Abstract
Warfarin is used in paediatric populations, but dosing algorithms incorporating pharmacogenetic data have not been developed for children. Previous studies have produced estimates of the effect of polymorphisms in CYP2C9 and VKORC1 on stable warfarin dosing, but data on time in therapeutic range, initial dosing and adverse effects are limited. Participants (n=97) were recruited, and routine clinical data and salivary DNA samples were collected from all participants, and analysed for CYP2C9*2, *3 and VKORC1-1639 polymorphisms.VKORC1 -1639 was associated with a greater proportion of the first six months’ treatment time spent within the target INR range, accounting for an additional 9.5% of the variance in the proportion of time. CYP2C9*2 was associated with a greater likelihood of INR values exceeding the target range during the initiation of treatment (OR [per additional copy] 4.18, 95% CI 1.42, 12.34). CYP2C9*2 and VKORC1-1639 were associated with a lower dose requirement, and accounted for almost 12% of the variance in stable dose. VKORC1-1639 was associated with an increased likelihood of mild bleeding complications (OR [heterozygotes vs homozygotes] 4.53, 95% CI 1.59, 12.93). These data show novel associations between VKORC1-1639 and CYP2C9*2 and INR values in children taking warfarin, as well as replicating previous findings with regard to stable dose requirements. The development of pharmacogenomic dosing algorithms for children using warfarin has the potential to improve clinical care in this population.
Keywords: Warfarin, Paediatric, Pharmacogenetic, VKORC1, CYP2C9, Haemorrhage
Introduction
Warfarin, a synthetic coumarin used for anticoagulation, has a narrow therapeutic index and exhibits wide inter-individual variation in the maintenance dose required (0.5 mg/day to 20 mg/day). Proportionally, far fewer children are treated with warfarin than adults. Indications for paediatric use include treatment of thromboembolic disorders, prophylaxis for heart valve replacement, following cavopulmonary shunts and completion of the Fontan circulation in patients with complex congenital heart disease 1. Warfarin requires regular therapeutic monitoring through the use of the International Normalised Ratio (INR). The INR required for successful anticoagulation varies by indication for anticoagulation (Supplementary Data Table 1), following national recommendations 2.
The pharmacokinetics and pharmacodynamics of warfarin in the young differ both from adults 1, 3, 4, and across childhood3, 5, 6. Within children, dose requirements have been shown to be affected by weight (directly associated), and age (inversely associated)3, 5-7. Drug interactions also contribute to dose variation5.
Polymorphisms in 2 genes, vitamin K epoxide reductase complex subunit 1 (VKORC1) and Cytochrome P450 2C9 (CYP2C9), in combination with environmental factors, have a significant effect on warfarin dosing in adults 8-10. Adult patients from a variety of ethnic backgrounds with the variant allele(s) in one of the recognised polymorphisms in the VKORC1 gene (rs9923231, VKORC1-1639), and/or a variant allele(s) in polymorphisms in the CYP2C9 gene (rs1057910 and rs1799853, CYP2C9*2 and *3 respectively) require lower doses of warfarin 9, 11-18. In adults, up to 30% of the dose variation can be explained by these 2 genetic factors 19, increasing to between 31.4 and 58.7% when clinical factors (including age, sex, drug interactions, ethnicity, BMI, etc.) are incorporated 19. Dosing algorithms that incorporate this pharmacogenetic data have been developed for adult patients 9, 20 and shown to be more effective than standard dosing when using an initial loading dose strategy 21.
Dosing algorithms have also been published for paediatric populations. Pharmacogenetic information is not included 3, 6, 22, 23, but paediatric haematologists are interested in updated algorithms including genetic factors 24. In children, the stable dose requirement is related to age, as well as patient height, genetic polymorphisms in VKORC1 and CYP2C9, and indication for warfarin 25-27. The effect estimates for stable dose vary considerably between publications, both for genetic factors [the estimates of VKORC1, contribution to warfarin stable dose requirements in children varies between 3.7-26.6%, for CYP2C9 0.4-12.8%], and total explained variability (38-72%) 25-27.
However, previous studies have not found a relationship between the time spent within INR range 26 and time to therapeutic range 27 and either CYP2C9 or VKORC1 polymorphisms. There are data suggesting a relationship between these genetic factors and INR exceeding the target range in the first week of therapy 28, but they have not been replicated, and there is no data on the risk of haemorrhage.
The aims of this study were therefore to examine the association between genetic (VKORC1 and CYP2C9) and non-genetic factors and stable maintenance dosing, and quantify the proportion of variability explained by these factors. We also aimed to examine the association between these genetic polymorphisms and time within therapeutic INR range, an INR above the recommended range in the first week of use, and haemorrhage.
Methods
Study design
This retrospective cohort study received full ethical approval (North West 3 Research Ethics Committee). Participants were recruited between November 2009 and January 2011. Eligibility criteria: warfarin prescribed for ≥ 3 months; age ≤ 18 years; therapeutic drug monitoring of INR undertaken by Alder Hey Children’s Hospital, UK, a tertiary referral centre; and written informed consent received from parent (participant age < 16 years) or participant (if age ≥ 16 years). Participant assent was sought from children and young people (each child assessed individually for level of understanding). Refusal of assent by a young person with good understanding of the study was an exclusion criterion.
The majority of children in Alder Hey Children’s hospital on warfarin are cared for in the community, using point of care (POC) INR testing. Management of children receiving warfarin using POC testing has been shown to be a safe and effective alternative to testing of INRs in clinic 29. Patients and families inform the cardiac liaison nurse team about the INRs and dose adjustment’s carried out 30, and all this data are recorded electronically. All data (INR, dose, clinical variables) were collected retrospectively from the cardiac liaison team electronic database and medical records, from the date of initiation of warfarin therapy to the date of recruitment. The current recommended levels of anticoagulation for children at Alder Hey children’s hospital are derived from British Society of Haematology guidelines (Supplementary Data Table 1) 2.
Data collection
Warfarin dose and INR values for each POC testing event is routinely collected for all patients using warfarin at our institution, and this data were extracted onto our study database. From this information, each patient’s full dose and INR history could be determined.
Data were also collected on indication for treatment, target INR, age, gender, height, weight, BMI, haemorrhagic complications, serial serum albumin concentration(s) (as many children were hypoalbuminaemic at onset of therapy), and height and weight measurements. Clinical data were collected from hospital notes and the cardiac liaison team database.
All variables were checked for completeness and accuracy. Where missing data were noted, an attempt was made to retrieve these data. To check data accuracy, range and consistency checks were undertaken.
DNA collection and extraction, and Genotyping
Genotyping of CYP2C9*2 (rs 1799853), CYP2C9*3 (rs 1057910) and VKORC1-1639 (rs 9923231) was undertaken. Full details are given in the online supplementary data section.
Outcomes
Study outcomes were as pre-specified in the protocol, as follows:
Primary outcome variable
The proportion of time in which INR measurements fell within the target range (PTIR) within the first six months. Linear interpolation 31 was used to estimate the proportion of time patients spent in therapeutic range between two test days.
Secondary outcome measures
INR exceeding the target range within the first week of treatment (Yes/No);
Stable dose (mg/day), defined as the mean daily dose required to achieve three consecutive INR measurements within the individual’s target range over a minimum period of four weeks, at the same daily dose;
Haemorrhagic complications: classified according to the methods of both Fihn et al. and Streif et al. 5, 32
Statistical analysis
All analyses were performed in R version 2.13 33. The following quality control (QC) for each single nucleotide polymorphism (SNP) was undertaken prior to analyses of association: checks for Hardy-Weinberg equilibrium (p <0.01 indicated deviation); checks for missingness per sample (excluded if >5% missing); checks for missingness per SNP (excluded if >5% missing); checks for SNP minor allele frequency (MAF) (exclude if MAF <1%).
For the purpose of calculating the outcome of stable dose, where a patient was on a dosing regimen that required different doses to be taken on different weekdays, the daily warfarin dose was taken as the average dose over a week (e.g. if a patient takes 2mg for 2 consecutive days then 3mg on the third day, the average weekly dose is 16mg, and therefore the mean daily dose was calculated as 2.3mg). For the purpose of estimating the INR value for a day that fell between two test days, the method of linear interpolation 34 was used.
In terms of tests of association, first, non-genetic variables (age, gender, height, weight, BMI, albumin, target INR group indication for treatment, and haemorrhagic complications), specified a priori as being of potential importance, were each individually tested for association with each outcome. Patients were grouped according to their target INR range, with groups based on the lower limit of the target INR range (rounded to the nearest 0.5).
Two different classifications for indication for treatment were used: two-group, in which patients were divided into non-Fontans and Fontans groups; or three-group, in which patients were divided into non-Fontans cardiac, Fontans cardiac and non-cardiac groups. For the primary outcome and the outcome of INR exceeding target range during the first week, values of these variables at the start of warfarin treatment were referred to. For the outcome of stable dose, values of these variables at the time stability was achieved were referred to. For the outcome of haemorrhagic complications, no tests for association with non-genetic variables were undertaken, since it was not possible to determine what time point should be referred to for obtaining the value of these variables in those not experiencing an event.
For some of the non-genetic variables there was a considerable amount of missing data (see Table 1). In order to minimise the impact of this, multiple imputation using chained equations 34, specifically the predictive mean matching method, was used to impute data for height, weight and albumin at the start of warfarin treatment (all variables had <30% missing observations). Multiple imputation was not used for these variables at the time stable dose was achieved, as the amount of missingness was deemed too high (>40%). Instead, these variables were excluded from the list of potential covariates.
Table 1.
Summary of participant characteristics at the start of warfarin therapy and at the time stable dose was achieved
| Variable | Start of therapy (n=97) | Time of stable dose (n=85)* | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Missing | Mean | SD | Missing | |
| Age (decimal yrs)# | 2.3 (median) | 4.2 (IQR) | 0 | 5.7 (median) | 8.0 (IQR) | 0 |
| Height (m) | 0.94 | 0.28 | 26 | 1.06 | 0.28 | 40 |
| Weight (kg) | 15.7 | 14.5 | 12 | 20.6 | 15.7 | 35 |
| BMI (kg.m−2)† | 16.1 | 3.1 | 26 | 16.5 | 2.5 | 40 |
| Albumin (g.L−1) | 38.3 | 5.3 | 30 | 39.2 | 6.2 | 62 |
12 patients did not achieve stable dose during follow-up (see supplementary table 2)
Age distribution was skewed, hence median and IQR range given
BMI = body mass index
To test for association with each individual SNP, two regression models were fitted. The first (the ‘baseline model’) included all non-genetic factors giving a p-value < 0.10 univariately and the second (the ‘genetic model’) was the same as the first but also included covariates to represent the SNP.
Both models were compared using the likelihood ratio test. Two analyses were carried out for each SNP: one with no underlying assumptions regarding the mode of inheritance (i.e. using two variables to represent the SNP – one representing heterozygotes, one representing mutant homozygotes) and another assuming an additive mode of inheritance (i.e. using a single variable to represent the SNP, with heterozyotes coded ‘1’ and mutant homozygotes coded ‘2’). Due to the multiple tests of association, the false discovery rate (FDR) controlling procedure was used, with an overall error rate of 5% 35. For the outcome of haemorrhagic complications and any outcome where none of the non-genetic variables were significant univariately, the baseline model was the null model, since it did not include any variables.
Finally, to investigate the total amount of variation explained by non-genetic and genetic factors combined, a multiple regression model was fitted for each outcome. Covariates included non-genetic factors giving p < 0.10 univariately as well as SNPs that were significant under FDR control. For continuous variables, multiple linear regression was performed and the adjusted R2 value was recorded. For binary variables, logistic regression was performed and pseudo R2 values calculated
Results
Participant characteristics
Participant characteristics are provided in Table 1. A total of 100 patients were recruited (Male n=55), all of whom took warfarin for ≥6 months. Ethnicity of the patients was reported as European Caucasian (n=97), Black (n=1), Indian (n=1) and Afghan (n=1). Further analyses were restricted to the 97 European Caucasians - 85 of these were anticoagulated following surgery for congenital heart disease (Fontan’s procedure n=62; other cardiac procedures n=23) and 12 were non-cardiac patients. The lower limit of the target INR range (rounded to the nearest 0.5) was 1.5 in 3 patients, 2.0 in 71 patients, 2.5 in 8 patients and 3.0 in 15 patients. The genotype frequencies of the patients were: CYP2C9*2 76 (78.4%), 19 (19.6%) and 2 (2.1%) (Homozygous wildtype, heterozygote, homozygote variant respectively); CYP2C9*3 79 (81.4%), 18 (18.6%), and 0 (0%); VKORC1-1639 40 (41.2%), 45 (46.4%), and 12 (12.4%). Genotype combinations are shown in supplementary table 3. All three SNPs were in Hardy-Weinberg equilibrium (p > 0.60).
Outcome data
The mean PTIR in the first six months of treatment was 0.50 (SD 0.27) and the mean stable warfarin dose was 2.70 (SD 1.65) mg/day. Thirty-nine (40.2%) patients had an INR above range in the first week, 49 (50.5%) did not, and insufficient data were available to determine this in nine (9.3%) patients. There were no reported major (Streif classification5), or severe (or greater) (Fihn Classification32) haemorrhagic complications. Sixty-nine (71.1%) patients experienced minor (Fihn classification32) or mild (Streif classification5 bleeds. A chi-square analysis between INR≥4 in week one and minor/mild bleeding did not show a significant association (p=0.19).
Analyses of association with each non-genetic variable
Data regarding associations between the non-genetic factors and outcome variables (with the exception of the haemorrhagic complications outcome) are shown in Table 3. It was not possible to test for association between height, weight, BMI or albumin levels and stable dose due to the significant amount of missing observations for these variables at the time stable dose was achieved. Target INR group and indication for treatment were both associated with proportion time in INR range during the first six months (p<0.10), and were therefore adjusted for in the SNP-association analyses with this outcome. None of the non-genetic variables were associated with the outcome of INR exceeding target range during the first week,whilst age and target INR group were associated with stable dose (p<0.10).
Table 3.
Individual SNP association analyses
| Outcome | SNP (assumption) | p-value from LRT |
|---|---|---|
|
Proportion of time in INR range (PTIR) (adjusted for indication for treatment and target INR group) |
CYP2C9*2 (additive) | 0.53 |
| CYP2C9*3 (NA†) | 0.95 | |
| VKORC1-1639 (additive) | 0.001 * | |
| INR above range in week 1 | CYP2C9*2 (additive) | 0.004 * |
| CYP2C9*3 (NA†) | 0.800 | |
| VKORC1-1639 (additive) | 0.020 | |
|
Stable dose (adjusted for age and target INR group) |
CYP2C9*2 (additive) | 0.008 * |
| CYP2C9*3 (NA†) | 0.049 | |
| VKORC1-1639 (additive) | 0.003 * | |
| Haemorrhagic complications | CYP2C9*2 (additive) | 0.423 |
| CYP2C9*3 (NA†) | 0.482 | |
| VKORC1-1639 (none) | 0.006 * |
FDR = false discovery rate; LRT = likelihood ratio test.
No mutant homozygotes so assumption regarding mode of inheritance irrelevant
Remains statistically significant following False discovery rate (FDR) adjustment
Full details of all modes of inheritance are shown in supplementary table
Analyses of association with each SNP
Results from undertaking likelihood-ratio tests comparing the baseline model to the genetic model for each outcome are provided in Table 3. As analyses were undertaken both without any assumptions regarding the underlying mode of inheritance and assuming an additive mode, only the lowest p-value is reported. SNPs that remained significant under FDR control were entered into the final multiple regression models (Table 4).
Table 4.
Final multiple regression models
| Outcome | Variables included | Adjusted/pseudo R2 | ||
|---|---|---|---|---|
| Non-genetic variables | Genetic variables | All variables | ||
| PTIR | Indication for treatment | 11.3% | 9.5% | 20.8% |
| INR group | ||||
| VKORC1-1639 (additive) | ||||
|
| ||||
| INR exceeding target range in week 1 | CYP2C9*2 (additive) | - | 6.8% | 6.8% |
|
| ||||
| Stable dose | Age | 29.2% | 11.9% | 41.4% |
| INR group | ||||
| CYP2C9*2 (additive) | ||||
| VKORC1-1639 (additive) | ||||
|
| ||||
| Haemorrhagic complications | VKORC1-1639 (none) | - | 8.7% | 8.7% |
PTIR = Proportion of time spent in target INR range.
Multiple regression models
The R2 values for multiple regression models built for each outcome, including the non-genetic variables with p <0.10 and genetic variables significant under FDR control, are provided in Table 4.
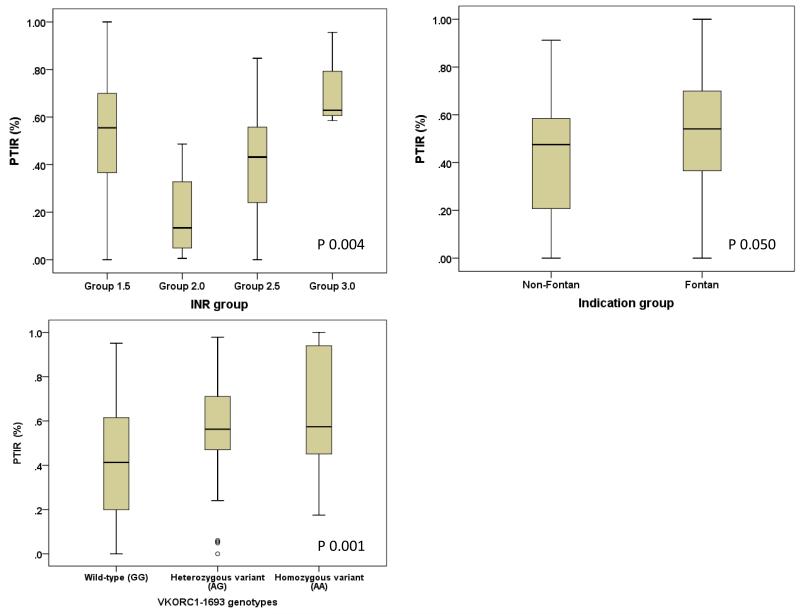
Together, the indication for treatment and target INR groupings accounted for approximately 11% of the variance in the PTIR during the first six. The presence of the variant allele VKORC1-1639 was associated with a greater time spent within range, with each additional variant allele associated with approximately 13% (95% CI 5% to 21%) more time spent within the target INR range in the first six months. VKORC1-1639 combined with the aforementioned non-genetic factors explained almost 21% variability in proportion of the first six months’ treatment time spent within the target INR range. Genetic data related to proportion of time INR within target range is shown in Figure 1. Patients heterozygous for CYP2C9*2 were approximately four times as likely, and mutant homozygotes around 17 times as likely, to have INR values exceeding the target range in the first week as wild-type homozygous patients.
Figure 1. Box and whisker plots showing the distribution of PTIR based on (a) target INR group, (b) indication treatment group, and (c) stratified by VKORC1 genotypes.
Boxes represent 25th-75th percentile (interquartile range) of PTIR, whiskers represent 5th-95th percentile, solid lines represent median dose. Open dots denote outliers (a value between 1.5 and 3 times the interquartile range away from the 25th or 75th percentile).
Overall, the patient’s age, target INR range and genotypes at CYP2C9*2 and VKORC1-1639 accounted for approximately 41% of the variability in the warfarin dose required to stabilise INR. The presence of the variant alleles, CYP2C9*2 and VKORC1-1639, was associated with a lower dose required to stabilise INR, with CYP2C9*2 variant alleles associated with an approximate decrease in daily dose of 0.82 mg and VKORC1-1639 variant alleles associated with an approximate decrease of 0.66 mg. Based on the change in adjusted R2 values with the addition of the genetic variables, VKORC1-1639 accounted for approximately 7% and CYP2C9*2 approximately 5% of the overall variance in stable dose.
The presence of one variant allele at VKORC1-1639 was associated with an increased likelihood of haemorrhagic complications, with heteroygotes having approximately 4.5 times the likelihood of a bleeding event as wild-type homozygotes. No significant association was found between having two copies of the mutant allele and haemorrhagic complications largely because these individuals were rare and we had insufficient power to detect an association.
Discussion
Many significant pharmacogenomic associations do not make clinical impact due to lack of replication. We are therefore pleased to report that these data replicate previous publications showing that VKORC1-1639 and CYP2C9*2 both result in decreased stable warfarin dose requirements. The estimates of R2 for daily dose requirement that we have derived for these variant alleles suggests that VKORC1-1639 and CYP2C9*2 contribute 7% and 5% respectively. Previously published R2 values for the effect of VKORC1-1639 on stable warfarin dose requirements have produced values of 3.7% 27, 18.2% 26 and 26.6% 25, putting our value of 7% at the lower end of the range. The R2 values previously published for the effects of mutant alleles of CYP2C9 on stable warfarin dose were 0.4% 27, 2.0 26 and 12.8% 25, and again our value of 5% is consistent. The total variability in stable warfarin dosing explained by the genetic factors above, as well as target INR range and age, in our cohort is 41.4%. This is similar to Nowak-Göttl et al. (38%), but lower than Biss et al. (72.4%) and Moreau et al. (69.7%). Possible reasons for the difference include the age of the children studied (younger than in previous studies) and the concomitant use of age as a surrogate for height/weight. Fifty-four percent of our study cohort were less than 6 years old when stable dose was reached, compared to 23% in the cohort studied by Biss et al 25. Similarly, in the study by Moreau et al.26 the mean age was 8.4 years (± 5.6) while in this study, it was 6.9 (± 4.5) years. It is possible that genetic factors may become more important as the child grows older and would be consistent with the increased expression of CYP2C9 seen through gestation and childhood 36. An alternative approach would be to conduct a meta-analysis of stable doses of warfarin in children which was considered by us. However, in the six previously published studies, there was a lack of reporting of key data (including mean dose and standard deviation per genotype group). Indeed, the only meta-analysis possible would have been limited to a single polymorphism (VKORC1-1639) and included only two out of the six studies, and so would not significantly have improved the evidence base. Ideally there is a need for an individual patient data meta-analysis, as well as prospective randomized studies similar to those undertaken in adults 21.
The finding that the PTIR is significantly associated with indication for treatment (Fontans or Non-Fontans) also corresponds with that of Moreau et al 26. However, our study showed that PTIR was also significantly associated with VKORC1 polymorphisms, with each additional variant allele associated with approximately 13% more time spent in range in the first six months. This contrasts with previous paediatric 25 and adult 37 studies, where those homozygous for VKORC1 variant alleles spent more treatment time above their INR therapeutic range (although not significantly in the paediatric study 25). In addition, Biss et al. 28 found no association between the CYP2C9 and VKORC1 genotypes and the proportion of INR values above the target range beyond the first month of therapy 25. This suggests that the adjustment of warfarin doses based on INR after the first month of therapy counteracted the influence of CYP2C9 and VKORC1 genotypes. A possible explanation is that carriers of VKORC1-1639 variant allele are more sensitive to warfarin and may have had more unstable INRs which may have led to more frequent monitoring by their POCT, and more frequent dose titration leading to a more stable INR over the duration of our follow up. However, we did not have the data to confirm this in our cohort. Nevertheless, it is consistent with the fact that POCT monitoring of INR is associated with safe anticoagulation 38. It may also explain some of the differences between our studies and those which have been published previously – only one of the previous studies 26 states that it included children who were monitored by POCT.
Clinically, the initiation of warfarin therapy is a time when large variations in the INR can occur as the stable dose has not yet been established. This replicates the previous data showing that possession of CYP2C9*2 alleles increase the likelihood of above therapeutic range INRs in the first week 28, although as this only explains 6.8% there are clearly other, more significant factors involved here. However, it does add to the evidence suggesting that inclusion of pharmacogenomic data in paediatric dosing algorithms could improve the management of children using warfarin.
This is also the first paediatric study to include minor bleeding complications, as we feel that INR variations are not necessarily associated with poor clinical outcomes. Heterozygotes for VKORC1-1639 were significantly associated with increased minor bleeding complications; however, unlike previous adult studies39, CYP2C9 variants were not. This may reflect the smaller sample size in our study. We believe these data also add to the clinical case for developing pharmacogenomic based dosing algorithms for children using warfarin, as it is the first evidence linking the lower dose required in children with VKORC1-1639 mutant alleles with the adverse effects of overtreatment with warfarin.
Although we recruited 100 children, analysis was limited to 97 to ensure identical ethnic origin, and we did not have any children homozygous for CYP2C9*3, limiting the analyses possible with these data. In addition, these data, although recorded systematically from a single site, in a prospective manner, by the team undertaking the POC testing, were collected retrospectively and so there were missing data that could not be determined. It was also not possible to ascertain compliance with medication in this cohort, and this may explain a significant part of the currently unexplained variability. However, this is a closely monitored population through the POC testing system used, and regular education of the patients and family is undertaken, minimizing this potential limitation as much as possible.
In conclusion, we have added to the evidence showing that variant alleles in CYP2C9 and VKORC1 are important determinants of warfarin dose requirements and anticoagulation stability in children. However, as with other studies in children, this is based on a retrospective cohort, which may introduce bias into the associations. Our data also indicate that age may be important in determining the relative contribution of genetic and non-genetic factors. Given all of these complexities, the development of dosing algorithms in children will be extremely complicated and will need to initially undertake an individual patient data meta-analysis as was undertaken in adults by the International Warfarin Pharmacogenetics Consortium 40. Furthermore, the dosing algorithms will have to be robust and take into account the pharmacokinetics of warfarin. The importance of dosing algorithms has been highlighted in two recent trials in adults where different results were obtained based on the dosing strategies utilized21, 41.
Supplementary Material
Table 2.
P values from tests of association of non-genetic variables with initial outcome measures
| PTIR in 1st 6 months |
INR exceeding target range in week 1 (Y/N) |
Stable dose (mg/day) |
|
|---|---|---|---|
| Age | 0.144 | 0.584 | <0.001 * |
| Height | 0.105 | 0.802 | - |
| Weight | 0.336 | 0.619 | - |
| BMI | 0.334 | 0.215 | - |
| Albumin | 0.982 | 0.897 | - |
| Gender | 0.468 | 0.173 | 0.351 |
| INR group | 0.004 * | 0.657 | 0.015* |
| Indication (2 groups) | 0.050 * | 0.543 | 0.379 |
| Indication (3 groups) | 0.144 | 0.771 | 0.575 |
PTIR: Proportion of time spent in target INR range. 2 Groups: Patients were divided into non-Fontan and Fontan group. 3 Groups: patients were divided into non-Fontan cardiac, Fontan cardiac and non-cardiac. - Variable excluded due to high amount of missingness (>30%).
Variables with p < 0.10 were included as covariates in subsequent analyses of association
Acknowledgements
This work was funded by the MRC Centre for Drug Safety Science (CDSS) at the University of Liverpool, and the Department of Health Chair in Pharmacogenetics.
Footnotes
The authors have not competing interests to disclose.
References
- 1.Monagle P, Michelson AD, Bovill E, Andrew M. Antithrombotic therapy in children. Chest. 2001;119(1):344S–370S. doi: 10.1378/chest.119.1_suppl.344s. [DOI] [PubMed] [Google Scholar]
- 2.Guidelines on oral anticoagulation. third edition. 1998. pp. 374–387. [DOI] [PubMed] [Google Scholar]
- 3.Andrew M, Marzinotto V, Brooker LA, Adams M, Ginsberg J, Freedom R, et al. ORAL ANTICOAGULATION THERAPY IN PEDIATRIC-PATIENTS - A PROSPECTIVE-STUDY. Thromb Haemost. 1994;71(3):265–269. [PubMed] [Google Scholar]
- 4.Buck ML. Anticoagulation with warfarin in infants and children. Ann Pharmacother. 1996;30(11):1316–1322. doi: 10.1177/106002809603001117. [DOI] [PubMed] [Google Scholar]
- 5.Streif W, Andrew M, Marzinotto V, Massicotte P, Chan AKC, Julian JA, et al. Analysis of warfarin therapy in pediatric patients: A prospective cohort study of 319 patients. Blood. 1999;94(9):3007–3014. [PubMed] [Google Scholar]
- 6.Bonduel M, Sciuccati G, Hepner M, Torres AF, Pieroni G, Frontroth JP, et al. Acenocoumarol therapy in pediatric patients. Journal of Thrombosis and Haemostasis. 2003;1(8):1740–1743. doi: 10.1046/j.1538-7836.2003.00256.x. [DOI] [PubMed] [Google Scholar]
- 7.Tait RC, Ladusans EJ, ElMetaal M, Patel RG, Will AM. Oral anticoagulation in paediatric patients: Dose requirements and complications. Arch Dis Child. 1996;74(3):228–231. doi: 10.1136/adc.74.3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7(2):99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 9.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MTM, et al. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. New England Journal of Medicine. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106(7):2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Schwarz UI, Ritchie MD, Roden DM, Stein CM, Kurnik D. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009;113(17):3925–3930. doi: 10.1182/blood-2008-09-176859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limdi N, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83(2):312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limdi NA, Karnett D, Goldstein JA, Beasley TM, McGwin G, Adler BK, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9(5):511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obayashi K, Nakamura K, Kawana J, Ogata H, Hanada K, Kurabayashi M, et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clin Pharmacol Ther. 2006;80(2):169–178. doi: 10.1016/j.clpt.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. New England Journal of Medicine. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 16.Veenstra DL, You JHS, Rieder MJ, Farin FM, Wilkerson HW, Blough DK, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15(10):687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 17.Cavallari LH, Momary KM, Patel SR, Shapiro NL, Nutescu E, Viana MAG. Pharmacogenomics of Warfarin dose requirements in Hispanics. Blood Cells Mol Dis. 2011;46(2):147–150. doi: 10.1016/j.bcmd.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen AL, Al-Zubiedi S, Zhang JE, Keniry A, Hanson A, Hughes DA, et al. Genetic and environmental factors determining clinical outcomes and cost of warfarin therapy: a prospective study. Pharmacogenet Genomics. 2009;19(10):800–812. doi: 10.1097/FPC.0b013e3283317ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan G-M, Wu E, Lam Y-Y, Yan BP. Role of warfarin pharmacogenetic testing in clinical practice. Pharmacogenomics. 2010;11(3):439–448. doi: 10.2217/pgs.10.8. [DOI] [PubMed] [Google Scholar]
- 20.You JHS, Wong RSM, Waye MMY, Mu YW, Lim CK, Choi KC, et al. Warfarin dosing algorithm using clinical, demographic and pharmacogenetic data from Chinese patients. J Thromb Thrombolysis. 2011;31(1):113–118. doi: 10.1007/s11239-010-0497-x. [DOI] [PubMed] [Google Scholar]
- 21.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, et al. A Randomized Trial of Genotype-Guided Dosing of Warfarin. N Engl J Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 22.Manco-Johnson MJ. How I treat venous thrombosis in children. Blood. 2006;107(1):21–29. doi: 10.1182/blood-2004-11-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne JH. Aspects of anticoagulation in children. British Journal of Haematology. 2010;150(3):259–277. doi: 10.1111/j.1365-2141.2010.08225.x. [DOI] [PubMed] [Google Scholar]
- 24.Thornburg CD, Jones E, Bomgaars L, Gage BF. Pediatric warfarin practice and pharmacogenetic testing. Thromb Res. 2010;126(2):E144–E146. doi: 10.1016/j.thromres.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 25.Biss TT, Avery PJ, Brandao LR, Chalmers EA, Williams MD, Grainger JD, et al. VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood. 2012;119(3):868–873. doi: 10.1182/blood-2011-08-372722. [DOI] [PubMed] [Google Scholar]
- 26.Moreau C, Bajolle F, Siguret V, Lasne D, Golmard JL, Elie C, et al. Vitamin K antagonists in children with heart disease: Height and VKORC1 genotype are the main determinants of the warfarin dose requirement. Blood. 2012;119(3):861–867. doi: 10.1182/blood-2011-07-365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak-Gottl U, Dietrich K, Schaffranek D, Eldin NS, Yasui Y, Geisen C, et al. In pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypes. Blood. 2010;116(26):6101–6105. doi: 10.1182/blood-2010-05-283861. [DOI] [PubMed] [Google Scholar]
- 28.Biss TT, Avery PJ, Williams MD, BrandÃO LR, Grainger JD, Kamali F. The VKORC1 and CYP2C9 genotypes are associated with over-anticoagulation during initiation of warfarin therapy in children. Journal of Thrombosis and Haemostasis. 2013;11(2):373–375. doi: 10.1111/jth.12072. [DOI] [PubMed] [Google Scholar]
- 29.Bradbury MJE, Taylor G, Short P, Williams MD. A comparative study of anticoagulant control in patients on long-term warfarin using home and hospital monitoring of the international normalised ratio. Arch Dis Child. 2008;93(4):303–306. doi: 10.1136/adc.2006.113886. [DOI] [PubMed] [Google Scholar]
- 30.Murray M, Keenan R, Billington R. Standard Operating Procedure (SOP) for the Management of Children on Warfarin at a Paediatric Regional Anticoagulation Centre. Alder Hey Children’s Hospital; Liverpool: 2005. [Google Scholar]
- 31.Rosendaal FR, Cannegieter SC, Van der Meer FJM, Briet E. A method to determine the optimal intensity of oral anticoagulant therapy. Thrombosis and Haemostasis. 1993;69(3):236–239. [PubMed] [Google Scholar]
- 32.Fihn SD, Callahan CM, Martin DC, McDonell MB, Henikoff JG, White RH. The risk for and severity of bleeding complications in elderly patients treated with warfarin. The National Consortium of Anticoagulation Clinics. Ann Intern Med. 1996;124(11):970–979. doi: 10.7326/0003-4819-124-11-199606010-00004. [DOI] [PubMed] [Google Scholar]
- 33.Team RDC, editor. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 34.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software. 45(3):1–67. [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. [Google Scholar]
- 36.Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, et al. Developmental Expression of Human Hepatic CYP2C9 and CYP2C19. Journal of Pharmacology and Experimental Therapeutics. 2004;308(3):965–974. doi: 10.1124/jpet.103.060137. [DOI] [PubMed] [Google Scholar]
- 37.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heneghan C, Ward A, Perera R, Bankhead C, Fuller A, Stevens R, et al. Self-monitoring of oral anticoagulation: systematic review and meta-analysis of individual patient data. The Lancet. 379(9813):322–334. doi: 10.1016/S0140-6736(11)61294-4. [DOI] [PubMed] [Google Scholar]
- 39.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: A HuGEnet[trade] systematic review and meta-analysis. Genet Med. 2005;7(2):97–104. doi: 10.1097/01.gim.0000153664.65759.cf. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen AL, FitzGerald RJ, Oyee J, Pirmohamed M, Williamson PR. Influence of CYP2C9 and VKORC1 on Patient Response to Warfarin: A Systematic Review and Meta-Analysis. PLoS ONE. 2012;7(8):e44064. doi: 10.1371/journal.pone.0044064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, et al. A Pharmacogenetic versus a Clinical Algorithm for Warfarin Dosing. N Engl J Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oragene·DNA. DNAgenotek; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.