Abstract
Members of a gene family expressed in a single species often experience common selection pressures. Consequently, the molecular basis of complex adaptations may be expected to involve parallel evolutionary changes in multiple paralogs. Here, we use bacterial artificial chromosome library scans to investigate the evolution of the voltage-gated sodium channel (Nav) family in the garter snake Thamnophis sirtalis, a predator of highly toxic Taricha newts. Newts possess tetrodotoxin (TTX), which blocks Nav’s, arresting action potentials in nerves and muscle. Some Thamnophis populations have evolved resistance to extremely high levels of TTX. Previous work has identified amino acid sites in the skeletal muscle sodium channel Nav1.4 that confer resistance to TTX and vary across populations. We identify parallel evolution of TTX resistance in two additional Nav paralogs, Nav1.6 and 1.7, which are known to be expressed in the peripheral nervous system and should thus be exposed to ingested TTX. Each paralog contains at least one TTX-resistant substitution identical to a substitution previously identified in Nav1.4. These sites are fixed across populations, suggesting that the resistant peripheral nerves antedate resistant muscle. In contrast, three sodium channels expressed solely in the central nervous system (Nav1.1–1.3) showed no evidence of TTX resistance, consistent with protection from toxins by the blood–brain barrier. We also report the exon–intron structure of six Nav paralogs, the first such analysis for snake genes. Our results demonstrate that the molecular basis of adaptation may be both repeatable across members of a gene family and predictable based on functional considerations.
Keywords: adaptation, coevolution, gene families, molecular evolution, predator–prey interactions, toxins
Introduction
One of the primary goals of evolutionary biology is to understand the genetic basis of adaptive phenotypic evolution. In recent years, great progress has been made toward this goal, particularly for traits that have a relatively simple genetic architecture (Rosenblum et al. 2004; Hoekstra et al. 2006; Hoekstra and Coyne 2007; Steiner et al. 2007; Stern and Orgogozo 2008), but dissecting the genetic basis of adaptation for complex polygenic phenotypes has proved somewhat elusive (Rockman 2012; Travisano and Shaw 2013). Multimember gene families, which consist of paralogs created by segmental or whole-genome duplication, are abundant across the diversity of life (Thornton and DeSalle 2000) and provide a potential middle ground between studies of single-gene and polygenic evolution. Paralogs within gene families may have tissue-specific expression but often retain sufficiently similar functional roles. Consequently, their evolutionary response to common selection pressures may be somewhat predictable (Jost et al. 2008). If the functions of paralogs are well-characterized, then candidate genes underlying adaptation may be readily identified and investigated, providing empirical tractability similar to that of studies of single-gene evolution. At the same time, the evolution of multiple paralogs may be necessary for adaptations at the level of the whole organism. As such, studies of gene families may provide simplified models of polygenic adaptation.
Voltage-gated sodium channels (Nav) are an excellent model system for studying both the evolution of gene families and the genetics underlying phenotypic adaptation (Liebeskind et al. 2011; Zakon 2012). These channels are formed by an α subunit consisting of four domains (DI–DIV), which together form a sodium-selective pore in the plasma membranes of excitable tissue such as nerves and muscle, and an optional β subunit that may modify its activity (Yu and Catterall 2003; Yu et al. 2005). Although invertebrate species typically have a single α subunit, vertebrates have at least four (Widmark et al. 2011; Zakon et al. 2011); these α subunits (Nav1) are encoded by genes in the SCNA family (Catterall et al. 2005). The original expansion of the SCNA family appears to derive from the two genome duplications early in vertebrate evolution (Dehal and Boore 2005; Blomme et al. 2006), but the family has expanded further in some lineages (Widmark et al. 2011; Zakon et al. 2011). In teleost fishes, a third whole-genome duplication has expanded the family to eight members, whereas in tetrapods, duplication along chromosomes has increased the family size to six (amphibians), nine (reptiles including birds), or ten (mammals) (Zakon et al. 2011).
Nav channels are the targets of numerous neurotoxins used as either predator venoms or prey defenses (Daly 1995; Cestèle and Catterall 2000; Fry et al. 2009). Both toxic species and their predators are expected to be under strong selection for resistance to these neurotoxins, making Nav a model system for the evolutionary genetics of predator–prey interactions (Yoshida 1994; Geffeney et al. 2005; Jost et al. 2008; Rowe et al. 2013). One of the best characterized neurotoxins is tetrodotoxin (TTX), which acts by binding to the Nav outer pore, which is formed by the “P-loop” region of each domain, and blocking the conductance of sodium ions across the membrane (Hanifin 2010; Moczydlowski 2013). Pufferfishes (Tetraodontidae), which employ TTX as an antipredator defense, have evolved resistance to the toxin in each of their eight Nav paralogs through a variety of amino acid substitutions that are often shared across paralogs (Jost et al. 2008). Resistant sodium channels have also evolved in predator species that consume tetrodotoxic prey. For example, some populations of the garter snake Thamnophis sirtalis have evolved resistant skeletal muscle sodium channels (Nav1.4) as a consequence of the ongoing coevolutionary arms race with Pacific newts (Taricha spp.) (Brodie and Brodie 1990; Brodie et al. 2002; Geffeney et al. 2005; Hanifin et al. 2008). Similar arms races have independently led to the convergent evolution of resistance in Nav1.4 in a number of other snake species that also consume tetrodotoxic prey (Feldman et al. 2009, 2012).
Although the evolution of TTX resistance in Nav1.4 has been well-characterized, little is known about how the rest of the Nav family has evolved in Th. sirtalis or other predator species. In this study, we used a bacterial artificial chromosome (BAC) library and 454 sequencing to obtain the sequences of six SCNA genes from an individual Th. sirtalis collected from a TTX-resistant population. Three of these channels, Nav1.1, Nav1.2, and Nav1.3 (encoded by the genes SCN1A, 2A, and 3A, respectively), are found primarily in the central nervous system (CNS) in mammals (Goldin 2001, 2002; Catterall et al. 2005). As TTX does not appear to cross the blood–brain barrier (Zimmer 2010), these channels should be sheltered from TTX, even when predatory snakes consume toxic prey. Thus, these channels should not be under selective pressure to evolve TTX resistance, and we do not expect to detect TTX resistance-conferring replacements in these loci. Three others, Nav1.4, Nav1.6, and Nav1.7 (encoded by the genes SCN4A, 8A, and 9A), are found additionally or exclusively in the muscles or peripheral nervous system (PNS) (Caldwell et al. 2000; Thornton and DeSalle 2000; Goldin 2001, 2002; Trimmer and Rhodes 2004; Dib-Hajj et al. 2013). These tissues will be exposed to TTX when snakes consume tetrodotoxic prey, and should be under selective pressure to evolve TTX resistance. We predict that these channels will contain amino acid substitutions necessary to support TTX resistance at the organismal level and that, as in pufferfishes, TTX resistance should evolve in parallel across paralogs; that is, paralogs are expected to evolve resistance independently through similar or identical amino acid substitutions. Because our methods generated high-quality genomic sequence for most paralogs, we also report their exon–intron structure, the first such analysis for snake genes.
Results
Gene Structure
Sequences were obtained from a BAC library constructed using an individual Th. sirtalis from Benton County, OR, a population with medium-high resistance to TTX that coexists with the most highly tetrodotoxic population of Taricha newts known (Brodie et al. 2002; Hanifin et al. 2008). Complete or nearly complete genomic sequences were obtained for three paralogs: SCN4A (at least 65.1 kb from start codon to stop codon), SCN8A (73.8 kb), and SCN9A (56.8 kb) (fig. 1). Despite the recent publication of two snake genomes (Castoe et al. 2013; Vonk et al. 2013), to our knowledge these sodium channel genes represent the first snake genes to be presented and analyzed at the genomic sequence level. Full coding sequences (CDSs) were obtained for SCN3A (6,009 bp, 2,003 amino acids), 4A (5,625 bp, 1,875 amino acids), 8A (5,961 bp, 1,987 amino acids), and 9A (5,892 bp, 1,964 amino acids) (supplementary table S1, Supplementary Material online). The CDS obtained for SCN4A was identical to the previously published sequence of the same gene from Benton County, OR, except for a single nucleotide substitution (A3427C) that led to an amino acid difference (I1143L) (Geffeney et al. 2005). Other published SCN4A sequences of Th. sirtalis from other populations have C3427/L1143 as well (Feldman et al. 2010). The CDS lengths of these four genes were comparable to their orthologs in Rattus and Anolis. Partial CDSs were obtained for SCN1A (5,542 bp, 1,847 amino acids) and 2A (5,747 bp, 1,915 amino acids). Alignment of the SCN1A and 2A CDS with Rattus and Anolis orthologs suggested that 483–486 bp (161–162 amino acids) and 279 bp (93 amino acids), respectively, were missing from the 3′-ends of the snake version of these genes.
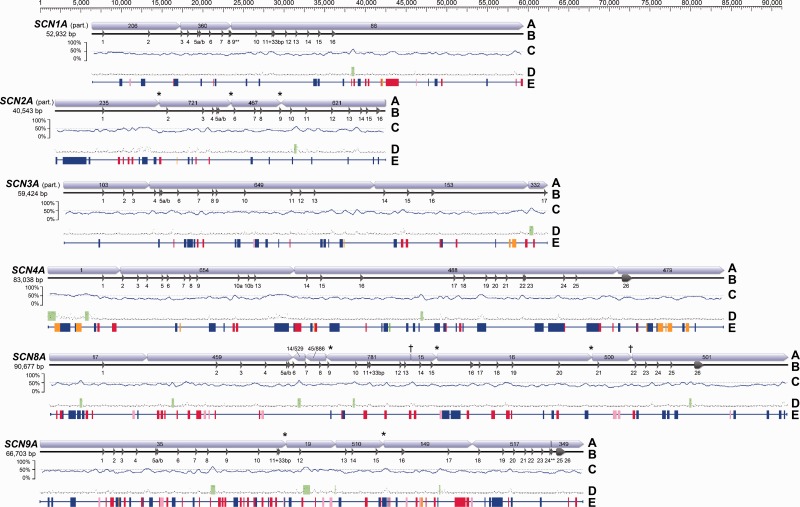
Fig. 1.
Gene structure for the six SCNA paralogs detected using BAC library scanning. To facilitate comparison, genes are aligned by the start codon in exon 1. Total base pair length represents the concatenated length of the contigs shown and may include untranscribed regions. (A) Concatenated contigs from 454 sequencing. (* indicates that contigs share an edge; † indicates that two contigs share an edge with a third small contig.) (B) Exon/intron structure. Partial CDS was obtained for three paralogs. (** indicates an incomplete exon.) (C) GC content using a 500-bp sliding window. (D) CpG islands (green) predicted by Geneious. (E) Simple repeats or TEs identified by Censor (green = satellite; red = DNA transposon; pink = endogenous retrovirus; orange = LTR retrotransposon; blue = non-LTR retrotransposon).
The exon–intron structure of these six genes was similar to that found in other species (fig. 1). SCN8A and 9A showed the 26-exon structure typical for this gene family (Widmark et al. 2011). The regions of SCN1A, 2A, and 3A for which we obtained genomic sequence also show typical exon–intron structure. As in most nonmammalian vertebrates, SCN4A has 25 exons, with two exons in place of the exons numbered 10–12 in other paralogs (Widmark et al. 2011). We have named these exons, which encode the intracellular loop linking DI and DII, 10a and 10b. Other SCN4A exons are numbered 1–9 and 13–26 by their homology to other paralogs following Widmark et al. (2011).
All introns with complete sequence displayed standard GT–AG ends, with exceptions in two locations. All paralogs had an AT–AC intron between exons 2 and 3 except SCN4A, which had an AT–AA intron in this location. There was also an AT–AC intron between exons 23 and 24 in SCN4A, 8A, and 9A. (We obtained only cDNA sequence for this region in the other paralogs; see Materials and Methods.) Both AT–AC introns are found in vertebrate orthologs and are thought to have an ancient origin (Widmark et al. 2011). The AT–AA intron in SCN4A has not been previously noted, although it is also present in Anolis. The lengths of completely sequenced introns ranged from 89 to 11,139 bp (median = 1,391 bp, mean = 1,837 bp). Although these lengths are typical for vertebrate genes (Hong et al. 2006), extremely long introns may have overlapped contig boundaries, biasing the mean downward slightly. The intron between exons 16 and 17 appears to be quite long in at least four paralogs (SCN1A, 3A, 4A, and 9A). Although we did not detect exon 17 in SCN1A, contig 88 suggests that this intron may be at least 23 kb in length (fig. 1).
Putative splice variation was also present. Except for SCN4A, all paralogs had two copies of exon 5 separated by a short (95–147 bp) intron (fig. 1). These copies, which we call 5a and 5b, differed by 1–2 amino acids depending on the paralog. The amino acid sequence of 5b is identical across all paralogs. This exon encodes the extracellular loop between transmembrane segments S3 and S4 in domain I, and the duplication of the exon allows for alternative splicing (Copley 2004). In one paralog (SCN5A), this splice variation has been shown to lead to alternative neonatal and adult channels with different physiological properties (Onkal et al. 2008). Critically, this region does not seem to affect TTX sensitivity (Onkal et al. 2008). In addition, three paralogs (SCN1A, 8A, and 9A) have a 33-bp segment that may or may not be spliced from the end of exon 11 (fig. 1). Both types of splice variation appear to influence interaction with the β subunit (Farmer et al. 2012).
The GC content for all paralogs was approximately 40% (fig. 1), which is close to the genome-wide mean base composition for several reptiles (Shedlock et al. 2007; Alföldi et al. 2011). However, all paralogs contained at least one CpG island, usually within introns (fig. 1). At least four paralogs (SCN1A, 3A, 4A, and 9A) had a CpG island in the long intron between exons 16 and 17. CpG islands were also detected upstream of exon 1 in two paralogs (SCN4A and 8A), suggesting a possible role in gene regulation (Deaton and Bird 2011). Transposable elements (TEs) were common in introns, making up 8–19% of noncoding regions (fig. 1, table 1). The TE profile across paralogs was comparable to that of published reptile genomes (Shedlock 2006; Alföldi et al. 2011; Castoe et al. 2013). Paralogs differed substantially in TE content (table 1). SCN4A had the highest TE content, which was primarily due to an abundance of non-LTR (long terminal repeat) retrotransposons, particularly CR1, L2, and R4, the last of which was absent from other paralogs (table 1). DNA transposons, particularly hAT and Mariner/Tc1, were most common in SCN9A, which showed the second highest TE content (table 1).
Table 1.
Simple Repeats and TEs in SCNA Paralogs as Detected by CENSOR.
|
SCN1A |
SCN2A |
SCN3A |
SCN4A |
SCN8A |
SCN9A |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Repeat Class | No. | Length | No. | Length | No. | Length | No. | Length | No. | Length | No. | Length |
| Simple repeat | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 33 | 0 | 0 | 0 | 0 |
| Satellite | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 33 | 0 | 0 | 0 | 0 |
| SAT | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 33 | 0 | 0 | 0 | 0 |
| TE | 30 | 5,199 | 26 | 5,770 | 35 | 4,979 | 74 | 16,119 | 63 | 7,292 | 81 | 10,298 |
| DNA transposon | 11 | 2,277 | 8 | 948 | 11 | 986 | 13 | 1,990 | 29 | 2,213 | 37 | 4,083 |
| Dada | 0 | 0 | 1 | 189 | 0 | 0 | 1 | 52 | 0 | 0 | 0 | 0 |
| Harbinger | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 119 | 0 | 0 | 1 | 39 |
| Mariner/Tc1 | 5 | 1,811 | 1 | 73 | 5 | 473 | 4 | 1,285 | 12 | 922 | 17 | 2,545 |
| Polinton | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 55 | 0 | 0 | 0 | 0 |
| hAT | 6 | 466 | 6 | 686 | 4 | 416 | 5 | 426 | 13 | 845 | 18 | 1,428 |
| Endogenous retrovirus | 2 | 181 | 0 | 0 | 1 | 80 | 0 | 0 | 9 | 809 | 9 | 1,118 |
| ERV1 | 2 | 181 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 573 | 5 | 608 |
| ERV2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 162 | 3 | 410 |
| ERV3 | 0 | 0 | 0 | 0 | 1 | 80 | 0 | 0 | 1 | 74 | 1 | 100 |
| LTR retrotransposon | 1 | 93 | 1 | 38 | 3 | 571 | 12 | 2,217 | 0 | 0 | 2 | 233 |
| DIRS | 0 | 0 | 1 | 38 | 1 | 102 | 2 | 153 | 0 | 0 | 0 | 0 |
| Gypsy | 1 | 93 | 0 | 0 | 2 | 469 | 9 | 2,020 | 0 | 0 | 2 | 233 |
| Non-LTR retrotransposon | 16 | 2,648 | 17 | 4,784 | 19 | 3,304 | 49 | 11,912 | 25 | 4,270 | 33 | 4,864 |
| CR1 | 3 | 598 | 5 | 3,666 | 2 | 484 | 10 | 3,601 | 2 | 258 | 9 | 2,042 |
| L1 | 3 | 282 | 2 | 99 | 0 | 0 | 8 | 792 | 1 | 72 | 3 | 203 |
| L2 | 2 | 353 | 4 | 403 | 3 | 197 | 12 | 2,398 | 3 | 212 | 5 | 911 |
| Penelope | 0 | 0 | 1 | 208 | 4 | 808 | 1 | 117 | 2 | 163 | 3 | 231 |
| R4 | 0 | 0 | 0 | 0 | 3 | 693 | 10 | 4,020 | 0 | 0 | 0 | 0 |
| RTE | 3 | 557 | 0 | 0 | 0 | 0 | 1 | 29 | 5 | 1,979 | 1 | 63 |
| RTEX | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 127 | 0 | 0 | 0 | 0 |
| Rex1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 510 | 1 | 55 | 0 | 0 |
| SINE | 5 | 858 | 5 | 408 | 7 | 1,122 | 4 | 246 | 9 | 1,380 | 11 | 1,299 |
| SINE2/tRNA | 4 | 549 | 4 | 327 | 4 | 632 | 4 | 246 | 5 | 397 | 7 | 937 |
| SINE3/5S | 0 | 0 | 0 | 0 | 1 | 80 | 0 | 0 | 1 | 150 | 1 | 81 |
| Tx1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 72 | 1 | 53 | 0 | 0 |
| Vingi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 115 |
| Total | 30 | 5,199 | 26 | 5,770 | 34 | 4,979 | 75 | 16,152 | 63 | 7,292 | 81 | 10,298 |
| % of total sequence | 9.8 | 14.2 | 8.4 | 19.5 | 8.0 | 15.4 | ||||||
Putative TTX Resistance and Interpopulation Variation
Sodium channels expressed predominantly in the CNS (Nav1.1–1.3) showed no evidence of TTX resistance evolution (fig. 2). Across all three channels, the amino acid sequence of the four P-loops was identical to the sequence from either rat or lizard. In contrast, all three channels expressed in the periphery (Nav1.4, 1.6, and 1.7) showed evidence of TTX resistance. There was no significant evidence of gene conversion between resistant paralogs, suggesting that TTX resistance initially arose through independent mutational events. However, gene conversion does appear to occur among SCN1–3/9A, which are found on the same chromosome in other amniotes (Zakon et al. 2011). Across these four paralogs, 13 CDS fragments found in seven distinct locations and averaging 120 bp in length showed evidence of gene conversion. Only one of these gene conversion events occurred in a P-loop region, the DI P-loop of SCN2/3A.
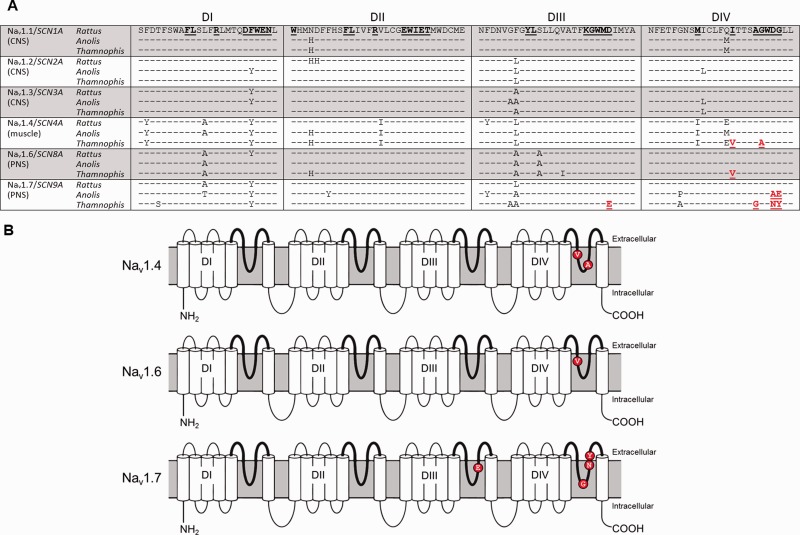
Fig. 2.
(A) Amino acid substitutions in the pore regions (or “P-loops”) for six Nav in Thamnophis sirtalis. All sequences are shown relative to rat Nav1.1, with residues involved in TTX binding underlined. Substitutions putatively conferring TTX resistance are shown in red. (B) Locations of putative resistance-conferring substitutions in Nav1.4, 1.6, and 1.7.
As previously described, the skeletal muscle channel Nav1.4 of Benton County Th. sirtalis contains two substitutions in DIV: an Ile–Val (I1561V) and a Gly–Asp (G1566A). Together, these substitutions decrease the binding affinity of TTX approximately 10-fold, providing moderate TTX resistance to skeletal muscle tissue (Geffeney et al. 2002, 2005) (fig. 2).
PNS channels Nav1.6 and 1.7 showed evidence of parallel evolution of TTX resistance. In Nav1.6, DIV contained an Ile–Val substitution (I1709V) identical to the one seen in Nav1.4 (fig. 2). On its own, this substitution is expected to confer 2-fold resistance to TTX (Geffeney et al. 2005). In garter snakes from Warrenton, OR, Nav1.4 displays this Ile–Val and no other TTX-resistance substitutions, providing low to moderate levels of resistance to skeletal muscle (Geffeney et al. 2002, 2005). This finding suggests that PNS axons in Th. sirtalis should also display low to moderate TTX resistance.
Nav1.7 contained four substitutions that are likely to confer resistance to TTX: An Asp–Glu (D1393E) in DIII and an Ala–Gly (A1681G), an Asp–Asn (D1684N), and a Gly–Tyr (G1685Y) in DIV (fig. 2). The Asp–Glu substitution has been previously described in Nav1.4 from two snake species, the resistant Th. atratus (Feldman et al. 2009) and the putatively resistant Amphiesma pryeri (Feldman et al. 2012), and replacements at this site lead to minor changes in TTX-binding affinity (Terlau et al. 1991; Choudhary et al. 2003), but this specific replacement has not been empirically tested for its effects on TTX binding. The Ala–Gly substitution occurs at the channel’s selectivity filter, and is also found in four different paralogs in TTX-bearing pufferfishes, where it provides 1.5-fold resistance to TTX and 11-fold resistance to saxitoxin (Jost et al. 2008). Despite its location, this substitution does not affect ion selectivity, but delays recovery from slow inactivation (Wu et al. 2013), suggesting the possibility that it evolved to compensate for other changes to the channel’s pore. The Asp–Asn substitution occurs in a highly TTX-resistant (600X) isoform of Nav1.4 found in California Th. sirtalis (Geffeney et al. 2005) and on its own should confer approximately 30- to 40-fold resistance to TTX (Penzotti et al. 1998; Choudhary et al. 2003). The Gly–Tyr substitution is previously undescribed and untested, but substitutions in this location are often found in TTX-resistant species (Geffeney et al. 2005; Jost et al. 2008; Feldman et al. 2012). This residue is located in the outer portion of the pore, and the replacement of glycine’s hydrogen with tyrosine’s large aromatic side chain may conceivably interfere with the ability of TTX to access its binding site in the pore. Taken together, these substitutions suggest that Nav1.7 in Th. sirtalis should be highly resistant to TTX, protecting the function of the various tissues in which this channel is expressed (Dib-Hajj et al. 2013).
Amino acid sequences of the P-loops from Nav1.6 DIV, and Nav1.7 DIII and DIV obtained from 16 individual snakes from two additional populations were uniformly identical to those obtained from BAC library sequencing. These two populations differ markedly in the levels of TTX exposure individuals may experience. At Willow Creek, CA, Th. sirtalis coexists with Taricha newts of moderate toxicity (Hanifin et al. 2008) and snake resistance phenotypes are a mixture of high resistance (64%) and low resistance (36%) (Feldman et al. 2009, 2010), whereas at Mountain Lake, VA, Notophthalmus viridescens newts are mildly toxic (Yotsu-Yamashita and Mebs 2001) and snakes have low-resistance phenotypes (Feldman et al. 2009). Based on these P-loop sequences, all snakes were predicted to have resistant peripheral nerves regardless of their geographical origin (eastern or western North America) or their level of organismal resistance to TTX, suggesting that resistant peripheral nerves may be fixed in Th. sirtalis. In contrast, various alleles of Nav1.4 associated with distinct resistance levels are found within and among populations (Geffeney et al. 2005; Feldman et al. 2009, 2010).
Incidentally, Anolis carolinensis Nav1.7 also shows evidence of TTX resistance. Differences putatively conferring TTX resistance occur in two of the same sites as in Th. sirtalis, but with different amino acid substitutions. The Asp–Ala substitution in A. carolinensis DIV has previously been shown to provide approximately 150-fold resistance to TTX (Choudhary et al. 2003), and the Gly–Glu substitution adds a negative charge in the adjacent site, a feature often seen in TTX-resistant channels (Feldman et al. 2012).
Discussion
Our results indicate that the evolution of organismal resistance to ingested TTX in Th. sirtalis involves substitutions in at least three genes, SCN4A, SCN8A, and SCN9A, which encode three Nav paralogs (Nav1.4, 1.6, and 1.7, respectively) expressed in the periphery. Two peripheral nerve channels, Nav1.6 and 1.7, show evidence of TTX-resistance evolution that parallels that previously described for skeletal muscle channel Nav1.4 (Geffeney et al. 2005; Feldman et al. 2009, 2010). In two cases, the amino acid substitution leading to putative TTX resistance is identical across paralogs. Nav1.7 has additional substitutions not seen in either of the other paralogs that are also expected to interfere with TTX binding. In a limited sample of individuals from across a wide geographic range (OR, CA, and VA), we found no evidence of variation in the TTX-binding region of either Nav1.6 or Nav1.7, suggesting either dramatically coincident replacements across populations in these genes, or more likely, that the substitutions in these two genes are fixed across the species’ range. Because Nav1.4 varies both within and among populations (Geffeney et al. 2005; Feldman et al. 2009, 2010), the latter scenario suggests that resistance in peripheral nerves is of more ancient origin than resistance in muscle. Three Nav paralogs found primarily in the CNS, Nav1.1–1.3, showed no evidence of TTX resistance, consistent with protection of these channels from toxins by the blood–brain barrier.
Ingested TTX has dramatic effects on peripheral nerves (Kao and Fuhrman 1963; Kiernan et al. 2005), suggesting that both Nav1.6 and 1.7 are likely to experience strong selection for resistance in predators that consume tetrodotoxic prey. In humans exposed to pufferfish toxin, sensory symptoms (numbness and paresthesia) occur in low-grade poisoning, with higher-grade poisoning leading to motor symptoms such as paralysis and slurred speech (Isbister and Kiernan 2005). Sensory symptoms in response to low doses of TTX likely derive from the blockade of Nav1.7, which sets the threshold for the initiation of action potentials in sensory neurons, particularly those involved in olfaction and nociception (Dib-Hajj et al. 2013). Later motor symptoms are likely mediated by effects on Nav1.6, which is found mainly at nodes of Ranvier, where it is involved in propagation of action potentials along axons (Caldwell et al. 2000). Human deaths from severe TTX poisoning are usually caused by respiratory failure, and evidence from laboratory mammals suggests that this effect derives from direct blockade of sodium channels in the diaphragm, which seems to be more sensitive to the toxin than most muscles (Cheng et al. 1968). However, squamates lack a diaphragm and instead power respiration with a variety of axial muscles (Rosenberg 1973; Carrier 1989). It is probable that the disruption of the motor neurons innervating this musculature contributes to respiratory failure in squamates with vulnerable Nav1.6. Another source of mortality in TTX poisoning, especially in laboratory mammals, is severe hypotension (Zimmer 2010), which is due in part to the blockade of sympathetic nerves (Feinstein and Paimre 1968). Nav1.7 is highly expressed in the sympathetic nervous system (Dib-Hajj et al. 2013), suggesting that TTX-induced hypotension may contribute to selection for TTX resistance in this channel.
Both western (OR, CA) and eastern (VA) populations of Th. sirtalis have substitutions in Nav1.6 and 1.7 that are predicted to confer TTX resistance to peripheral nerves, but as of yet, individuals with resistant Nav1.4 have only been detected on the west coast where the species overlaps with highly toxic Taricha newts (Geffeney et al. 2005). This result suggests that the origin of TTX resistance in nerves likely antedates that of TTX resistance in muscle. In general, nerves appear to be impaired by lower TTX concentrations than required to impair muscles. TTX-sensitive nerves are typically blocked by nanomolar concentrations of TTX (Kao and Fuhrman 1963), whereas blocking action potentials in muscle fibers, which tend to have diameters an order of magnitude wider than the axons that innervate them, requires concentrations an order of magnitude higher (Geffeney et al. 2002). Consequently, resistant peripheral nerves alone are likely to provide sufficient protection to allow ingestion of very small amounts of TTX, such as a garter snake might encounter when consuming the mildly toxic eastern newt N. viridescens (Yotsu-Yamashita and Mebs 2001). However, muscle resistance will be necessary to provide protection against the higher concentrations of TTX seen in many Taricha populations (Hanifin et al. 2008).
Although we suggest that resistance in Nav1.6 and 1.7 has evolved specifically in response to selection mediated by TTX in prey, it is also possible that either or both nerve channels evolved resistance as a byproduct of selection for other functional modifications. In general, TTX-resistant substitutions are associated with a decrease in conductance and/or sodium selectivity (Lee et al. 2011; Feldman et al. 2012). Although it is unlikely that reduced conductance or selectivity would be favored by selection in the absence of another selective agent such as TTX, other properties of sodium channels are also affected by the pore region and may be more likely targets of selection. For example, one of the substitutions in garter snake Nav1.7 is seen in pufferfishes and a number of nontetrodotoxic fishes and has been shown to delay recovery from slow inactivation (Wu et al. 2013). Mammalian Nav1.7 is characterized by a slow recovery from inactivation when compared with Nav1.6 (Herzog et al. 2003), so it is possible that the Ala–Gly substitution seen in garter snakes reflects a refinement of this property. The Ile–Val substitution seen in both garter snake Nav1.4 and Nav1.6 has also been shown to affect slow inactivation, altering its voltage sensitivity (Lee et al. 2011). Interestingly, garter snake Nav1.6 also has a Val–Ile substitution located in the analogous site of the DIII P-loop that may be involved in compensating for this change in voltage sensitivity.
The lack of resistance evolution in CNS channels can be attributed to the effectiveness of the blood–brain barrier against TTX. The blood–brain barrier of reptiles is similar to that of mammals (Kenny and Shivers 1974; Cserr and Bundgaard 1984) and prevents the transport of many molecules, particularly hydrophilic compounds and those of relatively high molecular weight. TTX has a molecular weight of 319.27 and is highly polar and thus lipid-insoluble. Based on these properties, one simple model (Fu et al. 2008) predicts a log ratio of steady-state brain to plasma concentrations (log BB) of −3.54 for TTX, meaning that concentrations of TTX should be 3–4 orders of magnitude lower in the brain than in the plasma. To our knowledge, there has been no direct empirical quantification of the log BB of TTX, although one early study found low concentrations of injected TTX in the brain as compared with other tissues (Kao 1966). In accordance with this observation, unconsciousness occurs only in the most severe cases of TTX poisoning in humans (Isbister and Kiernan 2005). The snake from which our BAC library was constructed coexists with the most toxic newts known (Hanifin et al. 2008), suggesting that even these newts probably do not contain enough TTX to impose selection for resistance in CNS channels. However, we predict that predator species that regularly attain higher plasma concentrations of TTX, either because they consume more toxic prey or because they have lower body weight, may evolve one or more resistant CNS channels. Highly toxic prey species, such as Taricha, constitutively experience high concentrations of TTX, and thus are predicted to evolve TTX resistance in all Nav paralogs, as observed in pufferfishes (Jost et al. 2008).
We were unable to sequence Nav1.5, 1.8, and 1.9, which are TTX resistant in mammals, using our BAC library methods. These channels were previously thought to be ancestrally resistant to TTX in vertebrates because all three paralogs share the replacement of an aromatic side chain (Tyr or Phe) with either Ser or Cys in a critical site in the DI P-loop, but recent genomic evidence has shown that these DI substitutions are not present in currently characterized teleosts, amphibians, birds, and reptiles (Vornanen et al. 2011). Future work should examine the evolution of TTX resistance in these three channels. We predict that resistance should have evolved at least in Nav1.5, the primary sodium channel in cardiac muscle (Goldin 2001). Despite the lack of evidence of TTX resistance of this channel in the A. carolinensis genome, Vornanen et al. (2011) report TTX resistance of cardiac muscle in the common European adder (Vipera berus), suggesting that this channel may indeed be TTX-resistant in snakes. In contrast, it is possible that organismal resistance to ingested TTX does not require resistant Nav1.8 and 1.9. These channels are expressed in small-diameter sensory neurons and are primarily involved in nociception (Dib-Hajj et al. 2002; Fang et al. 2002; Zimmermann et al. 2007). Hypothetically, a predator with sensitive Nav1.8 and 1.9 but resistant Nav1.4–1.7 might experience an analgesic effect but not inhibited locomotion or respiration. At least one predator, grasshopper mice (Onychomys spp.), which prey upon venomous Centuroides scorpions, has amino acid substitutions that make the blockade of Nav1.8 by prey toxins more likely (Rowe et al. 2013). Scorpion toxin typically causes pain by activating Nav1.7, but in grasshopper mice, it also binds to and blocks Nav1.8, producing a compensatory analgesic effect (Rowe et al. 2013). This blockade has no known detrimental side effects, suggesting that the blockade of Nav1.8 with TTX may not lead to a decrease in fitness.
In summary, our results show that at least three Nav paralogs have evolved resistance to TTX, either in garter snakes or in their ancestors. Two of the amino acid substitutions conferring resistance to TTX in the two PNS channels described here are identical to those that confer skeletal muscle resistance in the same species, and two others have been described in either other garter snakes (Feldman et al. 2009) or pufferfishes (Jost et al. 2008). Our results add to the growing consensus that evolution is often somewhat predictable at the molecular level (Stern and Orgogozo 2008, 2009). This observation seems to be particularly true for the evolution of toxin resistance in Nav (ffrench-Constant et al. 1998; Jost et al. 2008; Feldman et al. 2009, 2012). As in pufferfishes (Jost et al. 2008), multiple paralogs in the Nav family have evolved resistance in parallel, but this predictability has an additional layer of complexity in garter snakes. Our results demonstrate that in predator species, TTX resistance evolves only in those channels that are routinely exposed to ingested TTX, whereas channels protected from exposure retain their sensitivity. In addition, the pattern of resistance evolution in the family suggests that channels expressed in more sensitive tissues, such as peripheral nerves, should evolve resistance prior to channels expressed in tissues whose function is impaired less severely by TTX. Future work directly testing this hypothesis will require a broader comparative approach but should reveal the ultimate origins of TTX resistance in the Nav family.
Materials and Methods
BAC Library Scan
To obtain the genomic sequences of sodium channel genes previously uncharacterized in Th. sirtalis, we used radiolabeled DNA probes to scan a BAC library, which had been previously constructed using an individual snake from Benton County, OR (http://www.genome.gov/11008350, last accessed April 2, 2014). This population consists of snakes with medium-high resistance to TTX and coexists with a highly toxic population of Taricha newts (Geffeney et al. 2005; Hanifin et al. 2008). Our approach is described only briefly below; see Janes et al. (2011) for an overview of our approach and Ross et al. (1999) for a detailed protocol.
Probes for BAC library scanning were designed using Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012) and four sequence resources: The transcriptome of a related species, Th. elegans (Schwartz et al. 2010), the CDS of SCN4A from Th. sirtalis (Geffeney et al. 2005), the CDS of an unknown SCNA paralog identified using degenerate polymerase chain reaction (PCR) of brain cDNA (Chuckalovcak 2010), and the universal probe database UProbe (Kellner et al. 2005; http://uprobe.genetics.emory.edu, last accessed April 2, 2014). These probes fell into four categories, each of which was designed to detect one of the four major branches in the radiation of the SCNA family: SCN1/2/3/9A, SCN4A, SCN5/10/11A, and SCN8A (supplementary table S2, Supplementary Material online). Each probe consisted of two DNA oligomers, or “overgos” (22 or 24 bp) with 8 bp of complementary sequence at the 3′-end (supplementary table S2, Supplementary Material online). These overgos were combined in solution and radiolabeled following Ross et al. (1999), resulting in probes 36–40 bp in length.
The first 144 plates of the Th. sirtalis BAC library were gridded onto high-density nylon filters using a QBot gridding robot (Genetix, Ltd.) (Ross et al. 1999). Hybridization of labeled probes to these filters was performed as in Ross et al. (1999) in two steps. First, all 31 probes were pooled and 99 putative positives were identified using a Typhoon imager (GE Healthcare). Next, these putative positives were gridded onto new filters, which were hybridized with 18 smaller pools of probes to confirm positive hits (supplementary table S2, Supplementary Material online). Following this step, 31 BAC colonies were selected to be sequenced. We also added nine BAC colonies that had been previously identified using screens of the same library with SCN4A-specific probes (Chuckalovcak JP, unpublished data), for a total of 40 putative positives (supplementary table S3, Supplementary Material online).
DNA Isolation, Sequencing, and Assembly
These 40 colonies were cultured and BAC DNA was isolated in duplicate using a Qiagen R.E.A.L. Prep 96 Plasmid Kit. DNA concentration was quantified using a Qubit fluorometer (Invitrogen). Samples were then diluted to equal concentration (16.67 ng/μl) and pooled for sequencing. A shotgun library was prepared for 454 sequencing using standard protocols and was sequenced on two separate half-plate runs using a Roche GS-FLX with Titanium reagents at the University of Virginia (UVA) Biology Genomics Core Facility. Sequences (138 Mb) from the two half-plate runs were combined for assembly using gsAssembler 2.3 (Roche). Reads matching the Escherichia coli genome or the pTarBAC2.1 vector were filtered, and default assembly settings were used. The assembly produced 812 contigs with an average size of 5,096 bp and an average read depth of 31×.
Gene Annotation
Contigs containing SCNA genes were initially identified with tBLASTn searches using amino acid sequences of Th. sirtalis Nav1.4 (Geffeney et al. 2005) and the unknown Th. sirtalis paralog (Chuckalovcak 2010) as queries and the 454 output as the database. A variety of other BLAST (Basic Local Alignment Search Tool) searches using publicly available SCNA sequences were used to confirm these results. In all, 32 contigs ranging from 1,471 to 39,713 bp in length (average 12,416 bp) and with an average read depth of 74× were identified as containing SCNA exons. Most contigs contained multiple exons and the intervening intronic sequence (fig. 1). Edges between some contigs were identified by gsAssembler, providing further evidence of contig affinity (fig. 1). In two cases, two exon-containing contigs shared an edge with a third small (∼20 bp) contig. Identified contigs included six versions of the first half of SCNA (exons 1–16), but only three versions of the second half (exons 17–26). Note that we use the numbering scheme 1–26 regardless of paralog, following Widmark et al. (2011).
To identify the specific paralog contained in each contig, we used BLAT (Kent 2002) searches against the A. carolinensis genome (Alföldi et al. 2011), whose paralogs were identified by Zakon et al. (2011). The results of these searches suggested that our sequences contained exons 1–16 of SCN1A (three contigs; exon 9 was incomplete), exons 1–16 of SCN2A (four contigs), exons 1–17 of SCN3A (four contigs), exons 1–26 of SCN4A (four contigs; complete CDS), exons 1–26 of SCN8A (11 contigs; complete CDS), and exons 1–26 of SCN9A (six contigs; nearly complete CDS, exon 24 was incomplete) (fig. 1). Two versions of contigs containing SCN8A exons 6–8 were present; these contained minor variation in intronic regions as well as a single synonymous difference in exon 8. We were unable to detect evidence of SCN5A, 10A, or 11A in our sequence. Exon–intron boundaries were identified by comparing sequences to published genomes (A. carolinensis and Homo sapiens). The exon–intron structure of contigs was annotated using Geneious 6.1.7 (Biomatters). We also used algorithms within Geneious 6.1.7 to measure GC content and to detect CpG islands. To explore the genomic content of intronic sequence, contigs were screened against a database of vertebrate simple repeats and TEs (Repbase) using Censor (Kohany et al. 2006).
Gap Filling
Gaps in the CDS for SCN1A, 2A, and 3A were filled using Sanger sequencing of brain cDNA, which was obtained from a captive adult female Th. sirtalis that had been previously captured from the highly TTX-resistant population in Willow Creek, CA. The CDSs for exons 14–26 of SCN1A (44 reads, 3,114 bp, 4.7× coverage, 92% of sequence from both strands), exons 16–26 of SCN2A (32 reads, 2,632 bp, 4.3×, 77% from both strands), and exons 17–26 of SCN3A (38 reads, 2,821 bp, 4.4×, 79% from both strands) were obtained using primers designed from the transcriptome of Th. elegans (Schwartz et al. 2010) (supplementary table S5, Supplementary Material online). We attempted 3′-RACE (Rapid Amplification of cDNA Ends) to obtain the sequences of the 3′-ends of transcripts, but this gave low-quality sequence in SCN1A and was unsuccessful in SCN2A; therefore, the sequences of exon 26 were incomplete for these paralogs (690 and 916 bp, respectively). The complete CDS of exon 26 (1,193 bp, including the stop codon) plus 222 bp 3′-UTR was obtained for SCN3A. The CDS for the missing segment of SCN1A exon 9 was obtained using primers designed from contigs 360 and 88 of our 454 run (supplementary table S5, Supplementary Material online). Finally, CDS for the missing segment of SCN9A exon 24 was obtained from previous degenerate PCR results (Chuckalovcak 2010), as BLAST searches against our 454 contigs showed that sequence containing exons 19–26 obtained through this method belonged to SCN9A. All PCR followed standard protocols, and post-PCR cleanup was achieved using a combination of exonuclease I and shrimp alkaline phosphatase (Exo-SAP, USB). Sequencing was carried out either on an ABI 3130 at UVA’s Genomics Core Facility or on an ABI 3730 at Yale University’s DNA Analysis Facility.
Paralog Identification
The identity of each paralog was confirmed by using BLAT searches of the entire CDS against the A. carolinensis genome. The best BLAT hit was invariably the paralog assigned using our preliminary BLAT searches of separate contigs (supplementary table S4, Supplementary Material online). We also constructed a protein phylogeny of the six Th. sirtalis paralogs and all nine A. carolinensis paralogs (Zakon et al. 2011, accession nos. BK007953–BK007961). Amino acid sequences were aligned using MUSCLE (Edgar 2004) and a maximum-likelihood phylogeny was constructed using PhyML (Guindon and Gascuel 2003). Each Th. sirtalis paralog was shown to be most similar to its assigned ortholog in A. carolinensis, and each of these nodes had a bootstrap value of 100 (supplementary fig. S1, Supplementary Material online). We tested for evidence of gene conversion among paralogs using a MUSCLE alignment of the six Th. sirtalis paralogs in GENECONV (Sawyer 1989).
Putative TTX Resistance and Interpopulation Variation
We scanned for putative TTX resistance by comparing the P-loop regions from CDS of the six paralogs we identified to reference sequence from both A. carolinensis and a mammal (the rat Rattus norvegicus). From paralogs whose sequences indicated putative TTX resistance, we also examined sequences from individuals from two populations: Willow Creek, CA, which consists of a mixture of low- and high-resistance individuals (Feldman et al. 2009, 2010) and Mountain Lake Biological Station, VA, which displays low resistance and occurs outside the range of Taricha but overlaps with the mildly toxic N. viridescens (Yotsu-Yamashita and Mebs 2001). We used tissue samples from 16 individual snakes whose SCN4A genotypes and organismal resistance were reported by Feldman et al. (2009): CA high-resistance n = 10 (SCN4A accession nos. FJ571047–FJ571056), CA low-resistance n = 1 (GQ154075), and VA n = 4 (FJ571057–FJ571060). Sample sizes for individual regions were as follows: SCN8A DIII and DIV, CA high-resistance n = 2, VA n = 2; SCN9A DIII CA high-resistance n = 10, CA low-resistance n = 1, VA n = 1; SCN9A DIV CA high-resistance n = 10, CA low-resistance n = 1, VA n = 4. Primers were designed from our 454 contigs (supplementary table S6, Supplementary Material online). PCR and cleanup were carried out as above, and sequencing was performed on an ABI 3730 at the Yale DNA Analysis Facility.
Accession Numbers
All new sequences have been deposited in GenBank under accession numbers KJ908861–KJ908938 and KM066119. The predicted CDSs for each paralog have been deposited as Third Party Annotations under accession numbers BK008860-BK008865.
Supplementary Material
Supplementary figure S1 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Michelle Sivilich and Megan Kobiela for assistance in the laboratory, Dan Sloan for advice on molecular techniques, Shana Geffeney and Charles Hanifin for helpful discussions, Nigel Delaney and Yoel Stuart for logistical support in Cambridge, and Angela Hornsby, Marjorie Matocq, and Kendra Sewall for comments on the manuscript. This material is based upon work supported by the National Science Foundation (grant numbers DEB 0922216 to E.D.B. III and DEB 1034686 to M.E.P.). Publication of this article was supported by Virginia Tech's Open Access Subvention Fund.
References
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong LS, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477(7366):587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7(5):R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie ED, III, Brodie ED., Jr Tetrodotoxin resistance in garter snakes: an evolutionary response of predators to dangerous prey. Evolution. 1990;44(3):651–659. doi: 10.1111/j.1558-5646.1990.tb05945.x. [DOI] [PubMed] [Google Scholar]
- Brodie ED, Jr, Ridenhour BJ, Brodie ED., III The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution. 2002;56(10):2067–2082. doi: 10.1111/j.0014-3820.2002.tb00132.x. [DOI] [PubMed] [Google Scholar]
- Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR. Sodium channel Nav1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Natl Acad Sci U S A. 2000;97(10):5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier DR. Ventilatory action of the hypaxial muscles of the lizard Iguana iguana: a function of slow muscle. J Exp Biol. 1989;143:435–457. doi: 10.1242/jeb.143.1.435. [DOI] [PubMed] [Google Scholar]
- Castoe TA, de Koning APJ, Hall KT, Card DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu WJ, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 2013;110(51):20645–20650. doi: 10.1073/pnas.1314475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57(4):397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- Cestèle S, Catterall WA. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie. 2000;82(9–10):883–892. doi: 10.1016/s0300-9084(00)01174-3. [DOI] [PubMed] [Google Scholar]
- Cheng K, Ling Y, Wang JC. The failure of respiration in death by tetrodotoxin poisoning. Q J Exp Physiol. 1968;53:119–128. doi: 10.1113/expphysiol.1968.sp001951. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Yotsu-Yamashita M, Shang L, Yasumoto T, Dudley SC. Interactions of the C-11 hydroxyl of tetrodotoxin with the sodium channel outer vestibule. Biophys J. 2003;84(1):287–294. doi: 10.1016/S0006-3495(03)74849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuckalovcak JP. Thamnophis sirtalis. 2010. Identification and characterization of a novel voltage-gated sodium channel gene in the tetrodotoxin resistant garter snake. [master’s thesis]. Charlottesville (VA): University of Virginia. [Google Scholar]
- Copley RR. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20(4):171–176. doi: 10.1016/j.tig.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cserr HF, Bundgaard M. Blood-brain interfaces in vertebrates: a comparative approach. Am J Physiol. 1984;246(3):R277–R288. doi: 10.1152/ajpregu.1984.246.3.R277. [DOI] [PubMed] [Google Scholar]
- Daly JW. The chemistry of poisons in amphibian skin. Proc Natl Acad Sci U S A. 1995;92(1):9–13. doi: 10.1073/pnas.92.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Gene Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLOS Biol. 2005;3(10):1700–1708. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj S, Black JA, Cummins TR, Waxman SG. NaN/Nav1.9: a sodium channel with unique properties. Trends Neurosci. 2002;25(5):253–259. doi: 10.1016/s0166-2236(02)02150-1. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Nav1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14(1):49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with highaccuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Djouhri L, Black JA, Dib-Hajj SD, Waxman SG, Lawson SN. The presence and role of the tetrodotoxin-resistant sodium channel Nav1.9 (NaN) in nociceptive primary afferent neurons. J Neurosci. 2002;22(17):7425–7433. doi: 10.1523/JNEUROSCI.22-17-07425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer C, Cox JJ, Fletcher EV, Woods CG, Wood JN, Schorge S. Splice variants of Nav1.7 sodium channels have distinct β subunit-dependent biophysical properties. PLoS One. 2012;7(7):e41750. doi: 10.1371/journal.pone.0041750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein MB, Paimre M. Mechanism of cardiovascular action of tetrodotoxin in the cat. Block of conduction in peripheral sympathetic fibers. Circ Res. 1968;23(4):553–565. doi: 10.1161/01.res.23.4.553. [DOI] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Jr, Brodie ED, III, Pfrender ME. The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proc Natl Acad Sci U S A. 2009;106(32):13415–13420. doi: 10.1073/pnas.0901224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Jr, Brodie ED, III, Pfrender ME. Genetic architecture of a feeding adaptation: garter snake (Thamnophis) resistance to tetrodotoxin bearing prey. Proc R Soc Lond B Biol Sci. 2010;277(1698):3317–3325. doi: 10.1098/rspb.2010.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Jr, Brodie ED, III, Pfrender ME. Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proc Natl Acad Sci U S A. 2012;109(12):4556–4561. doi: 10.1073/pnas.1113468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ffrench-Constant RH, Pittendrigh B, Vaughan A, Anthony N. Why are there so few resistance-associated mutations in insecticide target genes? Philos Trans R Soc Lond B. 1998;353(1376):1685–1693. doi: 10.1098/rstb.1998.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Fu XC, Wang GP, Shan HL, Liang WQ, Gao JQ. Predicting blood-brain barrier penetration from molecular weight and number of polar atoms. Eur J Pharm Biopharm. 2008;70(2):462–466. doi: 10.1016/j.ejpb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Geffeney S, Brodie ED, Jr, Ruben PC, Brodie ED., III Mechanisms of adaptation in a predator-prey arms race: TTX-resistant sodium channels. Science. 2002;297(5585):1336–1339. doi: 10.1126/science.1074310. [DOI] [PubMed] [Google Scholar]
- Geffeney SL, Fujimoto E, Brodie ED, III, Brodie ED, Jr, Ruben PC. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature. 2005;434(7034):759–763. doi: 10.1038/nature03444. [DOI] [PubMed] [Google Scholar]
- Goldin AL. Resurgence of sodium channel research. Annu Rev Phys. 2001;63:871–894. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- Goldin AL. Evolution of voltage-gated Na+ channels. J Exp Biol. 2002;205(5):575–584. doi: 10.1242/jeb.205.5.575. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hanifin CT. The chemical and evolutionary ecology of tetrodotoxin (TTX) toxicity in terrestrial vertebrates. Mar Drugs. 2010;8(3):577–593. doi: 10.3390/md8030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanifin CT, Brodie ED, Jr, Brodie ED., III Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 2008;6(3):471–482. doi: 10.1371/journal.pbio.0060060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog RI, Cummins TR, Ghassemi F, Dib-Hajj SD, Waxman SG. Distinct repriming and closed-state inactivation kinetics of Nav1.6 and Nav1.7 sodium channels in mouse spinal sensory neurons. J Physiol. (Lond.) 2003;551(3):741–750. doi: 10.1113/jphysiol.2003.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61(5):995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313(5783):101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23(12):2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- Isbister GK, Kiernan MC. Neurotoxic marine poisoning. Lancet Neurol. 2005;4(4):219–228. doi: 10.1016/S1474-4422(05)70041-7. [DOI] [PubMed] [Google Scholar]
- Janes DE, Valenzuela N, Ezaz T, Amemiya C, Edwards SV. Sex chromosome evolution in amniotes: applications for bacterial artificial chromosome libraries. J Biomed Biotechnol. 2011;2011(2011):132975. doi: 10.1155/2011/132975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost MC, Hillis DM, Lu Y, Kyle JW, Fozzard HA, Zakon HH. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol Biol Evol. 2008;25(6):1016–1024. doi: 10.1093/molbev/msn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY. Tetrodotoxin, saxitoxin and their significance in the study of excitation phenomena. Pharmacol Rev. 1966;18(2):997–1049. [PubMed] [Google Scholar]
- Kao CY, Fuhrman FA. Pharmacological studies on tarichatoxin, a potent neurotoxin. J Pharmacol Exp Ther. 1963;140(1):31–40. [PubMed] [Google Scholar]
- Kellner WA, Sullivan RT, Carlson BH, Thomas JW, Prog NCS. Uprobe: a genome-wide universal probe resource for comparative physical mapping in vertebrates. Genome Res. 2005;15(1):166–173. doi: 10.1101/gr.3066805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny TP, Shivers RR. Blood-brain barrier in a reptile, Anolis carolinensis. Tissue Cell. 1974;6(2):319–333. doi: 10.1016/0040-8166(74)90056-1. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan MC, Isbister GK, Lin CSY, Burke D, Bostock H. Acute tetrodotoxin-induced neurotoxicity after ingestion of puffer fish. Ann Neurol. 2005;57(3):339–348. doi: 10.1002/ana.20395. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Lee CH, Jones DK, Ahern C, Sarhan MF, Ruben PC. Biophysical costs associated with tetrodotoxin resistance in the sodium channel pore of the garter snake, Thamnophis sirtalis. J Comp Physiol A. 2011;197(1):33–43. doi: 10.1007/s00359-010-0582-9. [DOI] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108(22):9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczydlowski EG. The molecular mystique of tetrodotoxin. Toxicon. 2013;63:165–183. doi: 10.1016/j.toxicon.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Onkal R, Mattis JH, Fraser SP, Diss JKJ, Shao D, Okuse K, Djamgoz MBA. Alternative splicing of Nav1.5: an electrophysiological comparison of “neonatal” and “adult” isoforms and critical involvement of a lysine residue. J Cell Phys. 2008;216(3):716–726. doi: 10.1002/jcp.21451. [DOI] [PubMed] [Google Scholar]
- Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC. Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J. 1998;75(6):2647–2657. doi: 10.1016/S0006-3495(98)77710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution. 2012;66(1):1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg HI. Functional anatomy of pulmonary ventilation in the garter snake, Thamnophis elegans. J Morphol. 1973;140:171–184. doi: 10.1002/jmor.1051400205. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Hoekstra HE, Nachman MW. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 2004;58(8):1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. [DOI] [PubMed] [Google Scholar]
- Ross MT, LaBrie SL, McPherson J, Stanton VP., Jr Screening large-insert libraries by hybridization. Curr Protoc Hum Genet. 1999;21:5.6.1–5.6.52. [Google Scholar]
- Rowe AH, Xiao YC, Rowe MP, Cummins TR, Zakon HH. Voltage-gated sodium channel in grasshopper mice defends against bark scorpion toxin. Science. 2013;342(6157):441–446. doi: 10.1126/science.1236451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6(5):526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schwartz TS, Tae H, Yang Y, Mockaitis K, Van Hemert JL, Proulx SR, Choi J, Bronikowski AM. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics. 2010;11:694. doi: 10.1186/1471-2164-11-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM. Phylogenomic investigation of CR1 LINE diversity in reptiles. Syst Biol. 2006;55(6):902–911. doi: 10.1080/10635150601091924. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Botka CW, Zhao SY, Shetty J, Zhang TT, Liu JS, Deschavanne PJ, Edwards SV. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Natl Acad Sci U S A. 2007;104(8):2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLOS Biol. 2007;5(9):1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323(5915):746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Heinemann SH, Stuhmer W, Pusch M, Conti F, Imoto K, Numa S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 1991;293(1–2):93–96. doi: 10.1016/0014-5793(91)81159-6. [DOI] [PubMed] [Google Scholar]
- Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- Travisano M, Shaw RG. Lost in the map. Evolution. 2013;67(2):305–314. doi: 10.1111/j.1558-5646.2012.01802.x. [DOI] [PubMed] [Google Scholar]
- Trimmer JS, Rhodes KJ. Localization of voltage-gated ion channels in mammalian brain. Annu Rev Phys. 2004;66:477–519. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJR, Kerkkamp HME, Vos RA, Guerreiro I, Calvete JJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 2013;110(51):20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornanen M, Hassinen M, Haverinen J. Tetrodotoxin sensitivity of the vertebrate cardiac Na+ current. Mar Drugs. 2011;9(11):2409–2422. doi: 10.3390/md9112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmark J, Sundstrom G, Daza DO, Larhammar D. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;28(1):859–871. doi: 10.1093/molbev/msq257. [DOI] [PubMed] [Google Scholar]
- Wu MM, Ye N, Sengupta B, Zakon HH. A naturally occurring amino acid substitution in the voltage-dependent sodium channel selectivity filter affects channel gating. J Comp Physiol A. 2013;199(10):829–842. doi: 10.1007/s00359-013-0845-3. [DOI] [PubMed] [Google Scholar]
- Yoshida S. Tetrodotoxin-resistant sodium channels. Cell Mol Neurobiol. 1994;14(3):227–244. doi: 10.1007/BF02088322. [DOI] [PubMed] [Google Scholar]
- Yotsu-Yamashita M, Mebs D. The levels of tetrodotoxin and its analogue 6-epitetrodotoxin in the red-spotted newt. Notophthalmus viridescens. Toxicon. 2001;39(8):1261–1263. doi: 10.1016/s0041-0101(00)00263-4. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4(3):207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol Rev. 2005;57(4):387–395. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- Zakon HH. Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc Natl Acad Sci U S A. 2012;109(Suppl 1):10619–10625. doi: 10.1073/pnas.1201884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Jost MC, Lu Y. Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 2011;28(4):1415–1424. doi: 10.1093/molbev/msq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer T. Effects of tetrodotoxin on the mammalian cardiovascular system. Mar Drugs. 2010;8(3):741–762. doi: 10.3390/md8030741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann K, Leffler A, Babes A, Cendan CM, Carr RW, Kobayashi J, Nau C, Wood JN, Reeh PW. Sensory neuron sodium channel Nav1.8 is essential for pain at low temperatures. Nature. 2007;447(7146):855–858. doi: 10.1038/nature05880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.