Summary
Here we consider time-to-event data where individuals can experience two or more types of events that are not distinguishable from one another without further confirmation, perhaps by laboratory test. The event type of primary interest can occur only once. The other types of events can recur. If the type of a portion of the events is identified, this forms a validation set. However, even if a random sample of events are tested, confirmations can be missing nonmonotonically, creating uncertainty about whether an individual is still at risk for the event of interest. For example, in a study to estimate e cacy of an influenza vaccine, an individual may experience a sequence of symptomatic respiratory illnesses caused by various pathogens over the season. Often only a limited number of these episodes are confirmed in the laboratory to be influenza-related or not. We propose two novel methods to estimate covariate e ects in this survival setting, and subsequently vaccine e cacy. The first is a pathway Expectation-Maximization (EM) algorithm that takes into account all pathways of event types in an individual compatible with that individual’s test outcomes. The pathway EM iteratively estimates baseline hazards that are used to weight possible event types. The second method is a non-iterative pathway piecewise validation method that does not estimate the baseline hazards. These methods are compared with a previous simpler method. Simulation studies suggest mean squared error is lower in the e cacy estimates when the baseline hazards are estimated, especially at higher hazard rates. We use the pathway EM-algorithm to reevaluate the e cacy of a trivalent live-attenuated influenza vaccine during the 2003-2004 influenza season in Temple-Belton, Texas, and compare our results with a previously published analysis.
Keywords: EM algorithm, Missing data, Vaccine efficacy, Validation set
1. Introduction
Here we consider time-to-event data where an individual can experience two or more types of events not distinguishable from one another without further confirmation, perhaps by laboratory test. The event type of primary interest can occur only once. The other types of events can occur an unlimited number of times, so the individual stays in the risk set for them for as long as he is in the study. The scientific problem is to estimate the effect of a covariate, or covariates, on the event type of primary interest. Assuming the event types are independent, if the type of each event is identified, estimation of type-specific covariate effects is straightforward. However, if the type of only a fraction of the events is identified, forming a validation set, several problems arise. Even if a random sample of events are tested, the confirmations can be missing nonmonotonically. If a person has an untested event, it could have been the event type of interest, so the person is no longer in the risk set to have that event in the future, but we do not know that.
Magaret (2008) incorporated validation subsets into discrete proportional hazards models for mismeasured outcomes but assumed only one type of event. Joint modeling of longitudinal measurements and event time data (Tsiatis and Davidian 2004; Ibrahim, Chu and Chen 2010) allows one to infer the association between a longitudinal biomarker that may be measured intermittently and with error and time to event. Our situation differs from those generally dealt with in such joint modeling in that we have at least two distinct outcomes that are not distinguishable without further tests. If the data were completely observed, the models for the effect of treatment on the different outcomes would be independent of one another.
Our research is motivated by data of this form in a study evaluating the efficacy of a vaccine in preventing symptomatic influenza. During an influenza season, a susceptible person may have zero, one, or more episodes of acute respiratory illness. An episode may be caused by either influenza virus or one of several other pathogens that cause similar symptoms. Each year, only a small proportion of respiratory illnesses may be sampled for culture to meet influenza surveillance needs. For most symptomatic episodes, the pathogen information is missing. Estimation of vaccine efficacy based on nonspecific outcomes will bias the estimate toward the null, because an influenza vaccine generally would have no effect on other pathogens. Surveillance cultures can be used as a validation set to correct the bias in the estimate based on the nonspecific outcome alone.
Halloran et al. (2007) developed an estimate of efficacy based on estimated events per person-time that allowed for time-dependent covariates and varying baseline hazards by aggregating the time-to-event data into time intervals. In each stratum and time interval, the estimated fraction of influenza positive cultures and observed number of nonspecific cases were used to estimate the number of influenza cases. However, their approach did not take into account the dependency of the other events within each person or that a previous untested symptomatic episode might have been influenza.
Here we develop two new methods that take into account all possible sequences of event types within an individual, called pathways, compatible with that person's events and test results. The first is an expectation-maximization (EM) algorithm (Dempster et al 1977) in the survival setting, called pathway EM, that combines the complete data on nonspecific events and the incomplete validation set and involves estimation of the baseline hazards. The second, called pathway piecewise validation (PV) method does not involve estimating the baseline hazards. The new statistical procedures can be implemented in publicly available software such as R (R Core Team, 2013). In Section 2, we develop the pathway EM algorithm. In Section 3, the performance of the procedures is evaluated and compared using simulated epidemics. In Section 4 we re-analyze a study of the efficacy of an influenza vaccine.
2. Methods
2.1 Notation
Although the following methodological development is motivated by this application, the methods are applicable to more general situations where covariate effects on specific event types are of interest, and the event types cannot be distinguished without some further test. Consider a population of N susceptible individuals at the start of an influenza epidemic occurring from time t = 0 to t = T . Assume there are M cocirculating pathogens. Each pathogen has a baseline hazard of infection, λm0(t), m = 1, . . . , M. Let Xij(t) be the jth possibly time-dependent covariate of person i at time t, i = 1, . . . , N, j = 1, . . . , p, and Xi(t) = (Xi1(t), . . . , Xip(t))′ . We assume the effective hazard from pathogen m for person i at time t is , where βm is a p × 1 column vector of unknown parameters. If Xi1(t) is the vaccination status for person i at time t, vaccine efficacy (VE) for pathogen m is VEm = 1 − exp(β1m).
Some pathogens, such as influenza, induce long-term immunity, and therefore infect a person at most once per season, whereas infection from other pathogens may occur multiple times. We assume the event types are independent. Thus, we assume there is no competition or interference between pathogens.
In our motivating example, the nonspecific outcome is medically-attended acute respiratory illness (MAARI). We are concerned with clinically ascertained cases rather than infection, but the results are the same for both, just interpretation of the efficacy estimates differs. Let ti = (ti1, . . . , tini)′ denote the sequence of ni onset times of MAARI episodes of person i. We assume a proportion of MAARI episodes are randomly sampled for testing. We further assume sampling does not depend on the underlying pathogens, but it may depend on known variables, that is, tests are missing at random (MAR) (Little and Rubin, 2002). Let Yim(t) indicate whether person i is at risk of pathogen m at time t−, and Zim(t) whether pathogen m infects persons i at time t. For pathogens that can reinfect, Yim(t) = 1 for all i and t. The complete history of {(Yim(t), Zim(t)) : 0 ≤ t ≤ T} is determined by their values at the onset times and T . For simplicity, let ti0 = 0 and ti(ni+1) = T for all i. Extension of our methods to studies where ti0 and ti(ni+1) differ between individuals is straightforward. Define Y im = (Yim(ti1), . . . , Yim(ti(ni +1)))′ and Zim = (Zim(ti1), . . . , Zim(ti(ni +1)))′. We assume each infection is caused by one pathogen. Let V i = (Vi1, . . . , Vini)′ be the vector infecting pathogens.
Without loss of generality, let Yim(0) = 1 and Zim(T ) = 0 for all i and m, i.e., every person was at risk at time 0, and no infection occurred at the last follow-up time. Let Y i = {Y im : m = 1, . . . , M}, Zi = {Zim : m = 1, . . . , M}, and Xi = {Xi(t) : 0 ≤ t ≤ T}. Let β = {βm : m = 1, . . . , M} and λ0 = {λm0(t) : m = 1, . . . , M, 0 ≤ t ≤ T}. Let t = {ti : i = 1, . . . , N}, Y = {Y i : i = 1, . . . , N}, Z = Zi : i = 1, . . . , N}, and X = Xi : i = 1, . . . , N. We assume ti and Xi(t) are completely observed for all i. We use * to denote missing values in Y im, Zim, and V i. As the covariates are fixed, to simplify notation, in all likelihoods, Xi and X are suppressed.
2.2 Complete-data scenario
Given that person i is at risk at ti(k−1)−, the probability of i escaping from pathogen m during the period (ti(k−1), tik] is
If Y i and Zi are completely observed, the likelihood contribution of person i is given by
| (1) |
subject to two constraints: if pathogen m can infect at most once; and , j = 1, . . . , ni. The first constraint is due to the nature of pathogens that induce long term immunity, or more generally, for event types that occur just once; the second constraint ensures each MAARI episode is caused by only one pathogen, or more generally each episode is due to just one event type. Contribution (1) could be written more simply, but this expression anticipates the EM algorithm with incomplete data developed below.
If Y i and Zi are observed for all individuals and if the baseline hazards are of no inferential interest, the standard Cox proportional hazard model can be used to estimate the covariate effects. In reality, observation of an epidemic is on a daily basis, and time takes integer values, t = 1, . . . , T . To write the likelihood using this kind of data, we embed the discrete-time data in the continuous-time model by assuming all infections occur at the end of the day.
Let be the risk score associated with person i with respect to pathogen m at time t. The partial likelihood for complete data is
| (2) |
The expression for (2) with the Efron correction for ties (Efron, 1977) is in Web Appendix A. If inferential interest focuses only on a single pathogen m and no parameter is shared across multiple pathogens, the information about all other pathogens can be ignored, as the overall partial likelihood is simply a product of the partial likelihoods for individual pathogens.
2.3 Incomplete-data scenario
We now assume laboratory results are incomplete and develop a pathway EM algorithm. In the E-steps, we evaluate the expectation of Yim(tj)Zim(tj) and Yim(tj) 1 − Zim(tj)), conditional on the observed data as well as current estimates of the parameters and the baseline hazards . Let Y obs(i) and Zobs(i) represent the observed parts of Y i and Zi. Let Yobs = {Yobs(i) : i = 1, . . . , N} and Zobs = {Zobs(i) : i = 1, . . . , N}. Similarly define the missing data counterparts, Ymis(i) Zmis(i) for person i, and Y mis and Zmis for the population. Let Oi = {Xi, ti, Y obs(i), Zobs(i)} be the observed data of person i, and O = {Oi : i = 1, . . . , N}.
The component Σk Ykm(t)rkm(t) in (2) can be rewritten as Σk (Ykm(t)Zkm(t)+Ykm(t)(1 − Zkm(t)))rkm(t). The first-order approximation of the expected log partial likelihood conditional on the observed data would be simply to replace Yim(t)Zim(t) and Yim(t)(1 − Zim(t)) with their conditional expectations. Let and . Then in the absence of ties,
| (3) |
Expression (3) with ties is in Web Appendix A. Using the R software package survival (Therneau, 2012), this log-partial likelihood can be solved by organizing the data as time intervals between symptom onsets, (ti(j−1), tij] and assigning the conditional expectations and as weights for possible infection outcomes. That is, can be thought of as a weight the event was type m, and as a weight it was not an event of type m, that is, an escape. We refer to the quadruplet (ti(j−1), tij, m, s) as a survival interval, where s indicates infection status (1=infection, 0=escape).
2.3.1 Assigning weights to survival intervals
If t = tij and Vij = m for some j, that is, a symptom onset occurs at t and is confirmed to be pathogen m, then and . It might be thought that and for pathogen m′ ≠ m; however, this would be true only if person i is at risk to pathogen m′ at tij−. Similarly, if t ∉ ti or t = tik and Vik ≠ m for some k, i.e., either no MAARI onset at t or a MAARI onset is confirmed not to be pathogen m, and only if person i is at risk to pathogen m at t−.
When there is some uncertainty, we have t = tik and Vik = * for some k. The immunological constraints and observed data Oi together restrict the set of possible realizations of (Y i, Zi). Let Ci be the set of possible pathways of outcomes of person i restricted by Oi and the immunological constraints. For example, suppose an individual has three MAARI episodes at ti1, ti2 and ti3. The following are three possible combinations of laboratory tests, missing or with outcomes, together with the compatible realizations, or pathways, of infection outcomes (1, 2, and * denote influenza, nuisance, and missing) (More details in Web Appendix B):
V i = (*, 1, *): Ci = {(2, 1, 2)};
V i = (*, 2, *): Ci = (1, 2, 2),(2, 2, 1), (2, 2, 2)};
V i = (*, *, *): Ci = {(1, 2, 2), (2, 1, 2), (2, 2, 1), (2, 2, 2)}.
Define Eimj as the set of possible outcomes compatible with Yim(tij)Zim(tij) = 1, i.e., an infection of pathogen m occurred at time tij. Analogously, define Ēimj as the set of possible outcomes compatible with Yim(tij) (1 − Zim(tij)) = 1, i.e., an escape from pathogen m occurred at time t. The weights, , for pathogen m = 1, . . . , M and infection status k = 0, 1 at time tij, are given by
| (4) |
for j = 1, . . . , ni. If subject i is known to be not at risk at tij−, set . For the last interval (tini, T ), as Zim(T ) = 0 for all m by our assumption and For any M, the following relationship holds for j = 1, . . . , ni:
;
if pathogen m can infect only once; and
if pathogen m can reinfect. In the presence of discrete time-dependent covariates, presentation of survival data would involve time intervals defined not only by events but also by covariate values. If (tij, t] is an interval defined by change of a covariate value at t, then as no infection occurs, for pathogens that reinfect, and for pathogens that infect only once.
2.3.2 Estimating baseline hazards
Calculating the weights involves the likelihood (1), which in turn requires evaluation of the baseline hazards. We first replace the Breslow estimate of the cumulative hazard for each pathogen under complete data (Breslow, 1974),
by
when there is uncertainty in infection outcomes. This nonparametric estimate is then smoothed using an approach such as a smooth cubic spline or local polynomial regression (Ramsay and Silverman, 1997). The derivative of the smoothed curve serves as the baseline hazard function for pathogen m. We chose local polynomial (cubic) regression for both its automatic correction for boundary effects (Cheng, Fan and Marron, 1997) and the computational efficiency of the R package lpridge (Burkhardt Seifert; packaged for R by Martin Maechler, 2011). We also explored whether smoothing methods that enforce monotonicity in the cumulative hazard, such as in Ramsay (1998), improve inference. The corresponding R package is fda (Ramsay et al., 2011).
2.3.3 Variance estimation
Standard asymptotic variance estimation for a full likelihood can be applied to a partial likelihood as well (Therneau and Grambsch, 2000). The dimension of the missing data, the unobserved infection status of uncultured MAARI episodes, is small for each individual but tremendous for the whole population. Thus, we use the Monte Carlo version of Louis’ method (Louis, 1982; Tanner, 1992) to estimate the asymptotic variances:
| (5) |
where the square of the score vector is the product of column vector and its transpose. Alternatively, one can use the bootstrap to estimate the variances (Efron and Tibshirani 1993). Considering each individual as a cluster (Therneau and Grambsch, 2000), we drew a sample data set from the original data with replacement by individual and applied the pathway EM-algorithm to obtain a new set of point estimates of vaccine efficacies. This approach is computationally much less efficient than its asymptotic counterpart. We compared the coverage of the two variance estimation methods in different settings.
2.3.4 A non-iterative pathway piecewise validation procedure
In Web Appendix C, we describe a non-iterative pathway piecewise validation (PV) procedure. Similar to the pathway EM algorithm, the pathway PV approach assigns weights according to probabilities taking into account within an individual all pathways of infection outcomes compatible with the observed data. However, it is simpler in that the baseline hazards are not estimated and taken into account in the weights. This approach is similar to that in Halloran et al. (2007), called here the aggregation PV approach, in that both approaches divide the influenza season into time intervals, and use the interval-specific proportion of positive cultures to inform the probability that a MAARI onset in that interval is influenza.
2.3.5 Extension to frailty models
Susceptibility often varies across groups of individuals, and frailty models have been proposed (Longini and Halloran, 1996). The pathway EM and PV procedures can be readily extended to accomodate frailties, namely, , where h indicates the frailty group of persin i, and the frailty ξh follows a distribution with mean 1. This model can be solved using either an additional E step for the mean of ξh conditioning on observed data, current weights and parameters estimates or a penalized partial likelihood nested within the M-step of the pathway EM algorithm (Klein, 1992; Therneau et al., 2003). A modified pathway EM procedure with an additional E-step to handle gamma frailties is given in Web Appendix D.
3. Simulation
While our approach holds for any number of pathogens, it is often sufficient to assume M = 2 with influenza (m = 1) that can infect only once and a nuisance pathogen (m = 2) that can infect an unlimited number of times. Multiple nuisance pathogens can be replaced by a single hypothetical nuisance pathogen. Estimation of VE for influenza will not be affected by this simplification, if the influenza vaccine has no effect on the nuisance pathogens, or if VE is the same for all nuisance pathogens.
In a population of 500 people, of which 30% are vaccinated before time 0, we simulate influenza (pathogen 1) epidemics with a cocirculating nuisance pathogen (pathogen 2) that induces no immunity at all, each epidemic running for T = 100 days. Details on simulating cocirculating pathogens are in Web Appendix E. Two epidemic sizes were investigated by setting the rates of infection events as γ1 = γ2 = 0.003 per day and γ1 = γ2 = 0.01 per day, corresponding to baseline attack rates of 0.26 and 0.63. The former setting is similar to a typical seasonal human influenza epidemic. Without vaccination, the average numbers of infections by any pathogen (MAARI episodes) per individual are 0.26 + 100 × 0.003 = 0.56 and 0.63 + 100 × 0.01 = 1.63 in the two settings.
We consider two functional forms of the baseline hazards: constant and a beta-density shape that can mimic a unimodal, epidemic-like shape, For the beta-density shape, the baseline hazard is constant within day t and is given by , where F(·|am, bm) is the beta cumulative distribution function with shape parameters am and bm. Vaccine efficacy is set to 0.8 for influenza (VE1) and zero for the nuisance pathogen (VE2). We assume sampling proportions for laboratory testing depend on vaccine status, higher in the vaccine group, because high efficacy can substantially reduce the number of confirmed influenza events in the vaccinated group. Small baseline hazards or sampling proportions may lead to sparse data and therefore numerical problems in the VE estimate such as boundary values (−∞ or 1), undefined VE (zero inlfuenza-positive cultures in both vaccine and placebo groups), or non-convergence of the EM algorithm. We excluded simulation runs with numeric problems in the following performance assessments. Details about the handling of numeric problems and their frequencies under various settings can be found in Web Appendix F.
We first compared the inferential performance of the pathway EM approach to the non-iterative aggregation PV and pathway PV approaches (Table 1). For the two noniterative approaches, we divided the epidemic period into four intervals according to the quartiles of MAARI episodes sampled for culturing, so each interval was guaranteed to contain cultures. Both small (γ1 = γ2 = 0.003) and large (γ1 = γ2 = 0.01) epidemics were explored. The relative performance of the three approaches depends on the epidemic size, the shape of the hazard function, and the culture proportion. When both the epidemic size and the sampling proportions are low, the patheway EM algorithm is clearly superior with smaller bias and mean square error (MSE), in particular when the hazard function takes a beta density shape. The two noniterative approaches have similar performance, except that the pathway PV approach gives much smaller MSE for the nuisance pathogen. When the sampling proportions are high but the epidemic size remains low, all three approaches perform similarly well with nearly unbiased VE estimates and small MSE for both pathogens, although the pathway EM approach still outperforms the other two. When the epidemic size is large, where many individuals have multiple MAARI episodes, the two noniterative approaches have much more severe bias, in particular when the sampling proportions are low, whereas the pathway EM estimates remain unbiased and continue to have the lowest MSE. The pathway PV seems notably preferable to the aggregation PV for large epidemics and beta-shaped hazards. Simulations with a population size of 2000 individuals found that (1) the pattern of larger bias for larger epidemic size of the noniterative PV approaches does not change; and (2) the bias of the pathway EM algorithm observed with small epidemic size, constant hazards and low sampling proportion will disappear.
Table 1.
Average and mean square error (MSE) of vaccine efficacy estimates by the intensity and shape of baseline hazards, sampling probabilities for laboratory confirmation, and estimation method. The coverage probabilities of 95% CIs are given only for the pathway-EM approach. True parameters are (γ1, γ2) for average baseline hazards and (VE1, VE2) = (0.8, 0) for vaccine efficacies. Results are based on 500 simulations. Bootstrap variance estimates are based on 200 resamplings per simulation.
| Baseline Hazard γ1 = γ2 | Baseline Hazard Shape | Sampling Proportion (pu, pv) | Pathogen Type | Estimation Method |
|||||
|---|---|---|---|---|---|---|---|---|---|
| PV |
Pathway-PV |
Pathway-EM |
|||||||
| Est. | MSE | Est. | MSE | Est. | MSE | ||||
| 0.003 | cons. | (0.1, 0.3) | Flu | 0.74 | 0.029 | 0.75 | 0.027 | 0.76 | 0.023 |
| Nui. | −0.087 | 0.35 | 0.036 | 0.072 | −0.022 | 0.075 | |||
| (0.5, 0.7) | Flu | 0.78 | 0.0086 | 0.79 | 0.0082 | 0.80 | 0.0071 | ||
| Nui. | −0.019 | 0.038 | 0.029 | 0.032 | −0.018 | 0.037 | |||
| Beta | (0.1, 0.3) | Flu | 0.75 | 0.023 | 0.76 | 0.020 | 0.81 | 0.0082 | |
| Nui. | −0.052 | 0.14 | 0.047 | 0.067 | −0.012 | 0.045 | |||
| (0.5, 0.7) | Flu | 0.79 | 0.0082 | 0.79 | 0.0075 | 0.80 | 0.0067 | ||
| Nui. | 0.0022 | 0.034 | 0.035 | 0.032 | 0.0082 | 0.031 | |||
| 0.01 | cons. | (0.1, 0.3) | Flu | 0.71 | 0.020 | 0.73 | 0.015 | 0.80 | 0.0060 |
| Nui. | −0.034 | 0.030 | 0.093 | 0.021 | −0.014 | 0.017 | |||
| (0.5, 0.7) | Flu | 0.74 | 0.0067 | 0.75 | 0.0057 | 0.80 | 0.0025 | ||
| Nui. | −0.0099 | 0.013 | 0.070 | 0.015 | −0.010 | 0.012 | |||
| Beta | (0.1, 0.3) | Flu | 0.72 | 0.016 | 0.77 | 0.0068 | 0.81 | 0.0029 | |
| Nui. | −0.022 | 0.020 | 0.058 | 0.015 | −0.015 | 0.012 | |||
| (0.5, 0.7) | Flu | 0.76 | 0.0050 | 0.78 | 0.0029 | 0.80 | 0.0020 | ||
| Nui. | −0.014 | 0.011 | 0.032 | 0.011 | −0.0098 | 0.011 | |||
We compared estimates of standard deviations (SD) and coverage probabilities of confidence intervals (CI) in Web Tables 6 and 7. Bootstrap estimates were used for the noniterative PV approaches, whereas both asymptotic and bootstrap estimates were obtained for the pathway EM algorithm. With small epidemic sizes and low sampling proportions, the SDs of the VE estimates for influenza tend to be overestimated for the two noniterative PV approaches. The aggregation PV approach also substantially overestimates the SDs for the nuisance pathogen for beta-shaped hazard. Increase in either epidemic size or sampling proportion leads to much more satisfactory estimation of the SDs by the noniterative approaches. For the pathway EM algorithm, the asymptotic method tends to slightly underestimate, whereas the bootstrap method tends to slightly overestimate, the actual SD for influenza when the epidemic size is small or sampling proportions are low. In general, the pathway EM algorithm yields the smallest SD, followed by the pathway PV approach, and the differences are more evident with influenza and beta-shaped hazards.
For the two noniterative PV approaches, the CI coverage probabilities are reasonable when the epidemic size is small but tends to be low for large epidemics, a result of increased bias but shrunken variances. Neither PV approach is designed for consistent VE estimation as their weighting schemes are “ad hoc” in most settings. For the pathway EM algorithm, both asymptotic and bootstrap CIs provide reasonable coverage probabilities, except that asymptotic CIs tend to provide insufficient coverage for the nuisance pathogen when the culture proportions are low and the hazards are constant.
The deviation of the average VE estimate from the true value and the deviation of the CI coverage probability from 95% may be partially accounted for by stochasticity in addition to underlying bias. A simple quantification of the stochasticity will help us better assess the true scale of the bias and is provided in Web Appendix G.
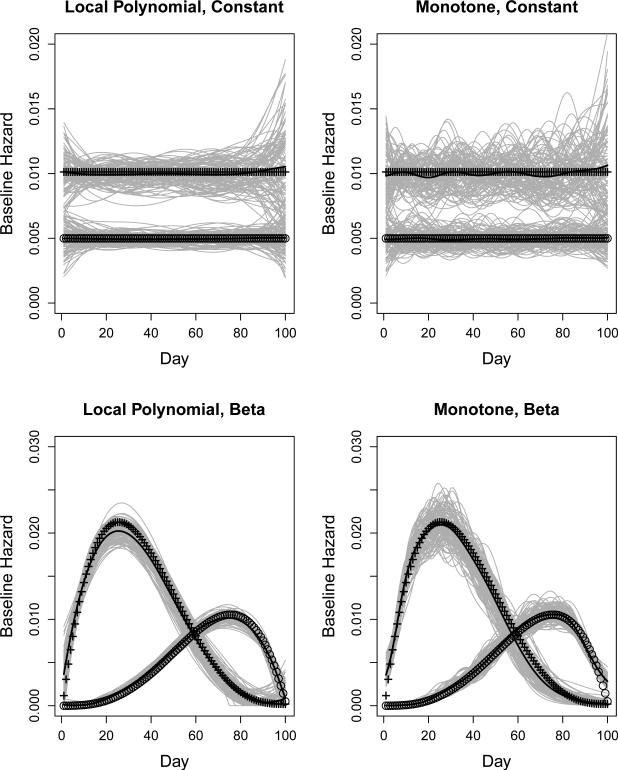
The simulation results are insensitive to choice of smoothing methods, with neither superiority or inferiority in bias or variation of the VE estimates, using the approach enforcing monotonicity (Ramsay 1998). Figure 1 displays the estimated baseline hazards and their mean curves for 100 simulations under the setting of low sampling proportions, by smoothing methods and the shape of hazards. To produce distinguishable estimated hazards, we used γ1 = 0.01 and γ2 = 0.005 for the two pathogens, and increased the population size to 2000. The estimates using local polynomial regression tend to be more stable than those based on enforcing monotonicity. A penalty weight of was used for the latter approach because it yielded reasonable balance between bias and local roughness in our empirical evaluation.
Figure 1.
Baseline hazard estimates from 100 simulations by hazard shape, smoothing methods, and pathogen. Solid lines are mean of the estimated hazard curves. Curves of symbols represent true baseline hazards, + for influenza and for the nuisance pathogen. Left panels use local polynomial regression and right panels use B-spline enforcing monotonicity for smoothing. The true baseline hazards are constant in top panels and have shapes of beta density in bottom panels.
We then compared three different schemes of sampling MAARIs to be cultured, given a fixed total number of samples:
Random sampling of MAARIs, stratified by vaccine status, the scheme used in Table 1.
This scheme assumes samples from individuals are collected and stored, so are available for selection for testing. That is, randomly select the maximum possible number of individuals and then randomly select one MAARI of each individual who has an event. Repeat this process until the quota is exhausted.
Maximize the number of individuals of whom all MAARIs are sampled. The first two options produce nonmonotone missing data, while the third produces monotone missing data. The MAR assumption holds for all three options. Results are shown in Table 2. The differences between scheme I and scheme II are negligible. As expected, scheme III is associated with larger bias and MSEs (Table 2) and higher frequencies of numeric di culties (Web Table 4), in particular when the epidemic size is small. Sampling scheme III samples all MAARIs from a limited number of individuals. Because any individual can have at most one influenza onset, and possibly none, this may not pick up many positive influenza cases. The bias given by scheme III with small epidemic size will be offset by a larger population size, e.g., a population size of 2000 yields an average VE estimate of 0.75 (MSE=0.025).
Table 2.
Average and mean square error of vaccine efficacy estimates by the intensity and shape of baseline hazards, sampling probabilities for laboratory confirmation, and design of sampling MAARI episodes for culture. True vaccine efficacies are set to (VE1, VE2)= (0.8, 0). Results are based on 500 simulations.
| Baseline Hazard γ1 = γ2 | Baseline Hazard Shape | Sampling Proportion (pu, pv) | Pathogen type | Sampling Scheme |
|||||
|---|---|---|---|---|---|---|---|---|---|
| I |
II |
III |
|||||||
| Est. | MSE | Est. | MSE | Est. | MSE | ||||
| 0.003 | constant | (0.1, 0.3) | Influenza | 0.76 | 0.024 | 0.76 | 0.024 | 0.54 | 0.21 |
| Nuisance | −0.017 | 0.076 | −0.0080 | 0.066 | 0.13 | 0.13 | |||
| (0.5, 0.7) | Influenza | 0.80 | 0.0072 | 0.80 | 0.0074 | 0.72 | 0.050 | ||
| Nuisance | −0.018 | 0.037 | −0.021 | 0.038 | 0.012 | 0.080 | |||
| Beta | (0.1, 0.3) | Influenza | 0.81 | 0.0081 | 0.80 | 0.0092 | 0.72 | 0.43 | |
| Nuisance | −0.020 | 0.046 | −0.0089 | 0.042 | −0.021 | 0.074 | |||
| (0.5, 0.7) | Influenza | 0.80 | 0.0067 | 0.80 | 0.0063 | 0.80 | 0.0097 | ||
| Nuisance | 0.0072 | 0.031 | 0.0019 | 0.033 | −0.014 | 0.047 | |||
| 0.01 | constant | (0.1, 0.3) | Influenza | 0.80 | 0.0061 | 0.79 | 0.0058 | 0.79 | 0.010 |
| Nuisance | −0.013 | 0.016 | −0.0062 | 0.014 | −0.0018 | 0.018 | |||
| (0.5, 0.7) | Influenza | 0.80 | 0.0025 | 0.80 | 0.0022 | 0.79 | 0.0041 | ||
| Nuisance | −0.010 | 0.012 | −0.0086 | 0.011 | −0.0059 | 0.013 | |||
| Beta | (0.1, 0.3) | Influenza | 0.81 | 0.0029 | 0.81 | 0.0026 | 0.81 | 0.0032 | |
| Nuisance | −0.015 | 0.012 | −0.0073 | 0.012 | −0.013 | 0.013 | |||
| (0.5, 0.7) | Influenza | 0.80 | 0.0020 | 0.80 | 0.0018 | 0.80 | 0.0023 | ||
| Nuisance | −0.010 | 0.011 | −0.0034 | 0.011 | −0.0049 | 0.011 | |||
In the influenza application, physicians might tend to choose children they believe to have influenza to culture. If a physician's reasons for choosing a person to sample were known, this could be built into the analysis. Otherwise, the estimates could be subject to selection bias. To evaluate the sensitivity of pathway-EM algorithm to the violation of the MAR assumption, we simulated data under missing not at random (MNAR) by letting the sampling probability differ between pathogens. For a range of scenarios of over-sampling influenza-induced MAARIs suggested by Scharfstein et al. (2006), biases in the VE estimates were observed for both pathogens but the magnitude is mild in general. For influenza, the bias is less than 10% in most settings (Web Appendix H).
Finally, we combine the methods with a frailty model, where the population is divided into 20 frailty groups and the frailties ξh ~ gamma(1, 1), h = 1, . . . , 20 (Web Table 2). If heterogeneity is ignored in inference, VE estimates for influenza are biased downwards, more so when the baseline hazards are constant, and in particular when the sampling proportions are low. Adjusting for frailties essentially eliminates the biases in the VE estimates and shrinks corresponding MSEs for both pathogens in all settings. The advantage of the frailty model is less obvious when the true value of VE is close to zero. If the number of frailty groups is too large such that not all frailties are well identified, adjusting for frailties could yield larger MSE than ignoring frailties. More simulation results and discussion regarding the frailty model are given in Web Appendix D.
4. Data Analysis
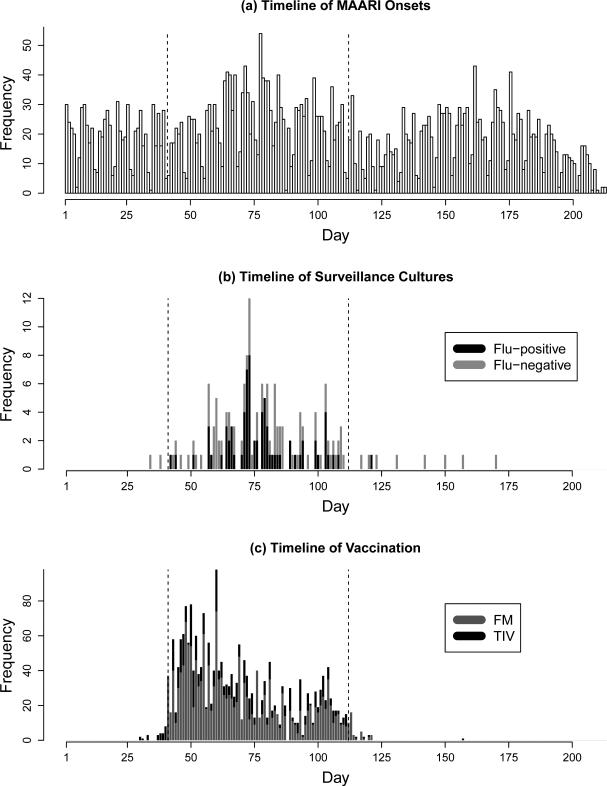
During the 2003-04 influenza season, a field study was conducted in Temple-Belton, Texas, to evaluate the efficacy of a live-attenuated trivalent influenza virus vaccine (LAIV-T) against influenza A (H3N2) virus. A total of 6403 age-eligible (18 months – 18 years) children who were enrolled in the Scott & White Health Plan (SWHP) and visited Scott & White clinics in Temple-Belton with MAARI presentation were identified in the SWHP database. Of these children, 1737 received LAIV-T and 552 received trivalent inactivated vaccine (TIV) over 11 weeks starting October 10, 2003. As in the primary analysis (Halloran et al., 2007), we restrict the analysis to the 10-week period defined as the primary influenza season October 10 – December 20, 2003 and exclude children receiving TIV as most had a contraindication for LAIV-T and were at higher risk for influenza. We ignore previous vaccination history as it was not found to be effective in the primary analysis. Among the remaining 5851 subjects, there were 1375 MAARIs, after any two MAARI episodes within five days of each other have been joined as one, and 139 cultures of which 66 are influenza-positive. Figure 2 shows the timelines of MAARI onsets, surveillance cultures and vaccination.
Figure 2.
Timelines of (a) MAARI onsets, (b) surveillance cultures, and (c) vaccinations of children enrolled in SWHP during the 2003-2004 Influenza A (H3N2) season. The vertical dashed lines are days 41 and 112, corresponding to October 10 and December 20, 2003, between which is the analysis period.
As in the primary analysis, we divide the population into two age groups, <10 years and 10– 18 years, and assume the two groups have proportional baseline hazards and different vaccine efficacies. This assumption is achieved by setting , where Xage,i is the age group and Xvac,i(t) is the vaccine status at time t of person i. As the culture data are fairly sparse, we also report the overall VE estimates via the main effect model . Influenza vaccine may take a few days to reach full effectiveness, and hence we performed the analysis for both one-day and seven-day delays of vaccine effectiveness. We report results based on the seven-day delay as our final findings, but compare the results based on one-day delay to the primary analysis, since the same assumption was adopted there. In the with-interaction model, the age effect is reported only for unvaccinated children. CIs are based on bootstrap samples. Potential heterogeneity in individual baseline hazards is not considered in this analysis due to the lack of cluster information.
The results indicate a significant overall VE, 64%(95% CI: 31%, 89%), against influenza-induced MAARIs and no efficacy against non-influenza-induced MAARIs (Table 3). Interestingly, compared to younger children, older children tended to have a higher hazard of influenza, though not significantly, and a lower hazard of non-influenza related illness. After stratification of influenza VE by age group, both age groups showed significant protection from the vaccine, VE = 0.82 (95% CI: 0.37, 0.90) for the younger and 0.58 (95% CI: 0.11, 0.87) for the older group. As expected, there was no protection against MAARIs induced by nuisance pathogens. The influenza VE estimates assuming a one-day delay were all lower, but statistical significance remained for the older group.
Table 3.
Estimates and 95% confidence intervals for LAIV-T efficacy and age effect against influenza A (H3N2) among children in Temple-Belton, Texas during the 2003-2004 season. Results are presented by model and the assumption about the delay of effectiveness after vaccination. 95% CIs are based on 500 bootstrap samples.
| Model | Factor | Category | Delay in Effectiveness after Vaccination |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 Day |
7 Days |
|||||||||
| Influenza |
Nuisance |
Influenza |
Nuisance |
|||||||
| Est. | 95% CI | Est. | 95% CI | Est. | 95% CI | Est. | 95% CI | |||
| Age-specific | ||||||||||
| VE | < 10 | 0.45 | (−0.33, 0.83) | 0.18 | (−0.11, 0.41) | 0.82 | (0.37, 0.90) | 0.078 | (−0.20, 0.34) | |
| 10 – 18 | 0.51 | (0.11, 0.83) | −0.008 | (−0.40, 0.33) | 0.58 | (0.11, 0.87) | −0.094 | (−0.45, 0.30) | ||
| Age | vac=0 | 1.40 | (0.86, 2.60) | 0.67 | (0.54, 0.90) | 1.24 | (0.81, 2.32) | 0.69 | (0.56, 0.92) | |
| Main Effect | ||||||||||
| VE | 0.47 | ( 0.14, 0.74) | 0.093 | (−0.16, 0.32) | 0.64 | (0.31, 0.89) | 0.0063 | (−0.29, 0.25) | ||
| Age | 1.35 | ( 0.86, 2.38) | 0.71 | (0.57, 0.91) | 1.25 | (0.82, 2.26) | 0.73 | (0.59, 0.94) | ||
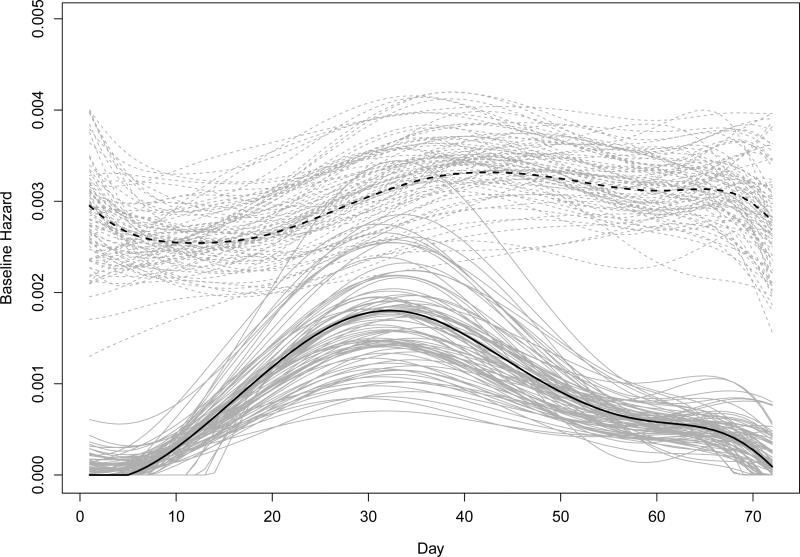
The estimates for age effects on the relative hazards of infection were not sensitive to the choice of vaccine effectiveness delay. The estimates for baseline hazards based on the main effect model and seven-day delay, together with estimates from 100 bootstrap samples, are shown in Figure 3. The patterns of the baseline hazards are consistent with the timeline of surveillance cultures in Figure 2(b), where the influenza hazard tended to be unimodal and the nuisance hazard seemed relatively flat. The bootstrap curves evince the uncertainty in the kurtosis of the mode of the influenza hazard.
Figure 3.
Baseline hazard estimates from data (black) and 100 bootstrap samples (gray) for both influenza-induced (dashed lines) and non-influenza-induced MAARIs (solid lines) among children in Temple-Belton, Texas during the 2003-2004 season. The estimates are based on the main effect model and seven-day delay of vaccine effectiveness.
Halloran et al. (2007) reported 0.56 (95% CI: 0.24, 0.84) for the overall VE, 0.66 (95% CI: −0.03, 1.0) among younger and 0.53 (95% CI: 0.12, 0.86) among older children. Those estimates lie between our pathway EM algorithm estimates assuming a one-day and a seven-day effectiveness delay. The overall estimate of VE against MAARI was 26% (95% CI: 11%, 39%), illustrating the attenuation in the estimate if only the nonspecific outcome is used.
5. Discussion
We have proposed the pathway EM and the pathway PV algorithms for estimating the effect of a covariate on an event type of interest that can occur only once in a sequence of nonspecific events with nonmonotonic confirmation of the event types. The methods take into account all pathways of event types compatible with the observed data in an individual. The methods perform well at a variety of baseline hazards and sampling proportions. In general, The pathway EM algorithm outperforms the noniterative PV approaches, and the pathway PV approach outperforms the aggregation PV approach, in particular when infection hazards are high. They perform similarly at low hazard rates. Reasonable performance of the pathway PV approach requires only that the hazards of nonrecurrent event types be small. Thus, one might consider using that approach when individuals tend to have just one or fewer events. The pathway EM algorithm also performs well under different sampling schemes.
Strengths of the methods are that they can handle any pattern of missing tests, time dependent covariates, different shapes of baseline hazards for different event types, and any number of event types. An advantage of the methods is the computations can be done using packages available in R. Although we chose to use an EM algorithm for our computation, Markov chain Monte Carlo methods could likely be used to solve the same problem.
A limitation of the methods is the assumption the laboratory tests are missing at random. Nevertheless, our method is relatively robust to the violation of MAR for a practical range of selection bias in the sampling of influenza-induced MAARIs. In the case of missing not at random, a selection model (Little and Rubin, 2002; Daniels and Hogan, 2008) that associates the probability of observing a laboratory result with the underlying pathogen and other covariates could be extended to the survival setting to perform sensitivity analyses. Scharfstein et al. (2006) proposed both frequentist and Bayesian approaches for estimating VE in the presence of selection bias. In the analysis of an influenza vaccine trial, they found that VE estimates based on the MAR assumption were higher than those based on elicited expert opinion on selection bias. However, under any plausible range of selection bias, the VE estimates based on the validation sample were much higher than the point estimate using just the nonspecific case definition.
We extended the methods to frailty models that account for unmeasured heterogeneity under a distributional assumption. For continuous distributions such as gamma and Gaussian, such extension is straightforward. However, difficulties may arise with other useful distributions, for example, a mixture with point mass at zero and a continuous distribution. In the context of infectious diseases, subjects infected before the study may not be susceptible to the disease during the study due to preimmunity. The proportion of preimmunity i.e., Pr(ξi = 0), is estimable given sufficient data (Longini and Halloran, 1996). If preimmunity is to be considered, a possible solution is to augment the data with a Bernoulli variable ηi such that ξi = 0 if ηi = 1 and ξi follows a continuous distribution otherwise. It would also be of interest to explore in the future how the VE estimation would be affected if such preimmunity exists but is ignored.
Some vaccine studies were designed based on close contact groups (clusters) rather than unrelated individuals, e.g., household studies. In that case, tracing back the risk history of each individual will be more complex, probably involving simultaneous estimation of the hazard of infection from the community at large and the hazard from infectious individuals within clusters. Yang et al. (2010) explored this setting in a Bayesian framework but assuming parametric hazard shapes for both community-to-person and person-to-person transmission. Both that and previous work found that a low culture proportion, as compared to much higher proportions, did not greatly compromise the accuracy of VE estimation. Kenah (2013) showed the feasibility of nonparametric estimation of person-to-person transmission hazard in the presence of complete data. It would be of interest to investigate how missing culture information may affect inference if hazards of both sources of transmission are to be estimated nonparametrically. Inference may be more sensitive to culture proportion in situations where the majority of individuals have multiple MAARI onsets, for instance, in the analysis of sequential outbreaks in multiple years, which is a subject of future research.
Supplementary Material
Acknowledgements
We thank Drs. Terry M. Therneau at Mayo Clinic and Ira M. Longini at University of Florida for their helpful discussion with us. We also thank the academic editor and the reviewers for their insightful comments which helped us improving this paper. This work was supported by the National Institute of Allergy and Infectious Diseases grant R37-AI032042. EK was supported by National Institute of Allergy and Infectious Diseases grant K99/R00-AI095302.
Footnotes
Supplementary Materials
Web Appendices referenced in Sections 2 and 3, together with the computer code, are available with this paper at the Biometrics website on Wiley Online Library.
References
- Breslow NE. Covariance analysis of censored survival data. Biometrics. 1974;30:89–99. [PubMed] [Google Scholar]
- Cheng M, Fan J, Marron JS. On automatic boundary corrections. Annals of Statistics. 1997;25:1691–1708. [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society B. 1977;39:1–38. [Google Scholar]
- Daniels MJ, Hogan JW. Missing Data in Longitudinal Studies: Strategies for Bayesian Modeling and Sensitivity Analysis. John Wiley; New York: 2008. [Google Scholar]
- Efron B. Efficiency of Cox's likelihood function for censored data. Journal of American Statistical Association. 1997;72:557–565. [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- Halloran ME, Piedra PA, Longini IM, Gaglani MJ, Schmotzer B, Fewlass C, Herschler GB, Glezen WP. Efficacy of trivalent, cold-adapted, influenza virus vaccine against influenza A (Fujian), a drift variant, during 2003-2004. Vaccine. 2007;25:4038–4045. doi: 10.1016/j.vaccine.2007.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim JG, Chu H, Chen LM. Basic concepts and methods for joint models of longitudinal and survival data. Journal of Clinical Oncology. 2010;28:2796–2801. doi: 10.1200/JCO.2009.25.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenah E. Nonparametric survival analysis of infectious disease data. Journal of the Royal Statistical Society B. 2013;75:277–303. doi: 10.1111/j.1467-9868.2012.01042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JP. Semiparametric estimation of random effects using the Cox model based on the EM algorithm. Biometrics. 1992;48:795–806. [PubMed] [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. John Wiley; New York: 2002. [Google Scholar]
- Longini IM, Halloran ME. A frailty mixture model for estimating vaccine efficacy. Applied Statistics. 1996;45:165–173. [Google Scholar]
- Louis TA. Finding the observed information matrix when using the EM algorithm. Journal of the Royal Statistical Society B. 1982;44:226–233. [Google Scholar]
- Magaret AS. Incorporating validation subsets into discrete proportional hazards models for mismeasured outcomes. Statistics in Medicine. 2008;27:5456–5470. doi: 10.1002/sim.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. URL http://www.R-project.org/ [Google Scholar]
- Ramsay JO, Silverman BW. Functional Data Analysis. New York: Springer. 1997 [Google Scholar]
- Ramsay JO. Estimating smooth monotone functions. Journal of Royal Statistical Society B. 1998;60:365–375. [Google Scholar]
- Ramsay JO, Wickham H, Graves S, Hooker G. fda: Functional Data Analysis. R package version 2.2.7. 2011 http://CRAN.R-project.org/package=fda.
- Scharfstein DO, Halloran ME, Chu H, Daniels MJ. On estimation of vaccine efficacy using validation samples with selection bias. Biostatistics. 2006;7:615–629. doi: 10.1093/biostatistics/kxj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert B, Maechler M. lpridge: Local Polynomial (Ridge) Regression. R package version 1.0-6. 2011 http://CRAN.R-project.org/package=lpridge.
- Tanner MA. Tools for statistical inference. 2nd edition Springer; New York: 1992. [Google Scholar]
- Therneau TM, Grambsch PM. Modeling survival data (extending the Cox model) Springer; New York: 2000. [Google Scholar]
- Therneau TM, Grambsch PM, Pankratz VS. Penalized survival models and frailty. Journal of Computational and Graphical Statistics. 2003;12:156–175. [Google Scholar]
- Therneau T. A Package for Survival Analysis in S. R package version 2.36-14. 2012.
- Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: An overview. Stat Sin. 2004;14:809–834. [Google Scholar]
- Yang Y, Halloran ME, Daniels M, Longini IM, Burke DS, Cummings DAT. Modeling competing infectious pathogens from a Bayesian perspective: Application to influenza studies with incomplete laboratory results. Journal of the American Statistical Association. 2010;105:1310–1322. doi: 10.1198/jasa.2010.ap09581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.