Abstract
A variety of cell types exhibit phenotype changes in response to the mechanical stiffness of the substrate. Many cells excluding neurons display increase in spread area, actin stress fiber formation and larger focal adhesion complexes as substrate stiffness increases in sparsely populated culture. Cell proliferation is also known to directly correlate with these phenotype changes/change in substrate stiffness. Augmented spreading and proliferation on stiffer substrates require nuclear transcriptional regulator YAP (Yes associated protein) localization in cell nucleus and is tightly coupled with larger traction force generation. In this study, we show that different types of fibroblasts can exhibit spread morphology, well defined actin stress fibers, and larger focal adhesion seven on very soft collagen gels (modulus in hundreds of Pascals)as if they are on hard glass substrate (modulus in GPa, several orders of magnitude higher). Strikingly, we show, for the first time, that augmented spreading and other hard substrate cytoskeleton architecture on soft collagen gels are not correlated with cell proliferation pattern and do not require YAP localization in cell nucleus. Finally, we examine the response of human colon carcinoma (HCT-8) cells on soft collagen gels. Recent studies show that human colon carcinoma (HCT-8) cells form multi cellular clusters by 2–3 days when cultured on soft polyacrylamide (PA) gels with a wide range of stiffness (0.5 – 50 kPa) and coated with extracellular matrix, ECM (collagen monomer/ fibronectin). These clusters show limited spreading/wetting on PA gels, form 3D structures at the edges, and eventually display a remarkable, dissociative metastasis like phenotype (MLP), i.e., epithelial to rounded morphological transition after a week of culture on PA gels only, but not on collagen monomer coated stiff polystyrene/glass where they exhibit enhanced wetting and form confluent monolayer. Here, we show that HCT-8 cell clusters also show augmented spreading/wetting on soft collagen gels and eventually form confluent monolayer as on rigid glass substrates and MLP is completely inhibited on soft collagen gels. Overall, these results suggest that cell-material interaction (soft collagen gels in this case) can induce cellular phenotype and cytoskeleton organization in a remarkably distinct manner compared to a classical synthetic polyacrylamide (PA) hydrogel cell culture model and may contribute in designing new functional biomaterials.
Keywords: Substrate stiffness, collagen gel, cell proliferation, Yes associated protein (YAP), cell mechanics
Introduction
In recent years it has become increasingly evident that mechanical micro-environment, i.e., substrate rigidity plays an important role in regulating cell functionalities. Cells can sense and respond to the substrate stiffness on which they are adhered to (as in two dimensional or 2D culture) or surrounded by (as in three dimensional or 3D culture).1–16 By doing so, cells can modulate their differentiation,3 morphology,4–6 migration/motility,9,13 bio-physical properties,16 growth,15 and other processes.10, 17
Many cell types such as fibroblasts,4 cardiac myocytes,18 and glioma cells19 show an increase in spread area as substrate elasticity increases in sparsely populated culture. In addition to morphology, cellular cytoskeleton organization at single cell scale is also mediated by substrate rigidity. Cells show well defined actin stress fibers on stiffer substrates only.4,19 Conversely, cortical actin is primarily observed on softer substrates.4,19 Further, cells show discrete, elongated and larger focal adhesion complexes on hard substrates.9,19 Whereas, small, punctate, and dot like focal adhesions are generally visualized on softer substrates.9,19
Cell proliferation rate also increases with increase of substrate modulus in general and is shown to be tightly coupled with enhanced traction force generation on stiffer substrates.15 Recent discovery demonstrates important role of nuclear transcriptional regulator YAP in cellular mechano transduction process.20 YAP is primarily localized in cytoplasmic region in less spread cells on soft polyacrylamide gels.20,21 Conversely, it becomes localized primarily in nucleus in well spread cells on stiffer substrates.20,21 Hence, one can infer that force dependent augmented cellular spreading, well defined actin stress fibers, and focal adhesions formation are directly correlated with higher cell proliferation rate and YAP localization in cell nucleus.
Recent experiment shows that fibroblasts can spread on soft fibrin gels of low modulus as if they are on substrates with very high modulus (glass).22 It has been hypothesized that cell mediated local stiffening of non-linear elastic fibrin gels result augmented spreading and hence the phenomenon is force dependent.21 This hypothesis is refuted by another recent paper which uses non-linear elastic material modeling to claim that non-linear strain stiffening alone cannot explain such spreading on fibrous soft gels.23 However, none of these studies explored the experimental correlation of cell proliferation and nuclear transcriptional regulator YAP activity with cell morphology and detailed cytoskeleton organization. It is conceivable that if augmented spreading and hard substrate cytoarchitectureon fibrous biological gels (e.g., fibrin/collagen) are force mediated, cell proliferation on these soft gels is expected to be higher. Also, YAP must be localized in cell nucleus. Otherwise, it can be implied that cells interact with these fibrous biological gels in a unique manner that induce hard substrate like cell morphology and cytoskeleton organization without the need for high force.
In addition to single cell spreading, multicellular aggregate spreading/wetting on ECM coated substrates is mediated by substrate stiffness as well.24 On softer substrates, cell clusters show less wetting, i.e. don’t spread well. Conversely, cell clusters show enhanced wetting/spreading on stiffer substrates presumably due to increased cell-substrate adhesions compared to cell-cell adhesions.24 The hypothesized mechanism is explained with the value of a single parameter, S = Wcs–Wcc, where Wcs and Wcc represent cell-substrate and cell-cell adhesions energy respectively. Complete wetting occurs for S > 0. For S < 0, partial wetting takes place. Recent studies show that human colon carcinoma (HCT-8) cells cultured on soft PA gels(0.5 kPa-50 kPa), functionalized by ECM, form multicellular clusters with well-defined boundaries within 2 – 3 days due to less wetting.1, 25 Cells from the cluster dissociate from one another after a week of culture, starting from the periphery. As this metastasis like phenotype (epithelial to rounded, E-R morphological transition) occurs, they reduce cell-cell and cell-ECM adhesion, and proliferate. However, on hard substrates, functionalized by ECM, HCT-8 cells form confluent monolayer and do not exhibit any metastaticphenotype transition. The formation of bounded clusters on soft gels and mono layers on hard substrates might be due to the difference between the cell-substrate wettability for the two types of substrates.1, 25 E-R transition might be a consequence of this wettability and not due to low force on soft substrates. If so, then HCT-8 cells on adhesive soft collagen gels may not show the transition as well.
Here, we show for the first time that fibroblasts can display hard substrate like cell morphology and cytoskeleton organization on very soft fibrous collagen gels, without YAP localization in cell nucleus. YAP is localized in cytoplasmic region and cell proliferation rate is low, as expected on soft substrates. Finally, we show that HCT-8 cells on soft collagen gels also display hard substrate like phenotype, i.e., augmented spreading, and confluent monolayer formation and no E-R transition. Overall, these results suggest that cell-material interaction (soft collagen gel in this case) can induce cellular phenotype and cytoskeleton organization in a remarkably distinct manner compared to a classical synthetic polyacrylamide hydrogel cell culture model and may contribute in designing new functional biomaterials.
Materials and methods
Collagen gel preparation
Formulation and synthesis of collagen gels were performed using a protocol described elsewhere.26
Briefly, collagen gels were synthesized using high concentration collagen-I from rat tail (BD Biosciences, San Jose, CA). Collagen-I was diluted to two final concentrations of 2 and 4 mg/mL as follows. Equal volume of collagen-I and 100 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer solution in 2X phosphate buffered saline, PBS (pH 7.3) were mixed to reach the final concentration. Gel solution was then placed on a 35 mm glass bottom petri dish (In vitro scientific, Sunnyvale, CA) and allowed to polymerize completely for 90 mins at 37°C and 5% CO2. Consequently, cells were seeded on polymerized gels and were incubated at 37°C and 5% CO2. The reported shear modulus values were ~104 and 391 Pa corresponding to final collagen concentrations of 2 and 4 mg/ml in precursor solution.26
PA gel and functionalized glass preparation
PA gel preparation and glass cover slip activation for the covalent attachment of gels were performed following the protocols described elsewhere.27–29 In brief, 2 kPa gel solution was obtained by mixing 5% w/v acryl amide (Sigma, St. Louis, MO) and 0.05% N, N′-methylenebisacrylamide (bis) solutions (Midwest Scientific, MO) in 10 mM HEPES-buffered saline (Lonza, Walkersville, MD).30 8% w/v acryl amide and 0.27% N, N′-methylenebisacrylamide (bis) solution in 10 mM HEPES-buffered saline were used for 20 kPa gels.30 Finally, 8% w/v acryl amide and 0.48% N, N′-methylenebisacrylamide (bis) solution in 10 mM HEPES-buffered saline were used for 40 kPa gels.30 1:200 ammonium persulfate (10% w/v) and 1:2000 N, N, N′, N′-tetramethylethylenediamine (TEMED) (both from Bio-Rad Laboratories, Hercules, CA) were used as the initiator and catalyst for polymerization process, respectively.
Glass cover slips were chemically activated to ensure covalent binding of the hydrogel as described earlier.28 Briefly, glass slips were treated with 3-Aminopropyltrymethoxysilane (ATS) from Sigma, MO, for 7 min at room temperature. Followed by the removal of the ATS completely with DI water rinse, cover slips were treated with 0.5% Glutaraldehyde (diluted in PBS from 70% Glutaraldehydestock solution from Polysciences, Inc,) solution for 30 min. A drop of 20 µL pre-polymer PA gel solution was deposited on the 12 mm2 activated glass cover slip. Another12 mm2 regular glass cover slip was placed (floated) on the drop. The drop spread between the cover slips due to capillarity and was sandwiched with uniform thickness. Curing of the PA gel was performed for 45 min sat room temperature. The top cover slip was manually peeled off using a single edge razor. During peeling, detachment proceeded from one edge of the sandwich.
PA gels and glass were functionalized with ECM molecules, rattail collagen (BD biosciences) at a concentration of 100 µg/ml. The surface functionalization protocol for binding collagen was described elsewhere.28 Briefly, substrates were incubated with pure hydrazine hydrate (Sigma-Aldrich, MO) overnight. Substrates were then washed with 5% acetic acid (Sigma-Aldrich, MO) and DI water for 1 hour. Collagen was deposited on top of substrates overnight at 4°C at a concentration of 100 µg/ml and rinsed with PBS on shaker for 10 minutes. All substrates were incubated at 37°C in culture media for 30 minutes before plating the cells.
Cell culture
3T3 fibroblasts (3T3Fs) and monkey kidney fibroblasts (MKFs) (ATCC, Manassas, VA) were cultured in DMEM medium(Cat. No. 30-2002, ATCC, Manassas, VA) supplemented with 10% serum (Cat. No. 30-2040, ATCC, Manassas, VA) and 1% Penicillin-Streptomycin (Cat. No. 30-2300, ATCC, Manassas, VA) of total solution. Human colon carcinoma HCT-8 cells (Cat. No. CCL-244, ATCC, Manassas, VA) were cultured in RPMI 1640 (Cat. No. 30-2001, ATCC, Manassas, VA) base medium supplemented with horse serum (Cat. No. 30-2040, ATCC, Manassas, VA) to a final concentration of 10% and Penicillin-Streptomycin (Cat. No. 30-2300, ATCC, Manassas, VA) to 1% of total solution.
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 15 min. Cells were then incubated with Image-iT FX signal enhancer (Invitrogen, CA) for 30 min. Next, cells were incubated in primary antibodiesagainst α-tubulin (Cat. No. A11126, Invitrogen, CA), vinculin (Cat. No. V9131, Sigma-Aldrich, MO), or YAP (H-9, Santa Cruz) for 45 min at room temperature, respectively. Following three times rinse in PBS, the samples were incubated for 30 min with secondary antibody (alexafluor™ 488 goat anti-mouse IgG/alexafluor™ 647 goat anti-mouse IgG (Invitrogen, CA)) at a 1:200 dilution in PBS at room temperature in each case. To visualize the F-actin structure, cells were incubated with tetramethylrhodamine (TRITC) phalloidin conjugates (Cat. No. P1951, Sigma-Aldrich, MO) at a concentration 50 µg/ml for 45 min at room temperature. To visualize cell nucleus, cells were finally incubated with 4', 6-diamidino-2-phenylindole, DAPI (1:1000) for 15 min at room temperature. All the samples were imaged either using the Zeiss LSM 700 confocal scanning laser microscope (Carl Zeiss, Inc.) or Olympus IX81 microscope.
Proliferation assay
Proliferation assay was performed using the Click-iTEdU (5-ethynyl-2’-deoxyuridine) cell proliferation assay kit (Invitrogen, CA) according to manufacturer’s instructions (http://tools.lifetechnologies.com/content/sfs/manuals/click_it_edu_imaging_kit_man.pdf).31
Briefly, MKFs were plated on soft collagen gels and glass from a confluent T25 flask culture., After 24 hrs of culture, cells were incubated with 10 µM EdU in complete media for 60 minutes.32 Then, cells were fixed, permeabilized, and incubated with alexafluor 488 azides to detect EdU. Finally, cells were counterstained with DAPI. Cells which show double fluorescence (both EdU and nuclei) were considered to be synthesizing new DNA (Deoxyribonucleic acid).
Image analysis and statistical analysis
ImageJ was used for determining cell spreading area.33 Average number of focal adhesions (FAs) per cell and average size of FAs are measured using ImageJ.34 Student’s t test was performed to evaluate statistical significance. Error was reported as standard deviation unless otherwise mentioned.
Results and discussions
Fibroblasts show augmented spreading, well defined actin stress fibers and larger focal adhesions on very soft collagen gels as if they are on stiff glass substrates
Cell spreading
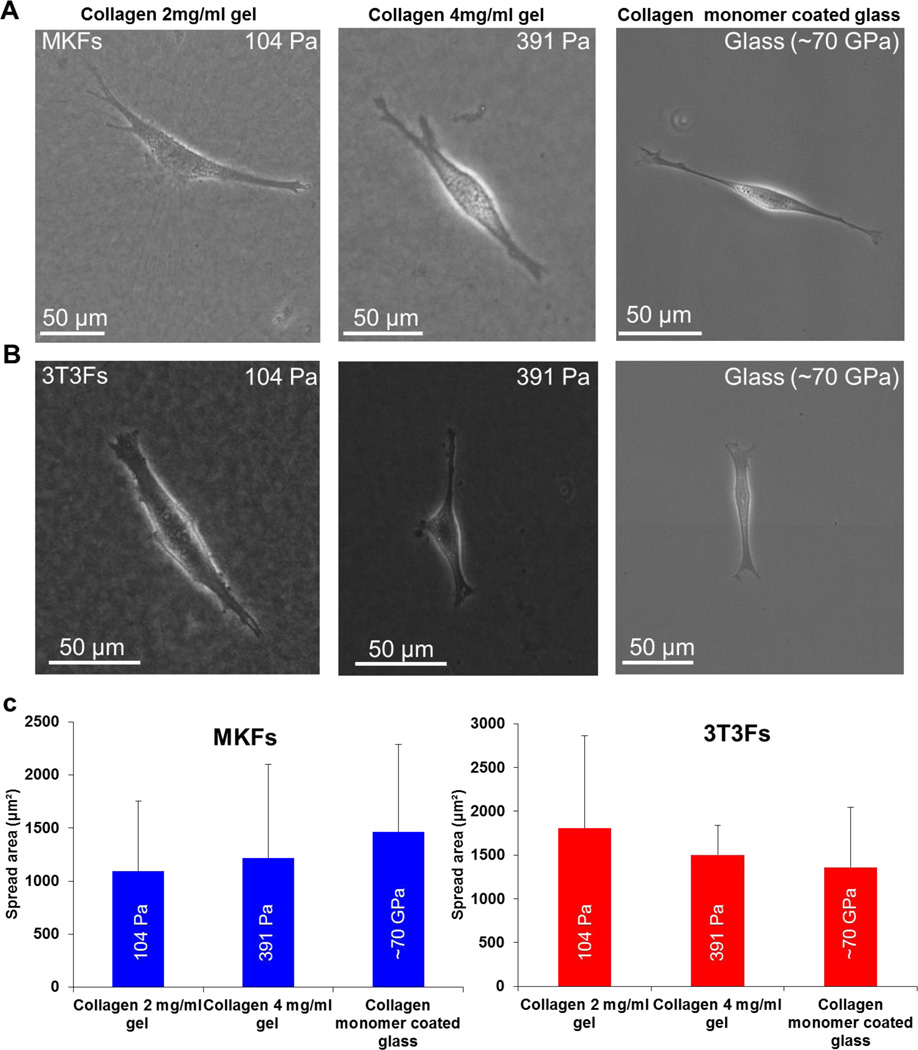
Cell spreading is a known readout of cellular mechano-sensitivity to substrate rigidity.35 Cells remain less spread/rounded on ECM functionalized polyacrylamide hydrogel substrates with low modulus (hundreds of Pa).4,19 Cells show a monotonic increase in area with increase in substrate rigidity unless it reaches a plateau. This maximum value and the corresponding substrate stiffness depend on cell type.4,32 In this study, we culture monkey kidney fibroblasts (MKFs) and 3T3 fibroblasts (3T3Fs) on collagen gels with modulus of 104 Pa and 391 Pa. The gels were~600 µm thick ensuring that the cells do not feel the underlying rigid glass substrate. Collagen monomer coated glass substrate (~ 70 GPa) is used as control. Interestingly, MKFs and 3T3Fs show well spread morphology on both soft collagen gels (Figure 1A-1B) as if they are on rigid glass substrates in sparsely populated culture (Figure 1A-1B).
Figure 1. Cells spread on soft collagen gels as if they are on rigid glass substrate.
(A-B) Phase contrast micrograph showing spread morphology of different types of fibroblasts on soft collagen gels and collagen monomer coated glass. (C) Spread area quantification for collagen gels and glass reveal no statistically significant difference for both MKFs and 3T3Fs (Student’s t test, p > 0.1). Total n = 63 and 67 cells were analyzed for MKFs and 3T3Fs, respectively. At least two independent experiments were performed per condition.
Further, we quantify cell spreading area using ImageJ. Both MKFs and 3T3Fs on soft collagen gels with two different moduli (104 Pa and 391 Pa) show similar quantitative spreading with no statistically significant difference (Figure 1C, Student’s t test, p > 0.1). No statistically significant difference with glass is observed compared to either formulation of collagen gels (Figure 1C, Student’s t test, p > 0.1), consistent with qualitative well spread morphology. This altered spreading behavior on soft collagen gels implies that the interaction between cells and soft collagen gels is distinct compared to ligand coated polyacrylamide hydrogels and glass substrates.
Visualization of Cytoskeleton organization
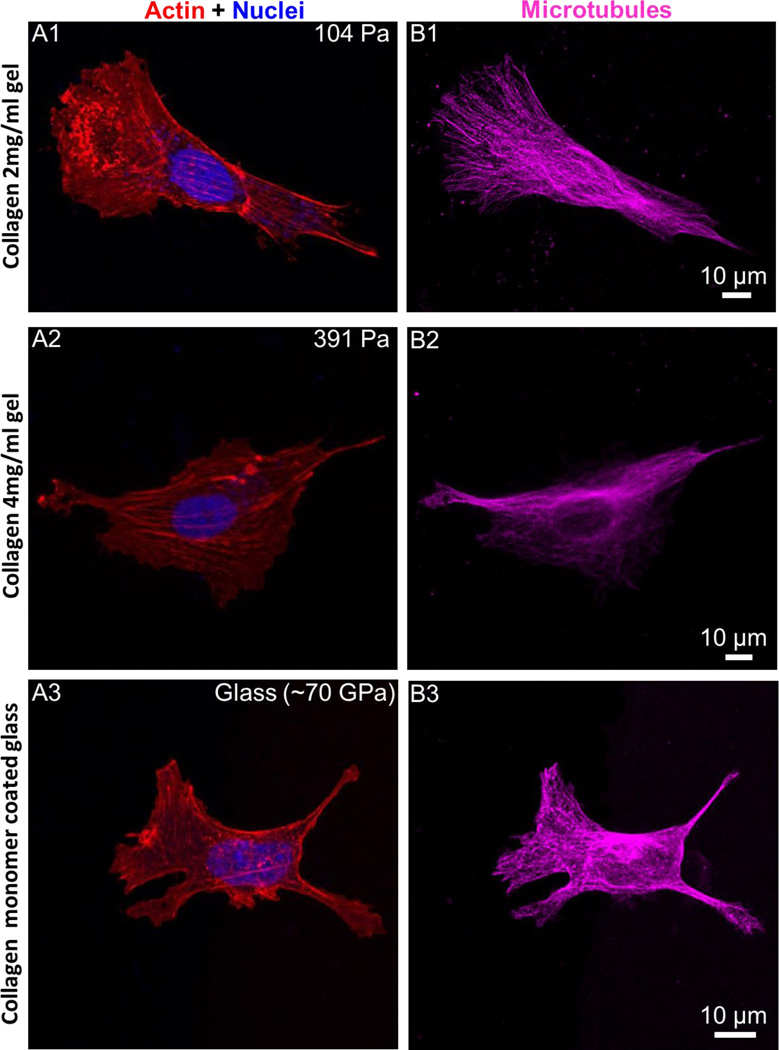
F-actin structure and microtubule organizations of fibroblasts adherent to thick collagen gels and collagen monomer coated glass substrates are visualized by staining with phalloidin and anti α-tubulin antibody respectively after fixation.
Confocal microscopy imaging results suggest that fibroblasts (MKFs) exhibit large and well-organized actin stress fibers on both soft collagen gels and glass (Figure 2A1-2A3). Further, microtubule organization on soft collagen gels also resembles hard substrate architecture (Figure 2B1-2B3). Previous studies suggest that fibroblasts on ECM functionalized compliant PA gels (in a range 0.5-5 kPa stiffness) show mostly cortical actin, but no actin stress fiber.16 Actin stress fibers become visible on stiff PA gels (~ 10 kPa) and larger, well organized stress fiber bundles are apparent on rigid glass substrate in a stiffness dependent manner.16 Here, we show that when we replace the underlying matrix material with soft biological material, namely, collagen gels of different compliance(yet of very low modulus, 104 Pa and 391 Pa), fibroblasts are able to display hard substrate cytoskeleton architecture in addition to augmented spreading as shown in Figure 1.
Figure 2. F-actin, and microtubules organization on soft collagen gels.
(A) Fibroblasts (MKFs)on soft collagen gels show actin stress fibers (A1-A2) similar to rigid glass substrates (A3), (B) microtubule organization on soft collagen gels (B1-B2) also resemble glass substrates (B3). At least three independent experiments were performed per condition.
Focal adhesions
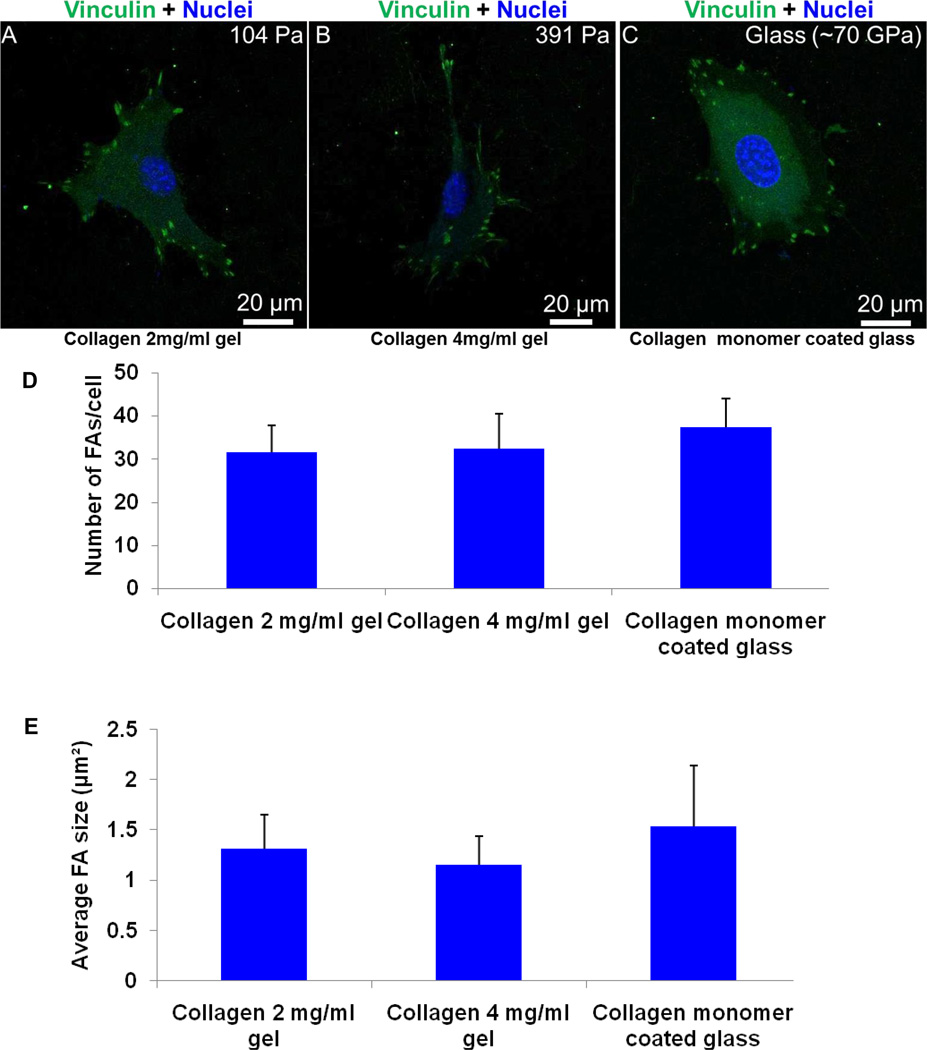
Focal adhesions (FAs) are sites of adhesion expressed by different types of cells in culture. They provide the linkage between the ECM components to intracellular cytoskeleton (F-actin) via integrin receptors. FAs are composed of a variety of proteins, such as vinculin, talin, and paxillin.36 However, vinculin depletion leads to dramatic changes in FA sizes and also in cell functionality.37–39 In addition, vinculin is the most abundant focal adhesion protein.40 Hence, adherent cells on soft collagen gels and collagen monomer coated glass are labeled with anti-vinculin antibody and are imaged via confocal microscopy.
Interestingly, fibroblasts (MKFs) on collagen gels show discrete, elongated focal adhesions as on rigid glass substrates (Figure 3A-3C). Focal adhesion (FA) size and number for each condition are assessed using ImageJ.34 Quantitative results suggest that there is no statistically significant difference in FA size and number (Figure 3D-3E, Student’s t test, p > 0.1), consistent with qualitative imaging results as shown in Figure 3A.
Figure 3. Elongated focal adhesions formation on collagen gels.
Fibroblasts (MKFs) show discrete, elongated focal adhesions on soft collagen gels with modulus of 104 Pa and 391 Pa (A-B) that are generally seen on very hard glass substrates (~70 GPa) (C). At least three independent experiments were performed per condition. (D-E) Quantitative results also suggest no statistically significant difference of FA size and number between each condition (Student’s t test, p > 0.1). Total n = 24 cells were analyzed.
Cells are known to exhibit small, dot like punctate vinculin structures on compliant PA gels.4,9,18 Conversely, discrete and elongated focal adhesions are characteristics of sparsely populated cell culture on very stiff gel or rigid glass substrates.4,9,19 These results imply that unusual fibroblast spreading and actin stress fiber formation on soft collagen gels (Figures 1 and 2) is orchestrated with discrete, elongated and mature focal adhesion formation (Figure 3).
Cell proliferation on soft collagen gels is not correlated with hard substrate morphological and cytoskeleton organization phenotype
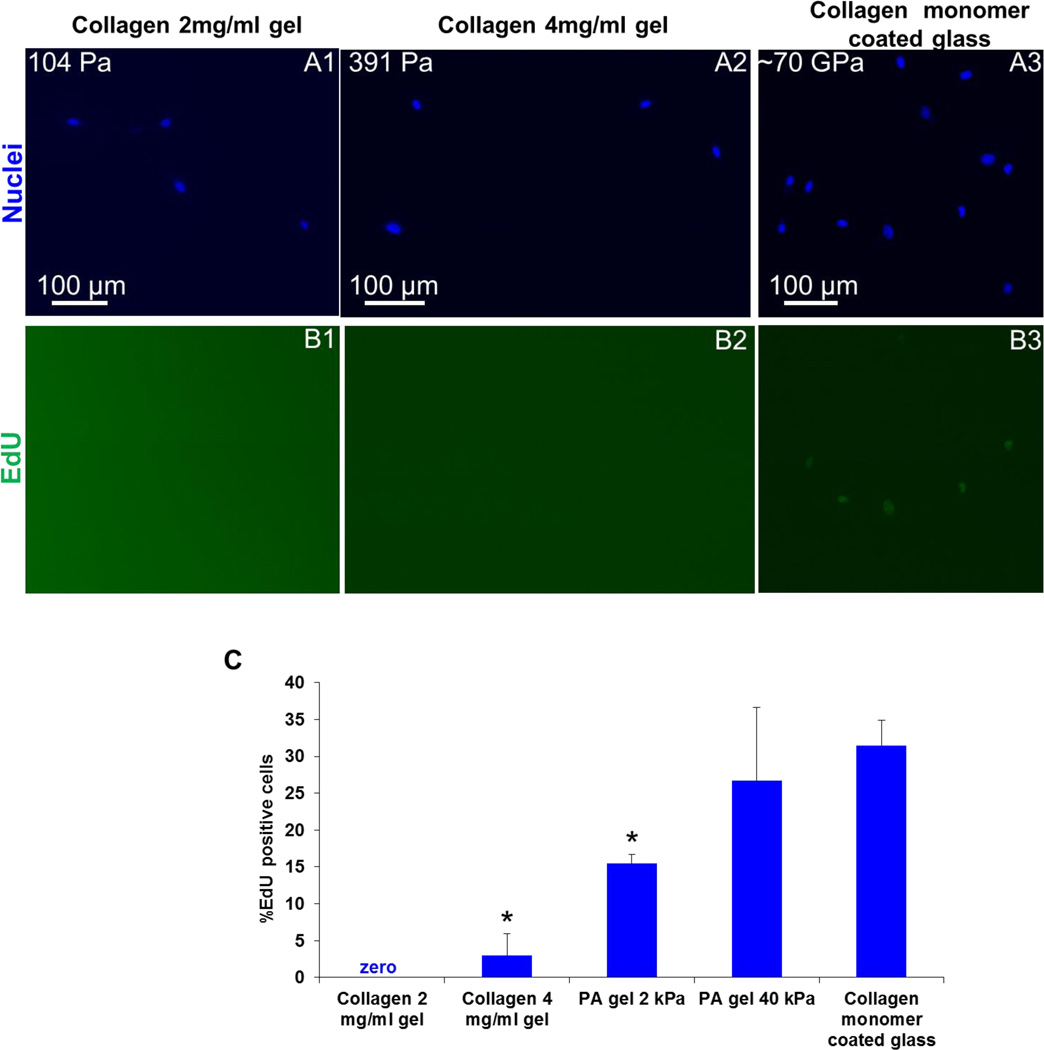
Substrate rigidity modulates cell proliferation rate, i.e., new DNA synthesis.15 It is inferred that cell proliferation rate is directly coupled to cell spreading and traction force generation.15 Hence, we ask the question whether augmented cell spreading and hard substrate cytoskeleton architectureon soft collagen gels can induce increased cell proliferation rate disregarding macroscale/globalmaterial softness. We examine MKFs cell proliferation rate on collagen gels of 104 Pa and 391 Pa, soft (2 kPa) and stiff PA gels (40 kPa), and glass after 24 hrs of cell plating. Our results show that fibroblast cell proliferation rate on collagen gels is very low unlike stiff polyacrylamide gels and glass substrates (Figure 4A-4C). On PA gels, cells show a higher proliferation rate as substrate rigidity increases (Figure 4C), consistent with previously published results.15.
Figure 4. Cell proliferation rate of fibroblasts is very low on soft collagen gels.
Fibroblasts (MKFs) show lower proliferation rate on soft collagen gels and PA gels. However, the proliferation rate is significantly higher on stiff PA gel and glass (Student’s t test, p < 0.05). Total n = 781 cells were analyzed. At least two independent experiments were performed per condition.
Augmented cellular spreading on soft collagen gels doesn’t require YAP localization in cell nucleus
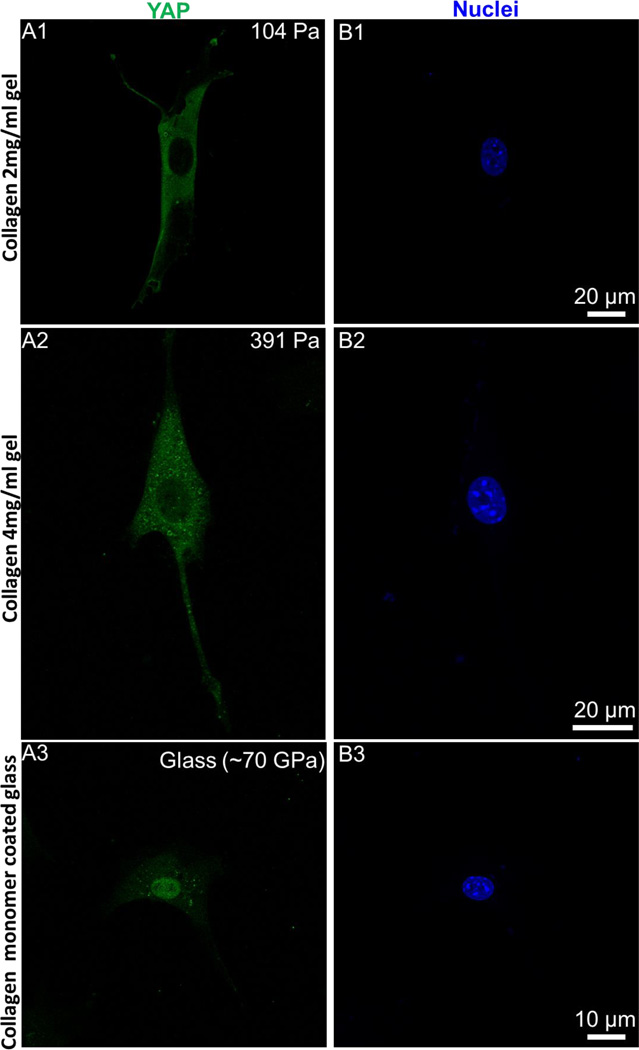
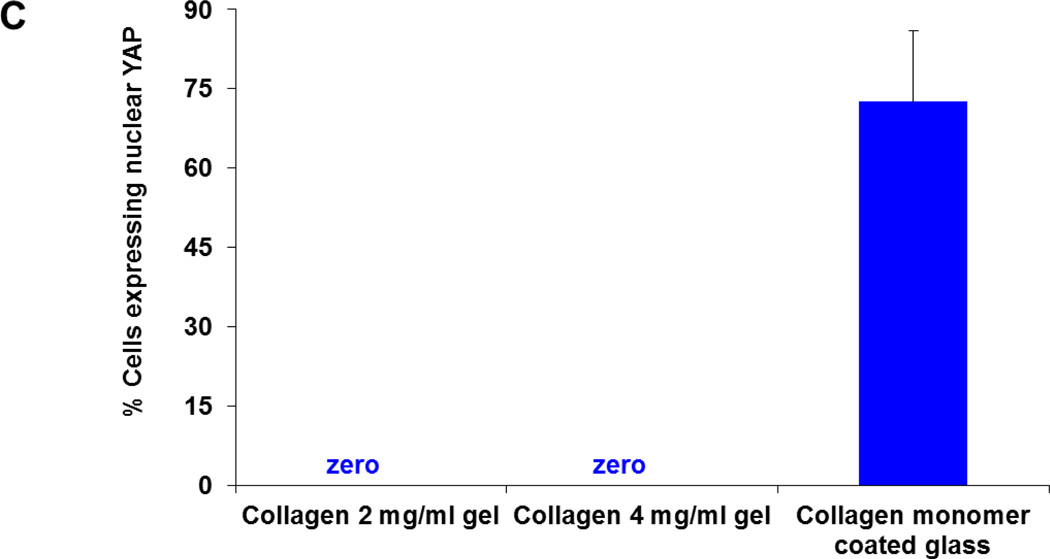
Recent discovery demonstrate that nuclear transcriptional regulator YAP plays an important role in cellular mechanotransduction.20 Traction force mediated augmented spreading either on stiffer substrates or on larger ECM micropatterns require YAP localization at cell nucleus.20 Conversely, for less spread cells that exert smaller traction at cell-substrate interface (either on soft PA gels or on smaller ECM micropatterns on hard substrate), YAP is primarily localized in cytoplasmic region.20 Thus intracellular YAP localization is also tightly coupled with traction force generation. Hence, to further explore the relationship between the spread cell morphology and traction force generation on collagen gels, we label the adherent cells on collagen gels and glass with anti YAP antibody. Imaging results suggest that YAP is primarily localized in the cytoplasmic region in well spread 3T3Fs on soft collagen gels (Figures 5A1-5A2). However, YAP is primarily localized in cell nucleus in well spread cells on glass, consistent with earlier observations.19,20 We further quantify the percentage of cells with YAP localized in nucleus. Consistent with qualitative observations, spread cells on collagen gels clearly don’t require YAP localization in cell nucleus unlike cells on glass (Figure 5C).
Figure 5. YAP is not localized in cell nucleus on soft collagen gels.
(A-B) Despite augmented spreading and hard substrate like cytoskeleton architecture, YAP is not localized in nucleus of fibroblasts (3T3Fs) on soft collagen gels (A1-A2 and B1-B2). Conversely, on hard substrates YAP is primarily localized in cell nucleus (A3-B3). (C) Quantification of percentage of cells expressing nuclear YAP. Consistent with qualitative imaging results, YAP is localized in cytoplasmic region of fibroblasts on soft collagen gels unlike glass substrates. Total n = 176cells were analyzed. At least two independent experiments were performed per condition.
HCT-8 cell clusters show increased wetting as on hard substrate and don’t show MLP on soft collagen gels
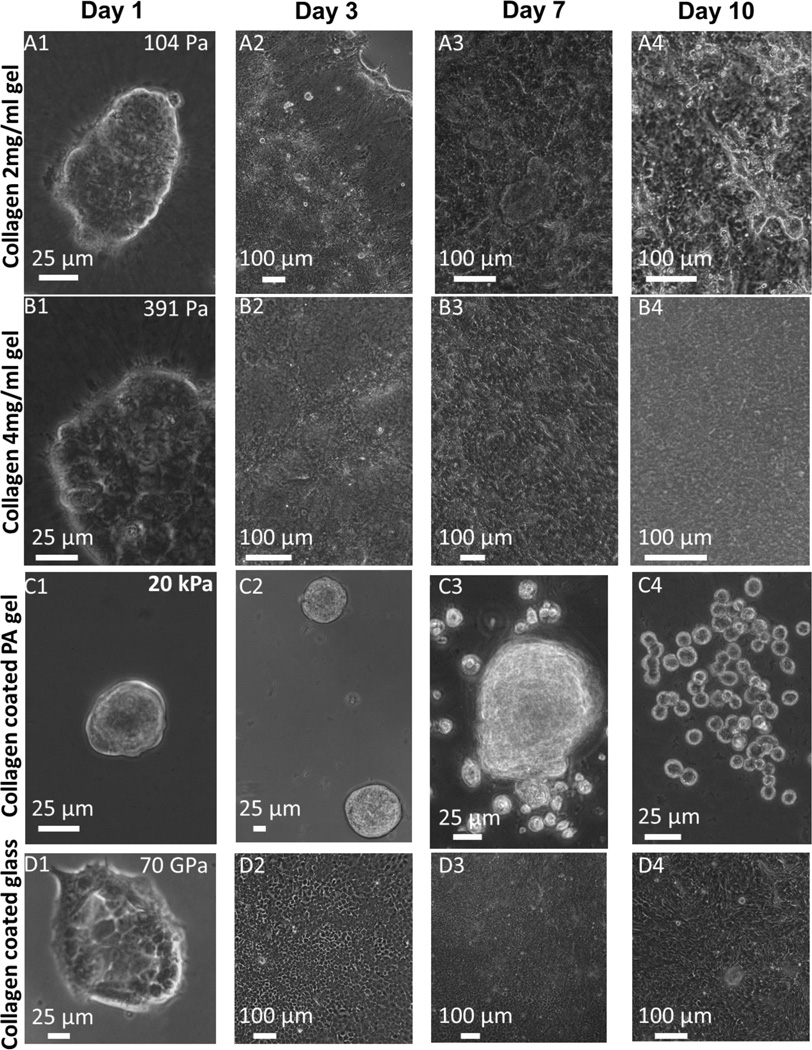
In addition to single cell spreading, cellular aggregate/cluster spreading or wetting is also shown to be regulated by substrate rigidity.24 Cell clusters exhibit poor wetting on ECM functionalized softer substrates.24 Increased wetting of cellular clusters is observed on stiffer substrates only.24 The hypothesized mechanism is explained with the value of a single parameter, S = Wcs–Wcc, where Wcs and Wcc represent cell-substrate and cell-cell adhesions energy respectively. Complete wetting occurs for S > 0. For S < 0, partial wetting takes place. Recent experiments with human colon carcinoma cells (HCT-8) show that these cells form tumor like cell clusters with peripheral 3D structures on ECM functionalized PA gels of stiffness ranging from 0.5 – 5 kPa in 2 – 3 days (Figure 6C1). These cell clusters show relatively poor wetting (Figure 6C2) and after a week of culture show a dissociative metastasis like phenotype (epithelial to rounded, E-R, morphological transition, Figures 6C3-6C4).1, 25 Conversely, HCT-8 cells cultured on glass spread (Figures 6D1-6D2), and eventually form a confluent monolayer by 2 – 3 days depending on initial seeding density (Figure 6D2). Cells on glass do not show any MLP over extended period of cell culture (Figures 6D3-6D4).1 Inspired from unusual single cell scale fibroblast spreading on soft collagen gels, we ask the question whether HCT-8 cell clusters show similar increased wetting on collagen gels at multicellular level and if so, is the MLP completely inhibited by soft collagen gels. Interestingly, cell clusters on both collagen gels (104 Pa and 391 Pa) show augmented wetting (Figures 6A1-6A2 and 6B1-6B2) forming confluent monolayer (Figures 6A2 and 6B2) as on hard substrate, and cells do not display MLP upon extended culture period (Figures 6A3-6A4 and 6B3-6B4). These results imply that soft collagen gel can induce increased wetting of multicellular aggregates as well.
Figure 6. Increased wetting of HCT-8 cells on soft collagen gels and inhibition of E-R transition.
Cell clusters on both collagen gels (104 Pa and 391 Pa) show augmented wetting (Figures 6A1-6A2 and 6B1-6B2), and confluent monolayer formation (Figures 6A2 and 6B2), and they do not display MLP upon extended culture period (Figures 6A3-6A4 and 6B3-6B4) as on very hard substrate like glass (Figures 6D1-6D4). On PA gels, these cells form tumor like cell clusters with peripheral 3D structures in 2-3 days (Figure 6C1). These cell clusters show relatively poor wetting (Figure 6C2) and after a week of culture exhibit a dissociative metastasis like phenotype (epithelial to rounded morphological transition, Figures 6C3-6C4).1, 24 At least three independent experiments were performed per condition.
Conclusions
To summarize, we have shown that fibroblasts can display morphological phenotype and cytoskeleton architecture similar to very hard glass substrate on very soft collagen gels. Yet, lower cell proliferation rate and YAP localization in cytoplasmic region on collagen gels are characteristics of being on soft substrates and are tightly coupled with lower cellular traction. Further, HCT-8 cell clusters also show increased spreading on soft collagen gels as on hard substrate and do not show metastasis like phenotype (epithelial to rounded morphological transition) on collagen gels that is otherwise observed on ECM functionalized soft PA gels (0.5-50 kPa). Overall, these results suggest that cell-material interaction (soft collagen gel in this case) or cell-substrate wettability may determine cell spreading and cytoskeletal architecture, independent of substrate softness.
Acknowledgements
This project was funded by the National Science Foundation ECCS grant10-02165, and the Interdisciplinary Innovation Initiative Program, University of Illinois grant 12035. M.Y. A. was funded at UIUC from NIH National Cancer Institute Alliance for Nanotechnology in Cancer ‘Midwest Cancer Nanotechnology Training Center’ Grant R25 CA154015A. Immnunostaining and confocal microscopy imaging were carried out at the Institute for Genomic Biology (IGB), UIUC. Assistance in gel preparation and image processing by Mr. Abdul Bhuiya of UIUC is gratefully acknowledged.
References
- 1.Tang X, Kuhlenschmidt TB, Zhou J, Bell P, Wang F, Kuhlenschmidt MS, Saif TA. Biophys. J. 2010;99:2460–2469. doi: 10.1016/j.bpj.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 5.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;3:299. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 6.Georges PC, Janmey PA. J. Appl. Physiol. 2005;98:1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 7.Guo W, Frey MT, Burnham NA, Wang Y. Biophys. J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X, Bajaj P, Bashir R, Saif TA. Soft Matter. 2011;7:6151. [Google Scholar]
- 9.Pelham RJ, Wang Yl. Proc. Natl. Acad. Sci. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker EL, Bonnecaze RT, Zaman MH. Biophys. J. 2009;97:1013–1021. doi: 10.1016/j.bpj.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE. J. Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Biophys. J. 2010;99:2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo CM, Wang HB, Dembo M, Wang YL. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King CA. Acta Biomater. 2013;9:4635–4644. doi: 10.1016/j.actbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Dembo M, Wang Y. Am. J. Physiol. Cell Physiol. 2000;279:C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 16.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys. J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou S-Y, Cheng C-M, Chen C-C, LeDuc PR. Soft Matter. 2011;7:9871. [Google Scholar]
- 18.Chopra A, Lin V, McCollough A, Atzet S, Prestwich GD, Wechsler AS, Murray ME, Oake SA, Kresh JY, Janmey PA. J. Biomech. 2012;45:824–831. doi: 10.1016/j.jbiomech.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulrich TA, de Juan Pardo EM, Kumar S. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 21.Chopra A, Murray ME, Byfield FJ, Mendez MG, Halleluyan R, Restle DJ, Raz-Ben Aroush D, Galie PA, Pogoda K, Bucki R, Marcinkiewicz C, Prestwich GD, Zarembinski TI, Chen CS, Puré E, Kresh JY, Janmey PA. Biomaterials. 2014;35:71–82. doi: 10.1016/j.biomaterials.2013.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winer JP, Oake S, Janmey PA. PLoS One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudnicki MS, Cirka HA, Aghvami M, Sander EA, Wen Q, Billiar KL. Biophys. J. 2013;105:11–20. doi: 10.1016/j.bpj.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douezan S, Dumond J, Brochard-Wyart F. Soft Matter. 2012;8:4578. doi: 10.1140/epje/i2012-12034-9. [DOI] [PubMed] [Google Scholar]
- 25.Ali MY, Saif MTA. Cell. Mol. Bioeng. 2014 [Google Scholar]
- 26.Baker EL, Srivastava J, Yu D, Bonnecaze RT, Zaman MH. PLoS One. 2011;6:e20355. doi: 10.1371/journal.pone.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YL, Pelham RJ. Methods Enzymol. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- 28.Damljanović V, Lagerholm BC, Jacobson K. Biotechniques. 2005;39:847–851. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Ali MY, Saif MTA. Soft Matter. 2012;8:7197–7206. doi: 10.1039/C2SM25533B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse JR, Engler AJ. Curr. Protoc. Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. Chapter 10, Unit 10.16. [DOI] [PubMed] [Google Scholar]
- 31.Leight JL, Liu WF, Chaturvedi RR, Chen S, Yang MT, Raghavan S, Chen CS. Cell. Mol. Bioeng. 2012;5:299–306. doi: 10.1007/s12195-012-0236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen JR, Ross ST, Davidson MW. J. Opt. 2013;15:094001. [Google Scholar]
- 33.Abràmoff MD, Hospitals I, Magalhães PJ, Abràmoff M. Biophoton. Int. 2004;11(7):36–41. [Google Scholar]
- 34.Chang SS, Guo W, Kim Y, Wang Y. Biophys. J. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chopra A, Tabdanov E, Patel H, Janmey PA, Kresh JY. Am. J. Physiol. Heart Circ. Physiol. 2011;300:H1252–H1266. doi: 10.1152/ajpheart.00515.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Otto JJ. Cell Motil. Cytoskeleton. 1997;36:101–111. doi: 10.1002/(SICI)1097-0169(1997)36:2<101::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 37.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat. Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 38.Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. J. Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR. Eur. J. Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Burridge K, Chrzanowska-Wodnicka M. Annu. Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]