Abstract
During cancer progression, malignant cells in the tumour invade surrounding tissues. This transformation of adherent cells to a motile phenotype has been associated with the epithelial mesenchymal transition (EMT). Here, we show that EMT-activated cells migrate through micropillar arrays as a collectively advancing front that scatters individual cells. Individual cells with few neighbours dispersed with fast, straight trajectories, whereas cells that encountered many neighbours migrated collectively with epithelial biomarkers. We modelled these emergent dynamics using a physical analogy to solidification phase transitions in binary mixtures, and validated it using drug perturbations, which revealed that individually migrating cells exhibit diminished chemosensitivity. Our measurements also indicate a degree of phenotypic plasticity as cells interconvert between individual and collective migration. The study of multicellular behaviours with single-cell resolution should enable further quantitative insights into heterogeneous tumour invasion.
Introduction
Heterogeneous tumour populations with differential capabilities for invasion, metastasis and therapeutic resistance often thwart existing anticancer treatments.1 One signature of malignant behaviour is the dissemination of individual cells from a tumour front.2 It has been hypothesized that these invasive cells arise from a coordinated program of phenotypic changes known as the epithelial-mesenchymal transition (EMT).3,4 During EMT, cells lose epithelial characteristics associated with collective invasion (strong cell-cell contacts and limited motility) while acquiring mesenchymal traits associated with individual invasion (weakened cell-cell contacts, increased motility and elongated spindle-like morphologies).5 Furthermore, EMT is associated with enhancements in drug resistance and anti-apoptosis pathways.6 EMT thus represents a significant therapeutic challenge by generating a mesenchymal subpopulation with a distinct fitness advantage in hostile microenvironments. This subpopulation may play a disproportionate role in driving metastasis, drug resistance and systemic tumour relapse.
The emergence of complex invasion behaviours associated with malignant tumour heterogeneity has been technically challenging to measure using existing assays.7,8 For instance, histological analysis only provides a static snapshot of tumour pathology.9 Intravital imaging in small animal models has revealed qualitative transitions from collective to individual invasion,10-12 but this is a low-throughput, resource-intensive technique. In comparison, in vitro assays have the potential for increased experimental control and higher-throughput measurements.13,14 Nevertheless, existing approaches are insufficient to characterize heterogeneous populations,8 and are specialized for either the individual migration of cells in Boyden chambers15 or the collective migration of multicellular sheets in wound healing assays.16-21 More recently, microfluidic devices have been developed to study individual cell migration in the presence of well-controlled chemokine gradients22 and 3D assays have explored the role of extracellular matrix architectures.23 In the context of EMT, these myriad in vitro migration assays have probed how the activation of pathways such as Snail24-26 or YAP27 affect migration in separate populations. However, existing assays cannot resolve heterogeneity within the same population. In order to construct an integrative, multiscale description of these complex phenomena, it is necessary to measure the activities of all the single cells that comprise the population.28
Here, we show that cells that have undergone EMT migrate through engineered microenvironments as a collectively advancing front from which individually migrating cells scattered. These dynamics were measured using automated tracking, enabling single cell resolution. Our measurements indicate that individually migrating cells display faster and straighter trajectories, enabling efficient dispersal away from their collectively advancing counterparts. Moreover, a collectively migrating subpopulation arose over the course of the experiment from cells that also overexpressed epithelial biomarkers. These complex phenomena were quantitatively modelled using a physical analogy with the solidification of binary mixtures. These behaviours were further perturbed using small molecule inhibitors, revealing that individually migrating cells exhibited diminished chemosensitivity compared to their collectively migrating counterparts.
Results
Cancer cell migration was characterized in enclosed microenvironments consisting of an array of polydimethylsiloxane (PDMS) micropillars coated with fibronectin (Fig. 1A). This architecture was designed to mechanically confine cells between micropillars, a bottom “floor” and a top “ceiling.” In this geometry, the periodic disruption of cell-cell interactions and enhancement of cell-surface interactions promoted individual cell scattering from a collectively advancing front. These microfabricated devices were also highly reproducible, allowing parallel measurements of multiple conditions simultaneously in a multiwell plate format.29 Cell nuclei were fluorescently labelled prior to experiments and cell plating into these devices. Their subsequent motion was imaged using time-lapse optical microscopy and tracked using the LAP algorithm, which accounts for division events by considering both daughter cells as new cells30 (Fig. 1B). Quantitative descriptors of migration were determined for each cell by averaging over their entire lifetime, which is representative of their overall behaviour, although transient dynamic are observed (Note SN1, Fig. S1-S4). These single cell descriptors were then classified using a Gaussian mixture model and independently assessed using principle component analysis (PCA), yielding qualitatively similar results (Note SN2). This functional classification of migration was further corroborated by immunostaining for biomarker expression.
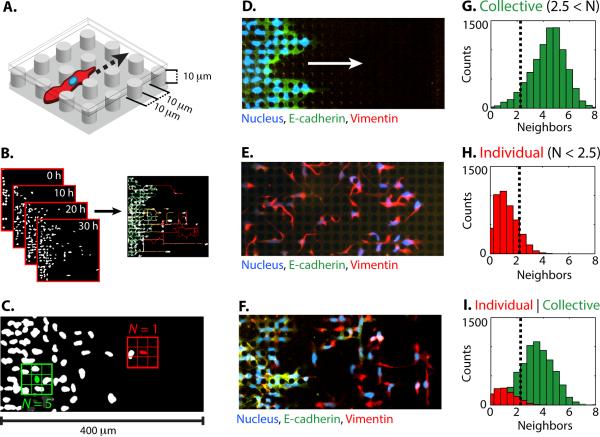
Fig. 1. Epithelial and mesenchymal cells migrate collectively and individually within enclosed micropillar arrays.
(A) Cells invaded an enclosed array of fibronectin-coated PDMS micropillars with height, diameter and spacing of 10 μm. (B) Cell migration was automatically tracked from time-lapse microscopy by segmenting fluorescently labelled nuclei. (C) A representative metric for collective or individual migration is based on the lifetime-average number of nearest neighbours within one pillar spacing. Immunofluorescent staining reveals biomarker expression associated with (D) epithelial phenotype (E-cadherin, green) in MCF-10A, (E) mesenchymal phenotype (vimentin, red) in MDA-MB-231, (F) both epithelial (E-cadherin, green) and mesenchymal phenotypes (vimentin, red) in MCF-10A Snail. Histograms of the number of lifetime averaged nearest neighbours per cell indicate (G) collective migration in MCF-10A, (H) individual migration in MDA-MB-231, and (I) collective and individual migration in MCF-10A Snail.
Profiling Collective and Individual Migration
One metric to classify collective or individual migration was based on the average number of nearest neighbours N (within one lattice spacing) encountered during its trajectory (Fig. 1C). Since the micropillars were arrayed as a regular lattice, the cell nuclei were approximately localized to discrete locations on a grid. In this geometry, each cell could have at most eight nearest neighbours. A baseline for collective migration was first established using a homogeneous population of mammary epithelial cells (MCF-10A) (Video S1). These cells expressed high levels of the epithelial biomarker E-cadherin (Fig. 1D) and cells tended to move with many nearest neighbours in a multicellular sheet, with a median of N ~ 4.5±1.3 (Fig. 1G). Next, a baseline for individual migration was established using a homogeneous population of breast adenocarcinoma cells (MDA-MB-231). These cells expressed high levels of the mesenchymal marker vimentin (Fig. 1E) and cells tended to move with few nearest neighbours, with a median of N ~ 1.1±0.8 (Fig. 1H). Based on this metric, collective and individual migration phenotypes could be quantitatively distinguished by a threshold of N ~ 2.5 nearest neighbours.
Next, EMT was rapidly induced in MCF-10A through a Snail-6SA (Snail) expression construct using a fused estrogen receptor (ER) response element.25 Treatment with tamoxifen (4-OHT) for 72 h resulted in complete phenotypic changes for the entire population, including elongated, spindle-like morphologies, down regulation of epithelial biomarkers (E-cadherin, P- cadherin, etc.) and up regulation of mesenchymal biomarkers (vimentin, fibronectin, serpin, etc.), consistent with EMT (Fig. S5).31 This approach allows consistent induction of EMT, compared to model systems based on prolonged exposure to growth factors or ectopic Snail 1. Nevertheless, while this MCF-10A Snail population initially displayed >95% mesenchymal biomarker expression and minimal epithelial biomarker expression, their dynamic migration behaviours over the course of the experiment (Video S2) were not consistent with the homogeneous migration behaviours observed with the purely epithelial MCF-10A or purely mesenchymal MDA-MB-231. Instead, the single cell migration dynamics were highly heterogeneous, with over half of the cells displaying transitions between collective migration (with many neighbours) and individual migration (with no neighbours) (Note SN1, Fig. S3). Other cells maintained a collective or individual migration phenotype throughout the experiment. To account for these transient periods of collective migration, the cut-off for the number of neighbours averaged over the lifetime of the trajectory was set higher at N ~ 2.5 rather than N ~ 1. For simplicity, always collectively and mostly collectively migrating cells were grouped together (N > 2.5), while always individually and mostly individually migrating cells were also grouped together (N < 2.5). This classification of two subpopulations was consistent with the biomarker expression at the conclusion of the experiment (60 h), with an epithelial subpopulation located towards the rear that highly expressed E-cadherin (Fig. 1F, left) and a mesenchymal subpopulation advancing towards the front that highly expressed vimentin (Fig. 1F, right). Overall, the distribution of lifetime averaged nearest neighbours N for MCF-10A Snail was much broader than those observed for MCF-10A or MDA-MB-231, with a wide distribution both spanning above and below N ~ 2.5 (Fig. 1I).
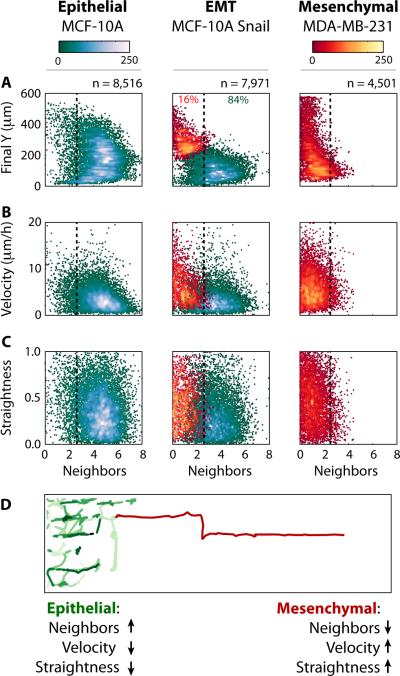
To classify these distinct collective or individual migration behaviours, a systematic comparison of the purely epithelial population (MCF-10A), the induced EMT population (MCF-10A Snail) and purely mesenchymal population (MDA-MB-231) was performed (Fig. 2, S6-9). For each population, the statistical distributions of four quantitative metrics were considered: (A) final Y position in the direction of migration; (B) lifetime-averaged number of nearest neighbours N; (C) lifetime-averaged velocity and (D) path straightness (displacement/integrated distance).
Fig. 2. Differences in migratory behaviour associated with collective or individual migration phenotypes were classified using a Gaussian mixture model.
The lifetime-averaged nearest neighbours of the migrating cells are compared to (A) the final Y position in the device, (B) averaged velocity, and (C) path straightness. Overall, at the completion of the experiment, individually migrating cells are observed to the front with collectively migrating cells at the rear. (D) Single cell tracking reveals that individually migrating cells scatter effectively due to increases in speed and straighter trajectories, relative to collectively migrating cells.
First, the distance migrated into the device (Final Y) was compared to the lifetime averaged nearest neighbours N (Fig. 2A). The purely epithelial population (MCF-10A) migrated ~400 μm with many neighbours (N > 2.5), indicative of collective migration. In contrast, the induced EMT population (MCF-10A Snail) was characterized by two subpopulations with distinct migration behaviours. One subpopulation was localized towards the rear, only advancing ~250 μm through collective migration (N > 2.5). A second subpopulation was localized towards the front, advancing to the end of the device (600 μm) through individual migration (N < 2.5). Finally, the purely mesenchymal population (MDA-MB-231) advanced to the end of the device (600 μm) through individual migration (N < 2.5).
Next, the lifetime-averaged velocity was compared to the lifetime averaged nearest neighbours N (Fig. 2B). The purely epithelial population (MCF-10A) migrated collectively with a median velocity of 3.3±2.5 μm/h. The induced EMT population (MCF-10A Snail) was characterized by a slower collectively migrating subpopulation (median velocity of ~ 2.9±2.4 μm/h), as well as a faster individually migrating subpopulation (median velocity ~5.8±4.9 μm/h). Finally, the purely mesenchymal population (MDA-MB-231) migrated individually with a faster median velocity of 5.5±3.6 μm/h. These differences correspond to a roughly two-fold increase in median velocity between collective and individual migration. This use of a median may be misleading as a measure of migration since it includes cells whose motion is hindered due to local crowding effects.18,19 Indeed, single cell migration velocities are highest at the invasion front and decrease with distance inward (Fig. S10), in agreement with wound-healing assays.18 Nevertheless, similar trends are observed for the fastest moving cells at the periphery (95th percentile). For the induced EMT population (MCF-10A Snail), this corresponds to a slower collective migration velocity of 7.8 μm/h, compared to a faster individual migration velocity of 15.3 μm/h.
Finally, the straightness index (Fig. 2C) was compared to the lifetime averaged nearest neighbours N. The purely epithelial population (MCF-10A) migrated collectively with a median straightness index of 0.34±0.22, corresponding to more tortuous trajectories. In comparison, the induced EMT population (MCF-10A Snail) was characterized by a collectively migrating subpopulation with more tortuous trajectories (0.27±0.21), compared to an individually migrating subpopulation with straighter trajectories (0.42±0.24). This difference represents a 50% longer route for a collectively migrating cell compared to an individually migrating cell. Similarly, the purely mesenchymal population (MDA-MB-231) migrated individually with straighter trajectories (0.44±0.24).
These systematic comparisons reveal that, over the course of the experiment, the induced EMT population (MCF-10A Snail) displays heterogeneous migration behaviours that represent a superposition of those observed in the collective migration of epithelial cells (MCF-10A) and the individual migration of mesenchymal cells (MDA-MB-231) (Fig. S6-9 and Note SN2). Classification of these distinct migration behaviours in the induced EMT population (MCF-10A Snail) using a Gaussian mixture model revealed that 16% of the total population migrated individually, while the remaining 84% migrated collectively. This profiling was then verified against immunofluorescent staining for 848 total cells at the completion of the experiment (Fig. S11), revealing 92% agreement between individual migration and vimentin, as well as 94% agreement between collective migration and E-cadherin (Table S1). Discrepancies were observed for cells located more than one lattice spacing away from their neighbours, perhaps in the process of breaking away.
These concurrent differences in migration behaviour promote the dispersion of individually migrating cells from a collectively advancing front (Video S2). In particular, individually migrating cells (MCF-10A Snail) utilize enhanced velocities and straighter trajectories for deep incursions into new territory (Fig. 2D). These differences between collective and individual migration were corroborated by principal component analysis (Note SN2), which grouped final Y position, velocity and straightness index for individual migration as well as a second grouping of lifetime-averaged nearest neighbours for collective migration (Fig. S12). These groupings indicate correlated differences between collective and individual migration phenotype that would not be captured with any of these parameters alone.
This scattering behaviour is strongly enhanced by the use of regularly spaced micropillars in enclosed geometries, which restricts the extent of cell-cell contact and increases the density of aligned actin stress fibers (Fig. S13), as observed elsewhere using channels of varying width.32,33 Scattering occurs optimally at a pillar spacing of 10 μm, which represents a characteristic “pore size” just large enough for one cell (nucleus) to proceed at a time.34 As the spacing is increased further, there is reduced individual scattering (N < 1) and more collective, multicellular “strand” migration (N ~ 2), analogous to those observed elsewhere,17 as well as differences in the relative proportions of collective and individual/strand subpopulations (Fig. S14). The complete absence of these pillars in 2D wound healing assays results in minimal mesenchymal scattering and increased reversion to epithelial phenotype (MET) (Fig. S15). Indeed, the migration behaviour of individually scattered cells in micropillars was the opposite of those observed in 2D wound-healing assays, where individually scattered (epithelial) cells displayed decreased straightness and were eventually recaptured by the collectively advancing front.21 In general, the differences in migratory behaviours between the induced EMT population (MCF-10A Snail) and the epithelial population (MCF-10A), including enhanced speed and straightness, are qualitatively consistent with measurements in 2D geometries, including cells at low density and wound healing assays, as well as with different ECM proteins (Fig. S16, 17). Overall, this individual scattering from a collectively migrating front occurs even after blocking proliferation with mitomycin C (Fig. S18, 19), albeit with slower speeds and reduced cell densities, consistent with previous wound healing experiments.16-18 This is likely a consequence of the relatively slow proliferation rate (division ~48 h) of induced EMT cells (MCF-10A Snail) (Fig. S20).
Binary Mixture Solidification Model
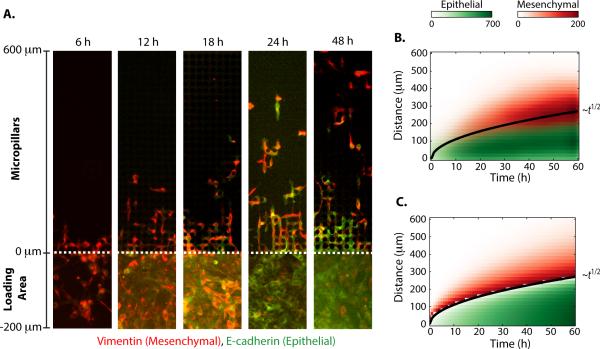
The dispersion of individually migrating cells from a collectively advancing front was then visualized by immunostaining (Fig. 3A). Prior to plating, Snail was induced for 72 h in the entire population, leading to elongated morphologies, high expression of mesenchymal biomarkers (vimentin) and low expression of epithelial biomarkers (E-cadherin) (Fig. S5). These cells were then plated into the loading area immediately adjacent to the micropillars (y < 0 μm). Immediately after plating (6 h), cells had just begun to advance into the micropillars (y > 0 μm). At this time point, all cells overexpressed vimentin and displayed elongated morphologies. At subsequent times (i.e. 18 h), individually migrating cells travelled deeper into the device (y > 100 μm) and continued to express vimentin with elongated morphologies. However, a dense, slower moving subpopulation of collectively advancing cells (y < 100 μm) was also observed at 18 h with increased E-cadherin and diminished vimentin. From 24 h through 48 h, the individually migrating cells traversed through the total length of the device (~600 μm) while maintaining vimentin and elongated morphologies. Nevertheless, the collectively migrating cells mostly expressed E-cadherin with limited expression of vimentin.
Fig. 3. The dynamics of individual scattering from a collectively migrating front can be understood as a dispersion phenomenon from a moving interface.
(A) Immunofluorescent staining of the pillar region (0 < y) reveals individually migrating mesenchymal cells detaching from a collectively migrating epithelial front. Cells in the rear loading region (y < 0) exhibit phenotypic plasticity and undergo a mesenchymal to epithelial transition (MET) over 24 h. (B) The measured spatial distributions of individual and collectively migrating cells are plotted as a function of time, showing an interface that propagates outward as the square root of time. (C) A solidification model for binary mixtures shows quantitative agreement with experimental data.
An examination of cells in the loading area (y < 0 μm) at 6 h also reveals initially high vimentin expression, minimal E-cadherin expression and elongated morphologies (Fig. 3A). Subsequently at 12 h and 18 h, more cells in the loading area display increased epithelial E-cadherin expression, reduced vimentin expression and more compact morphologies. By 24 h, most cells in the loading area primarily display E-cadherin. This behaviour suggests that the MCF-10A Snail cells display some phenotypic plasticity, whereby microenvironmental conditions such as enhanced cell-cell contact can modulate EMT phenotype (Fig. S21). This scenario has analogies with the physics of macrosegregation during the unidirectional solidification of a binary mixture.34 In this phenomenon, a solidification front advances and rejects excess solute ahead of it. This occurs as a consequence of differences in solubility of a molecular species between solid and melt phases. The solute accumulates ahead of the moving interface and may be transported away by diffusive processes. This physical scenario thus occurs as the competition of two physical mechanisms: the rejection of solute at a moving interface and the transport of solute away from the moving interface. During this process, the rejection of solute across the interface causes a cooling of the interface until it again exceeds the maximum concentration of allowed solute. These mechanisms correspond to the detachment of individual cells from a collectively migrating phase and the subsequent dispersal of individual cells by migrating away from the interface.
Based on this physical analogy, a minimal model was developed to capture essential elements of the experimental results. This scenario of binary mixture solidification in a semi-infinite domain is analytically tractable using a similarity solution (Note SN4) and exhibits two features:35 1) The position of the interfacial front moves diffusively with the square root of time, 2) the transient distribution of rejected solute is distributed as a complimentary error function. A comparison of the experimentally measured distributions of individual and collectively invading subpopulations (Fig. 3B) after EMT (MCF-10A Snail) with a best fit to the analytical model (Fig. 3C, S22) shows reasonable agreement (R2 = 0.90). The propagation of the collectively advancing front is well described by a square root of time dependence, with a collective migration coefficient DC = 306 μm2. The rejection of individually migration cells away from this front is also well described by a complimentary error function with an individual migration coefficient DI = 545 μm2. A third fitting parameter C0 = 622 depends on the differential solubility of the solute in solid and melt phase.
In a biological context, this model suggests that the scattering of individually migrating cells from a collectively advancing front occurs in part due to phenotypic plasticity as cells interconvert from individual to collective migration. Nevertheless, although some cells are incorporated into the collective sheet, other cells are released at the interface. This corresponds to a scenario of intermediate solubility where both phenotypic interconversion and segregation occur. The extent of this individual scattering varies with pillar spacing, indicating that phenotypic plasticity occurs, rather than pure segregation (Fig. S13, 14). Nevertheless, one difference with the physical model can be observed from the immunofluorescence measurements (Figure 3A), where the cells not only altered their functional phenotype from individual to collective migration, but also altered biomarker expression and morphology from mesenchymal to epithelial.
Differential Drug Sensitivity of Migrating Cancer Cells
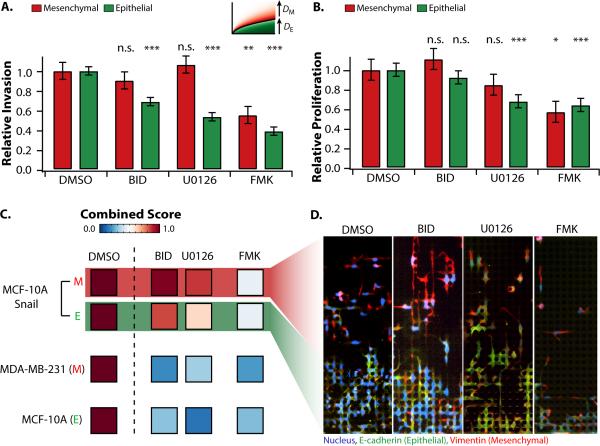
To further validate this multiscale analysis, collective and individual migration behaviours were perturbed using a panel of small molecule drug compounds. Previously, a genome-wide screen implicated the Rsk effectors in multiple downstream pathways of epithelial invasion.36 Inhibitors of this pathway, including the Rsk inhibitor BID1870, the MEK inhibitor MEK inhibitor U0126 and Rsk inhibitor FMKMEA were applied to an induced EMT population (MCF-10A Snail), a purely mesenchymal population (MDA-MB-231) and a purely epithelial population (MCF-10A).
The individual and collective migration coefficients were extracted from the best fits to the solidification model (Fig. 4A). Relative to control, BID-1870 and U0126 also had only slight effects on the individually invading subpopulation, but a ~30-40% reduction of collective migration. However, FMKMEA was responsible for a roughly 2-fold reduction in both individual and collective migration. The size of individual and collective subpopulations was used as a measure of proliferation rate (Fig. 4B, S22-25). Relative to the control, BID-1870 and U0126 only slightly affected the size of the individually invading subpopulation (< 20%), and caused a larger reduction in the size of the collectively invading subpopulation (20-30%). In contrast, FMKMEA led to a ~40% reduction in the size of both the individually migrating and collectively migrating subpopulations.
Fig. 4. Perturbation of invasion behaviours in different cell lines with different Rsk pathway inhibitors.
The individually migrating subpopulation of MCF-10A Snail displays low sensitivity to generic Rsk inhibitors, whereas the collectively migrating subpopulation of MCF-10A Snail shows higher sensitivity. Both subpopulations display sensitivity to FMKMEA. Unlike MCF-10A Snail, MCF-10A and MDA-MB-231 cell lines display sensitivity to all Rsk inhibitors tested. (A) Relative invasion based on based on best fits to the solidification model (dispersion coefficients). (B) Relative proliferation based on differences in subpopulation size. Error bars are standard error of the mean. * p < 0.05, ** p < 0.01, *** p < 10-3. (C) Combined scores incorporating invasion and proliferation. (D) Immunostaining of induced EMT population (MCF-10A Snail) indicates heterogeneous sensitivity of collective and individually invading subpopulations.
One additional metric addressed both migration and proliferation through a net flux of cells, accounting for both the number of cells as well as their respective speeds (Fig. 4C). These combined scores for the induced EMT population (MCF-10A Snail) were calculated separately for the individually and collectively migrating subpopulations, based on the classification of their respective behaviours. In contrast, the purely mesenchymal population (MDA-MB-231) only migrated individually and the epithelial population (MCF-10A) only migrated collectively, so the combined score was computed for the entire population.
The application of BID1870 and U0126 led to only a slight decrease in combined score (~10%) for the individually migrating EMT subpopulation (MCF-10A Snail), indicating minimal sensitivity (Fig. 4C). In contrast, the collectively migrating EMT subpopulation (MCF-10A Snail) displayed a roughly 2-fold decrease in combined score with BID1870 and U0126, corresponding to increased sensitivity. This is qualitatively consistent from immunostaining of these conditions, where individual migration is comparable between control and drug conditions, but collective migration shows some reduction. Migration was suppressed further for the homogeneous mesenchymal population (MDA-MB-231) and epithelial population (MCF-10A), where net migration decreased 3-5 fold.
The Rsk inhibitor FMKMEA37 caused a significant decrease in combined score for both the individually and collectively migrating EMT subpopulations, with a 2.5-fold reduction in migration score (Fig. 4C). This is qualitatively consistent with the immunostaining (Fig. 4D), where both the individual and collective migration are strongly diminished relative to control and qualitatively consistent with principal component analysis (Note SN3, Fig. S26). FMKMEA also led to a strong suppression of combined score for the homogeneous mesenchymal population (MDA-MB-231) and epithelial population (MCF-10A), where net migration decreased 3-5 fold, although this was comparable to the effect of BID1870 and U0126.
These analyses reveal that the overall decreases in net flux caused by Rsk inhibitors (Fig. 4C) is a consequence of a slowdown in migration speed (Fig. 4A) as well as proliferation rate (Fig. 4B). FMKMEA is highly effective since it results in reduced numbers and speeds for both collective and individually invading subpopulations (Video S3). In contrast, BID1870 and U0126 are less effective for highly invasive cells; resulting in reduced numbers and speed only for collectively migrating cells, but not for the individually migrating cells (Video S4).
Discussion
We report spatiotemporal measurements of collectively advancing fronts that scatter individually migration cells. This complex phenomenon has been associated with the activation of EMT pathways as well as invasion and dissemination of malignant tumours in vivo. Our approach utilized automated image analysis to resolve migration dynamics with single cell resolution within populations of tens of thousands. Based on their behaviours, cells were profiled into distinct collective or individual migration phenotypes, in good agreement with epithelial or mesenchymal biomarker expression. We find that individually migrating cells disperse efficiently because of concurrent differences in behaviour such as enhanced velocity and straighter trajectories.
The single cell dynamics were then linked to emergent multicellular behaviours by examining overall motion at the level of subpopulations. The dynamics of a collectively migrating subpopulation that rejects an individually migrating subpopulation were captured using a physical model based on the solidification of a binary mixture. As a consequence of this rejection, the interface between collectively and individually migrating subpopulations slows down (as the square root of time). Essentially, as the two subpopulations become increasingly dispersed and segregated, the forward motion of the collective front is impeded by the presence of the individually migrating cells. This is accounted for in the model, since the motion of the solidification front is limited by the rejection of solute and their transport away. This behaviour is not observed in an intact epithelial layer, where there is nothing ahead of the collective front that would impede its motion, allowing it to migrate forward at constant speed.17,18,20,21 Overall, this model captures the complex behaviours that may occur at a tumour invasion front.
Another essential feature of this model is the competition between a “sorting” mechanism and interconversion between collective and individual migration phenotypes, which captures the possibility of phenotypic plasticity during epithelial-mesenchymal and mesenchymal-epithelial transitions (EMT-MET).38 If the first mechanism is dominant, this model simplifies to the case of epithelial and mesenchymal subpopulations that display minimal phenotypic plasticity. In such a scenario, an initially random mixture of different subpopulations would self-segregate over time and the respective numbers of each subpopulation would remain conserved. This is qualitatively consistent with experimental studies of “cell sorting” based on the differential adhesion hypothesis.39 Nevertheless, the induced EMT (MCF-10A Snail) population in these experiments initially displayed >95% mesenchymal phenotype, including N-cadherin, Fibronectin, vimentin, etc. as well as reduced expression of E-cadherin and occlusion,31 but only ~16% were observed to migrate individually at the end. This mismatch between initial and final phenotypes for migratory behaviours and biomarker expression is consistent with an intermediate level of phenotypic plasticity, since some MCF-10A Snail cells migrated individually, while other MCF- 10A Snail cells reverted to epithelial phenotype and migrated collectively. Indeed, cells may display dynamic behaviours of initially collective migration transitioning to individual migration and vice-versa (Note SN1, Fig. S1-S5). The conversion to epithelial, collective phenotype is further supported by decreased vimentin and increased E-cadherin expression with increasing cell density (Fig.S11 and Table S1). Moreover, this behaviour can be “tuned” from individual scattering to multicellular strands by altering pillar spacing, further indicative of phenotypic plasticity of migration. These behaviours could arise from relative differences in Snail expression that modulate phenotypic plasticity, either permitting reversion to an epithelial, collective migration phenotype or maintaining a mesenchymal, individual migration phenotype that scatters due to “cell sorting.” A possible therapeutic strategy for suppressing individual invasion and dissemination could then be based on suppressing migration-associated pathways as well as enhancing a reverse mesenchymal to epithelial transition.6
It should be noted that there are several approximations in this model. First, there is the assumption of a sharp interfacial front, which appears rougher and more “diffuse” in the experimental data. These behaviours have been addressed in the fluid mechanics literature by including an intermediate “mushy zone.”40 However, solving this full system is mathematically cumbersome and may not add additional biological or physical insight. Future extensions of this model could incorporate interfacial instabilities that drive dendritic outgrowths and microsegregation during evaporation or solidification. These behaviours are qualitatively similar to the multicellular strand or finger invasion in malignant tumour histology, as well as viscous fingering in porous media.41 This effective “surface tension” could account for differences in pillar spacing and address the complexities of the in vivo tumour microenvironment. A second approximation of this analysis is the assumption that differences across single cells in the population are essentially captured by two distinct functional subpopulations.28 This approximation appears sufficient to explain the experimentally observed collective and individual migration behaviours, but undervalues the contributions of single cell outliers with exceptional phenotypes. Future work could utilize a multiscale approach where collective migration is coarse-grained as a subpopulation but individual migration is treated with single cell resolution.
Overall, this model establishes a convenient framework that is sufficient for quantitative comparisons of therapeutic perturbations on the migration and growth of heterogeneous populations. The Rsk inhibitor FMKMEA was highly effective for suppressing cancer cell migration, causing a significant decrease in migration and proliferation for both collective and individually migrating subpopulations. This widespread effectiveness may arise from the specific and irreversible binding of FMKMEA.37 It should be noted that the Snail construct was designed to be extremely stable, residing exclusively in the nucleus to trigger EMT,25 which may be responsible for the diminished chemosensitivity compared to other cell lines. The Rsk effector has been further implicated in affecting cell growth and survival, which makes it a promising therapeutic target.42
Finally, the use of microfabricated environments enables precise measurements of complex migration dynamics under well-controlled experimental conditions. Previous work has observed that EMT enhances individual scattering through integrin-dependent actomyosin contraction on cell-cell junctions in 2D.43 In this work, scattering is enhanced by the use of micropillars that periodically disrupt cell-cell contact, enhancing individual scattering and further maintaining this phenotype by limiting cell-cell contact. This detachment phenomena may thus have analogies with droplet breakup and curvature-based interfacial instabilities44,45 such as those a microfluidic orifice.46 Furthermore, since the overall length of the cell exceeds the pillar spacing, these regularly spaced pillars delineate a straight path that guides individually migrating cells in directionally persistent motion, analogous to experiments in confined channels.29,39,47
Overall, these enclosed geometries and architectures can be rationally designed, complimenting measurements in 3D where mechanics, microstructure and molecular signals exhibit complex, nonlinear couplings.23 One advantage of in vitro assays such as this one is the capability for sampling rare events while interrogating an extraordinary range of experimental conditions, including combinatorial screening of cancer cell lines and patient samples against potential therapeutic compounds.48 These capabilities compliment existing migration assays in vitro and in vivo. Ultimately, this integration of engineered microenvironments with single cell measurements will enable quantitative insights into tumour heterogeneity during metastasis.
Methods
For complete methods, see Supplementary Note SN5
Cell Culture
MCF-10A cells were cultured as described.26 MDA-MB-231 cells were cultured in DMEM (Invitrogen) with 10% fetal bovine serum (FBS) and 1% penicillin-streptavidin. To generate a potent reversible EMT-inducing stimulus in MCF-10A cells, a Snail-1 retroviral expression construct was used, with a fused estrogen receptor (ER) response element and a six amino acid substitution that confers constitutive activity through resistance to the inactivating GSK3b phosphorylation.25 Infection of non-transformed, immortalized mammary epithelial MCF10A cells with ER-Snail 6SA, followed by treatment with tamoxifen (4-OHT), triggered morphological and biomarker characteristics of EMT.26 For imaging and visualization, cells were labelled by incubation with nuclear-staining 1:500 10 mg/mL Hoechst 33258 (Invitrogen) in MCF-10A growth media for 1 h at 37°C.
Device Fabrication
Polydimethylsiloxane (SYLGARD 184; Dow Corning) devices were fabricated using standard soft lithography techniques and bonded to 24-well glass bottom plates (MatTek), as described.29 Devices were immediately functionalized with 100 ng/mL fibronectin (Sigma), rinsed and then stored in phosphate buffered saline (PBS) at 4°C until use.
Migration Assay
Prior to use, devices were prefilled with growth media for at least one hour. Approximately 30,000 cells were loaded into each device. Each well was then filled with growth media. Additional solutions include 10 μM BID-1870 (Stemgent), 10 μM U0126 (EMD Biosciences) and 10 μM FMK-MEA (a gift from J. Taunton, UCSF) and 5 ug/mL CDH1.
Time-Lapse Microscopy
Fluorescently-labelled cells were imaged using an inverted microscope (Nikon TiE) under environmentally controlled conditions (37° C and 5% CO2, humidified). Each device in the multiwall plate was scanned every 15 minutes using a 10X (NA 0.45) long-working distance objective, while maintaining a consistent focus (Perfect Focus System, Nikon Elements). Fluorescence illumination was provided by a light-guide-coupled mercury lamp illumination source with built-in shutter (Nikon Intensilight). Emission spectra were captured using DAPI (Chroma 31000v2), FITC (Nikon 96320) and TRITC (Nikon 96305) filter sets. All images were acquired with 12-bit resolution using a cooled-CCD camera (Photometrics CoolSnap HQ2). Care was taken to ensure that all images were recorded with identical acquisition parameters (exposure time, camera gain/gamma control and microscope aperture settings).
Automated Cell Tracking
Cell detection was accomplished using the DAPI fluorescent signals of cellular nuclei. Time-lapse images in the DAPI channel were run though a pipeline developed in Cell Profiler version 10415 (Broad Institute). The resultant intensities and spatial positions of the detected nuclei were loaded into u-track,30 a MATLAB based multi-particle tracking software, to determine the trajectory of each object.
Migration Analysis
Quantitative metrics were computed for each cell trajectory using custom MATLAB code. Only cell trajectories longer than 12 time frames (4 hrs) were considered. The number of nearest neighbours was computed for each segmented object in each time frame by counting the other segmented objects within a 28 μm search radius (one lattice spacing). This value was then averaged over the lifetime of each cell trajectory. The velocity vi (i = x, y) was calculated from the position xi at time τ and (τ-4), respectively as: vi = [xi(τ) – xi(τ – 4)] / 4. Since images were taken every 15 min, this corresponded to the average velocity every hour, which was chosen to reduce motions due to nuclei shape changes rather than cell migration. Note that cells were restricted to move on a square lattice, so that lifetime averaged velocities were were computed as vr = |vx| + |vy|, tather than . The straightness index (directional persistence) was computed as the ratio of the distance between initial and final positions for each cell, divided by the integrated distance travelled. . Statistical distributions were checked for significance using two-sample Kolmogorov-Smirnov tests as well as n-way ANOVA in MATLAB.
Classification of Collective or Individual Migration
Lifetime-averaged single cell migration metrics were normalized by the maximum value and clustered into distinct collective or individual migration phenotypes using a Gaussian mixture model in MATLAB (gmdistribution.fit). These migratory phenotypes were checked visually against biomarker expression using immunofluorescent staining.
Immunofluorescent Staining
After time-lapse microscopy, immunofluorescent staining was performed to verify biomarker expression. Briefly, cells were fixed for 1 hour at 4° C with 4% formaldehyde in 1X PBS and blocked with 10% goat serum in PBS for 2 hours. Cells were then incubated overnight at 4° C with primary antibodies: 1:500 250 µg/mL E-cadherin (BD Biosciences), 1:200 vimentin (Cell Signaling) and 1:600 phalloidin (Life Technologies) with 1% milk in 1X PBS. Cells were then incubated for 1 hour at room temperature in the dark with appropriately matched secondary antibodies: 1:500 2mg/mL Alexa Fluor 488 (Invitrogen), 1:500 2 mg/mL Alexa Fluor 555 (Invitrogen) and 1:500 2 mg/mL Alexa Fluor 647 (Invitrogen) in 1X PBS with 1% milk. Finally, 1:500 10 mg/mL Hoechst 33258 (Invitrogen) in PBS was added for half an hour at room temperature.
Supplementary Material
Acknowledgements
We thank J. Taunton for the gift of FMKMEA, A. Besser and G. Danuser for assistance with cell tracking as well as O. Hurtado for assistance with fabrication. This work was supported by the Merck Fellowship of the Damon Runyon Cancer Research Foundation (DRG-2065-10) to IYW, as well as the National Institute of Health under CA129933 to D.A.H, EB002503 (BioMEMS Resource Center) to M.T. , and CA135601 and GM092804 to D.I.
Footnotes
Author Contributions:
D.A.H. and D.I. conceived project; I.Y.W., S.J., D.A.H., M.T. and D.I. designed research; I.Y.W., S.J. and E.A.W. performed experiments; I.Y.W. and E.A.W. performed image analysis; I.Y.W., E.A.W. and S.P. performed data analysis; I.Y.W. and M.T. developed theoretical model; I.Y.W., M.T. and D.I. wrote the manuscript with feedback from S.J., E.A.W., S.P. and D.A.H. All authors reviewed the data and final manuscript.
References
- 1.Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217:998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- 2.Willis RA. The spread of tumours in the human body. 3d edn Butterworths; 1973. [Google Scholar]
- 3.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. doi:10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. doi:10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 5.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. doi:10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. doi:10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. doi:10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 8.Polacheck WJ, Zervantonakis IK, Kamm RD. Tumor cell migration in complex microenvironments. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1115-1. doi:10.1007/s00018-012-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson EW, Newgreen DF, Tarin D. Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res. 2005;65:5991–5995. doi: 10.1158/0008-5472.CAN-05-0616. discussion 5995, doi:10.1158/0008-5472.CAN-05-0616. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- 11.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem. Cell Biol. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. doi:10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 12.Giampieri S, et al. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. doi:10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson KJ, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. doi:10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 14.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–3281. doi: 10.1101/gad.1725808. doi:10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farooqui R, Fenteany G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J Cell Sci. 2005;118:51–63. doi: 10.1242/jcs.01577. [DOI] [PubMed] [Google Scholar]
- 17.Poujade M, et al. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. doi:10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol. 2012 doi: 10.1083/jcb.201207148. doi:10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tambe DT, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–475. doi: 10.1038/nmat3025. doi:10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindschadler M, McGrath JL. Sheet migration by wounded monolayers as an emergent property of single-cell dynamics. J Cell Sci. 2007;120:876–884. doi: 10.1242/jcs.03395. doi:10.1242/jcs.03395. [DOI] [PubMed] [Google Scholar]
- 21.Nnetu KD, Knorr M, Strehle D, Zink M, Käs JA. Directed persistent motion maintains sheet integrity during multi-cellular spreading and migration. Soft Matter. 2012;8:6913–6921. doi:10.1039/c2sm07208d. [Google Scholar]
- 22.Irimia D. Microfluidic technologies for temporal perturbations of chemotaxis. Annu Rev Biomed Eng. 2010;12:259–284. doi: 10.1146/annurev-bioeng-070909-105241. doi:10.1146/annurev-bioeng-070909-105241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 2011;3:267–278. doi: 10.1039/c0ib00095g. doi:10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 24.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. doi:10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 25.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. doi:10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 26.Javaid S, et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 2013;5:1679–1689. doi: 10.1016/j.celrep.2013.11.034. doi:10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. doi:10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. doi:10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol (Camb) 2009;1:506–512. doi: 10.1039/b908595e. doi:10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Meth. 2008;5:695–702. doi: 10.1038/nmeth.1237. doi:10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javaid S, et al. Dynamic Chromatin Modification Sustains Epithelial-Mesenchymal Transition following Inducible Expression of Snail-1. Cell Reports. 2013 doi: 10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung WC, et al. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. doi:10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc Natl Acad Sci USA. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. doi:10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf K, et al. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. doi:10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worster MG. Solidification of an alloy from a cooled boundary. J. Fluid Mech. 1986;167:481–501. [Google Scholar]
- 36.Smolen GA, et al. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev. 2010;24:2654–2665. doi: 10.1101/gad.1989110. doi:10.1101/gad.1989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. doi:10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. doi:10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 39.Balzer EM, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. doi:10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worster MG. Convection in mushy layers. Annu Rev Fluid Mech. 1997;29:91–122. [Google Scholar]
- 41.Guiot C, Pugno N, Delsanto PP, Deisboeck TS. Physical aspects of cancer invasion. Phys. Biol. 2007;4:P1–6. doi: 10.1088/1478-3975/4/4/P01. doi:10.1088/1478-3975/4/4/P01. [DOI] [PubMed] [Google Scholar]
- 42.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. doi:10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 43.de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. doi:10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mark S, et al. Physical Model of the Dynamic Instability in an Expanding Cell Culture. Biophys J. 2010;98:361–370. doi: 10.1016/j.bpj.2009.10.022. doi:10.1016/j.bpj.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolli CG, et al. Switchable adhesive substrates: Revealing geometry dependence in collective cell behavior. Biomaterials. 2012;33:2409–2418. doi: 10.1016/j.biomaterials.2011.12.012. doi:10.1016/j.biomaterials.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Nunes JK, Tsai SSH, Wan J, Stone HA. Dripping and jetting in microfluidic multiphase flows applied to particle and fiber synthesis. Journal of Physics D: Applied Physics. 2013;46 doi: 10.1088/0022-3727/46/11/114002. doi:10.1088/0022-3727/46/11/114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Impact of tumor cell cytoskeleton organization on invasiveness and migration: a microchannel-based approach. PLOS ONE. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. doi:10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. doi:10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.