI060561 to L.B.D., K08AI075031 to E.D.C.), Center for Translational Science Award (Pilot Grant CCL3001018) (E.D.C.), and Center for Translational Science Award (grant UL1 RR024143, to Rockefeller University), from the National Center for Research Resources, a component of National Institutes of Health. GML and AYK are supported by grants from the National Institutes of Health, DA033541 AI066345, AI082630. The work was supported by a Wellcome Trust SCF grant (WT076991MA) to SIK. The study was funded by AIRC (Associazione Italiana per la Ricerca sul Cancro), by ITT (Istituto Toscano Tumori) and by Fondazione Cassa di Risparmio di Firenze; Dr LG was supported by a fellowship from Fondazione Umberto Veronesi.
Footnotes
Authorship and Conflict of Interest Statements
ALZ, PC, MV, MC, AM, EC, LG, BT, VP, LBD, SIK, MPB, GML, AYK, LA, DLT collected data. DLT, GLW, PD, RL designed the research analysis and performed the statistical analysis. RL and DT designed and performed the targeted genotyping. DLT, GLW, ALZ, LG, and PD wrote the manuscript. All authors interpreted the data and revised the manuscript.
P Cacoub is a board member at BMS, Boehringer Ingelheim, GSK, Gilead, Janssen, MSD, Roche, Servier, Vifor; Speaker for Astra Zeneca, BMS, Boehringer Ingelheim, GSK, Gilead, Janssen, MSD, Roche, Servier, Vifor; and he receives grants from BMS, GSK, Gilead, MSD, Roche, Servier, Vifor. No other authors declare a conflict of interest.
References
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57(4):1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Zignego AL, Giannini C, Monti M, Gragnani L. Hepatitis C virus lymphotropism: lessons from a decade of studies. Dig Liver Dis. 2007;39(Suppl 1):S38–S45. doi: 10.1016/s1590-8658(07)80009-0. [DOI] [PubMed] [Google Scholar]
- 3.Terrier B, Cacoub P. Cryoglobulinemia vasculitis: an update. Curr Opin Rheumatol. 2013;25(1):10–18. doi: 10.1097/BOR.0b013e32835b15f7. [DOI] [PubMed] [Google Scholar]
- 4.Zignego AL, Giannini C, Gragnani L. HCV and lymphoproliferation. Clin Dev Immunol. 2012;2012:980–942. doi: 10.1155/2012/980942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monti G, Pioltelli P, Saccardo F, Campanini M, Candela M, Cavallero G, et al. Incidence and characteristics of non-Hodgkin lymphomas in a multicenter case file of patients with hepatitis C virus-related symptomatic mixed cryoglobulinemias. Arch Intern Med. 2005;165(1):101–105. doi: 10.1001/archinte.165.1.101. [DOI] [PubMed] [Google Scholar]
- 6.Sansonno D, De Vita S, Iacobelli AR, Cornacchiulo V, Boiocchi M, Dammacco F. Clonal analysis of intrahepatic B cells from HCV-infected patients with and without mixed cryoglobulinemia. J Immunol. 1998;160(7):3594–3601. [PubMed] [Google Scholar]
- 7.Zignego AL, Gragnani L, Giannini C, Laffi G. The hepatitis C virus infection as a systemic disease. Intern Emerg Med. 2012;7(Suppl 3):S201–S208. doi: 10.1007/s11739-012-0825-6. [DOI] [PubMed] [Google Scholar]
- 8.Ferri C, Zignego AL, Bombardieri S, La Civita L, Longombardo G, Monti M, et al. Etiopathogenetic role of hepatitis C virus in mixed cryoglobulinemia, chronic liver diseases and lymphomas. Clin Exp Rheumatol. 1995;13(Suppl 13):S135–S140. [PubMed] [Google Scholar]
- 9.Cicardi M, Cesana B, Del Ninno E, Pappalardo E, Silini E, Agostoni A, et al. Prevalence and risk factors for the presence of serum cryoglobulins in patients with chronic hepatitis C. J Viral Hepat. 2000;7(2):138–143. doi: 10.1046/j.1365-2893.2000.00204.x. [DOI] [PubMed] [Google Scholar]
- 10.Ferri C. Mixed cryoglobulinemia. Orphanet J Rare Dis. 2008;3:25. doi: 10.1186/1750-1172-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferri C, Sebastiani M, Giuggioli D, Cazzato M, Longombardo G, Antonelli A, et al. Mixed cryoglobulinemia: demographic, clinical, and serologic features and survival in 231 patients. Semin Arthritis Rheum. 2004;33(6):355–374. doi: 10.1016/j.semarthrit.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Ferri C, Monti M, La Civita L, Longombardo G, Greco F, Pasero G, et al. Infection of peripheral blood mononuclear cells by hepatitis C virus in mixed cryoglobulinemia. Blood. 1993;82(12):3701–3704. [PubMed] [Google Scholar]
- 13.Pietrogrande M, De Vita S, Zignego AL, Pioltelli P, Sansonno D, Sollima S, et al. Recommendations for the management of mixed cryoglobulinemia syndrome in hepatitis C virus-infected patients. Autoimmun Rev. 2011;10(8):444–454. doi: 10.1016/j.autrev.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Saadoun D, Asselah T, Resche-Rigon M, Charlotte F, Bedossa P, Valla D, et al. Cryoglobulinemia is associated with steatosis and fibrosis in chronic hepatitis C. Hepatology. 2006;43(6):1337–1345. doi: 10.1002/hep.21190. [DOI] [PubMed] [Google Scholar]
- 15.De Vita S, Quartuccio L, Isola M, Mazzaro C, Scaini P, Lenzi M, et al. A randomized controlled trial of rituximab for the treatment of severe cryoglobulinemic vasculitis. Arthritis Rheum. 2012;64(3):843–853. doi: 10.1002/art.34331. [DOI] [PubMed] [Google Scholar]
- 16.Cacoub P, Terrier B, Saadoun D. Hepatitis C virus-induced vasculitis: therapeutic options. Ann Rheum Dis. 2014;73(1):24–30. doi: 10.1136/annrheumdis-2013-203883. [DOI] [PubMed] [Google Scholar]
- 17.Dammacco F, Sansonno D. Therapy for hepatitis C virus-related cryoglobulinemic vasculitis. N Engl J Med. 2013;369(11):1035–1045. doi: 10.1056/NEJMra1208642. [DOI] [PubMed] [Google Scholar]
- 18.Bianchettin G, Bonaccini C, Oliva R, Tramontano A, Cividini A, Casato M, et al. Analysis of hepatitis C virus hypervariable region 1 sequence from cryoglobulinemic patients and associated controls. J Virol. 2007;81(9):4564–4571. doi: 10.1128/JVI.02104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Invernizzi R, Nano R, Ippoliti G, Girino M, Gerzeli G. Cytochemical study of tetrahydrofolate dehydrogenase in chronic lymphocytic leukemia cells. Haematologica. 1983;68(6):742–749. [PubMed] [Google Scholar]
- 20.Monti G, Saccardo F, Pioltelli P, Rinaldi G. The natural history of cryoglobulinemia: symptoms at onset and during follow-up. A report by the Italian Group for the Study of Cryoglobulinemias (GISC) Clin Exp Rheumatol. 1995;13(Suppl 13):S129–S133. [PubMed] [Google Scholar]
- 21.Lamprecht P, Gause A, Gross WL. Cryoglobulinemic vasculitis. Arthritis Rheum. 1999;42(12):2507–2516. doi: 10.1002/1529-0131(199912)42:12<2507::AID-ANR2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Pozzato G, Burrone O, Baba K, Matsumoto M, Hijiiata M, Ota Y, et al. Ethnic difference in the prevalence of monoclonal B-cell proliferation in patients affected by hepatitis C virus chronic liver disease. J Hepatol. 1999;30(6):990–994. doi: 10.1016/s0168-8278(99)80251-7. [DOI] [PubMed] [Google Scholar]
- 23.Ossi E, Bordin MC, Businaro MA, Marson P, Bonadonna P, Chiaramonte M, et al. HLA expression in type II mixed cryoglobulinemia and chronic hepatitis C virus. Clin Exp Rheumatol. 1995;13(Suppl 13):S91–S93. [PubMed] [Google Scholar]
- 24.Lenzi M, Frisoni M, Mantovani V, Ricci P, Muratori L, Francesconi R, et al. Haplotype HLA-B8-DR3 confers susceptibility to hepatitis C virus-related mixed cryoglobulinemia. Blood. 1998;91(6):2062–2066. [PubMed] [Google Scholar]
- 25.Hwang SJ, Chu CW, Huang DF, Lan KH, Chang FY, Lee SD. Genetic predispositions for the presence of cryoglobulinemia and serum autoantibodies in Chinese patients with chronic hepatitis C. Tissue Antigens. 2002;59(1):31–37. doi: 10.1034/j.1399-0039.2002.590106.x. [DOI] [PubMed] [Google Scholar]
- 26.Amoroso A, Berrino M, Canale L, Cornaglia M, Guarrera S, Mazzola G, et al. Are HLA class II and immunoglobulin constant region genes involved in the pathogenesis of mixed cryoglobulinemia type II after hepatitis C virus infection? J Hepatol. 1998;29(1):36–44. doi: 10.1016/s0168-8278(98)80176-1. [DOI] [PubMed] [Google Scholar]
- 27.Cacoub P, Renou C, Kerr G, Hüe S, Rosenthal E, Cohen P, et al. Influence of HLA-DR phenotype on the risk of hepatitis C virus-associated mixed cryoglobulinemia. Arthritis Rheum. 2001;44(9):2118–2124. doi: 10.1002/1529-0131(200109)44:9<2118::AID-ART364>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 28.Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med. 2013;158(4):235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Re V, Caggiari L, De Vita S, Mazzaro C, Lenzi M, Galli M, et al. Genetic insights into the disease mechanisms of type II mixed cryoglobulinemia induced by hepatitis C virus. Dig Liver Dis. 2007;39(Suppl 1):S65–S71. doi: 10.1016/s1590-8658(07)80014-4. [DOI] [PubMed] [Google Scholar]
- 30.Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tazi-Ahnini R, Cork MJ, Wengraf D, Wilson AG, Gawkrodger DJ, Birch MP, et al. Notch4, a non-HLA gene in the MHC is strongly associated with the most severe form of alopecia areata. Hum Genet. 2003;112(4):400–403. doi: 10.1007/s00439-002-0898-9. [DOI] [PubMed] [Google Scholar]
- 32.Kochi Y, Yamada R, Kobayashi K, Takahashi A, Suzuki A, Sekine A, et al. Analysis of single-nucleotide polymorphisms in Japanese rheumatoid arthritis patients shows additional susceptibility markers besides the classic shared epitope susceptibility sequences. Arthritis Rheum. 2004;50(1):63–71. doi: 10.1002/art.11366. [DOI] [PubMed] [Google Scholar]
- 33.Valdes AM, Thomson G, Consortium TDG. Several loci in the HLA class III region are associated with T1D risk after adjusting for DRB1-DQB1. Diabetes Obes Metab. 2009;11(Suppl 1):46–52. doi: 10.1111/j.1463-1326.2008.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.