Abstract
Objective
CD248 (tumor endothelial marker 1/endosialin) is found on stromal cells and is highly expressed during malignancy and inflammation. Studies have shown a reduction in inflammatory arthritis in CD248-knockout (CD248−/−) mice. The aim of the present study was to investigate the functional effect of genetic deletion of CD248 on bone mass.
Methods
Western blotting, polymerase chain reaction, and immunofluorescence were used to investigate the expression of CD248 in humans and mice. Micro-computed tomography and the 3-point bending test were used to measure bone parameters and mechanical properties of the tibiae of 10-week-old wild-type (WT) or CD248−/− mice. Human and mouse primary osteoblasts were cultured in medium containing 10 mM β-glycerophosphate and 50 μg/ml ascorbic acid to induce mineralization, and then treated with platelet-derived growth factor BB (PDGF-BB). The mineral apposition rate in vivo was calculated by identifying newly formed bone via calcein labeling.
Results
Expression of CD248 was seen in human and mouse osteoblasts, but not osteoclasts. CD248−/− mouse tibiae had higher bone mass and superior mechanical properties (increased load required to cause fracture) compared to WT mice. Primary osteoblasts from CD248−/− mice induced increased mineralization in vitro and produced increased bone over 7 days in vivo. There was no decrease in bone mineralization and no increase in proliferation of osteoblasts in response to stimulation with PDGF-BB, which could be attributed to a defect in PDGF signal transduction in the CD248−/− mice.
Conclusion
There is an unmet clinical need to address rheumatoid arthritis–associated bone loss. Genetic deletion of CD248 in mice results in high bone mass due to increased osteoblast-mediated bone formation, suggesting that targeting CD248 in rheumatoid arthritis may have the effect of increasing bone mass in addition to the previously reported effect of reducing inflammation.
CD248 (also known as endosialin or tumor endothelial marker 1) is a single-pass transmembrane receptor whose ligands are reported to be extracellular matrix molecules (fibronectin and type I/IV collagen) (1). CD248 is widely expressed on mesenchymal cells in the developing embryo and is required for proliferation and migration of pericytes and fibroblasts (2). Expression of CD248 is dramatically reduced in adults, but can be up-regulated during fibrosis, inflammation, and malignancy (3-6). This distinctive temporal presentation, exclusive expression on stromal cells (2,5), structural similarity to other family members (including thrombomodulin and C1qRp), association with matrix metalloproteinase activity (1,6), and expression on mesenchymal stem/stromal cells (MSCs) (7) suggest a role for CD248 in tissue remodeling and repair.
CD248 is required for the progression of certain tumors. Abdominal tumors implanted into CD248-knockout (CD248−/−) mice exhibit a reduction in growth, invasiveness, and metastasis (8). This defect is thought to be due to the role of CD248 in perivascular cells and tumor-associated fibroblasts, which support the vascularization of the tumor and facilitate malignant cell migration through the basement membrane. These findings have led to the development of anti-CD248 antibodies for use as therapeutic agents in cancer (9). In human kidney disease, the level of CD248 expression is predictive of fibrosis and disease outcome (4). CD248 is also highly expressed in the rheumatoid synovium. In particular, it has been shown to exacerbate inflammatory arthritis in mice, although the molecular basis of this phenotype has not been fully elucidated (6).
CD248 has also been identified as an MSC marker (3). Given this finding, and considering that findings from in silico data-mining of an online gene expression database have shown that CD248 is very highly expressed on osteoblasts (10), we explored the function of CD248 in bone formation using mice deficient in CD248. We found that CD248 acts as a negative regulator of bone formation, since mice lacking CD248 demonstrated increased bone formation in vivo and in vitro. Moreover, we were able to identify the molecular mechanism for this bone phenotype, determined to be an impairment of platelet-derived growth factor (PDGF) signaling in osteoblasts lacking CD248.
MATERIALS AND METHODS
Mice
The generation and genotyping of CD248−/− mice have been described previously (8). Mice on the C57BL/6 and 129SvEv genetic backgrounds were used for in vivo experiments. All studies in vitro were carried out on cells derived from the C57BL/6 mouse strain. Each experiment was performed in accordance with UK laws and with the approval of the local ethics committees. Mice with green fluorescent protein-labeled Col1a1 were generated and validated on a C57BL/6 background, as previously described (11,12). Briefly, 3.2 kb of the type I collagen α1 promoter was cloned, as were DNase I-hypersensitivity sites with enhancer activity from 7 kb and 8 kb 5′ of the promoter.
In vivo calcein labeling
Ten-week-old mice were injected intraperitoneally with 20 mg/kg calcein in saline. Two injections were given 7 days apart, and mice were killed 3 days after the second injection. Samples were fixed overnight in 4% formaldehyde and transferred into 70% ethanol, before dehydration through graded ethanol. Samples were then transferred into methyl methacrylate (MMA; Sigma-Aldrich). Processing took place at 4°C, and the schedule included a vacuum step for the infiltration of MMA. Samples were embedded in MMA at 37°C overnight. The blocks were cut into 4-μm sections using a tungsten carbide knife on a Leica 2165 motorized microtome. The sections were placed in a press overnight at 37°C.
Immunofluorescence
Tissue from newborn mice was snap-frozen and cut into 5-μm-thick sections on a cryostat. The sections were fixed for 10 minutes in acetone at 4°C. Sections were stained overnight at 4°C with rabbit anti-CD248 PI3 (10 μg/ml; generated in-house) (5), anti–PDGF receptor α (anti–PDGFRα) (clone APS5; eBioscience), and ELF 97 for detection of endogenous phosphatase activity (Invitrogen), as detailed in the manufacturers’ instructions (1-minute incubation at room temperature). The anti-mouse CD248 antibody has been previously described and shown to be CD248-specific through extensive testing against CD248−/− mouse tissue (2,5). Nuclei were stained with 20 μg/ml bisbenzimide dye (Hoechst 33258 Fluorochrom; Riedel De Haenag). Sections were mounted in 2.4% DABCO (Aldrich) in glycerol (Fissons Scientific) at pH 8.6. Images were captured using a Zeiss LSM 510 confocal microscope, with reference to cultures containing antibody only as secondary controls.
Histomorphometry
Ten-week-old mice were injected intraperitoneally with 20 mg/kg calcein AM (Sigma-Aldrich) in saline on day 1 and day 7. On day 8, the mice were killed and the tibiae were dissected and fixed in 4% buffered formalin/saline (pH 7.4) prior to embedding in methylmethacrylate. Longitudinal 1.5-μm sections were prepared and stained with toluidine blue to identify cells, and von Kossa’s stain was used to highlight ossified regions. Images were obtained on a Leica DM6000 microscope, with data analyzed using ImageJ software.
The data obtained were the lengths of the single- and double-labeled regions, and the distance between labels in double-labeled regions. The mineral apposition rate (MAR; in μm per day) was calculated as the distance between labels/7. The mineralized surface/bone surface (MS/BS; in %) was calculated as (cumulative length of double labels/[cumulative length of single labels + cumulative length of double labels]) × 100. The bone formation rate/bone surface (BFR/BS; in %) was calculated as MS/BS × MAR.
Mechanical testing
Tibiae from 10-week-old WT and CD248−/− mice were tested to the point of failure in the 3-point bending test, as described in a study by Huesa et al (13). Briefly, bones were positioned, for stability, with the fibula insertion placed upward and the test span, adjusted to accommodate the shortest bone, set at 10.0 mm. The load head was set to descend at 1 mm/minute, and force and displacement data were recorded. These data, comprising stiffness (gradient of the rising portion), load to failure (maximum load supported), and work required to fracture (determined from the area under the curve), were calculated from the resulting plot.
Material properties
The material properties of the bone were measured utilizing methods previously described by Somerville et al (14). Bone density was measured using Archimedes’ principle, comparing the weight of the bone in air with the weight when suspended in water. Density measurements were combined with the speeds of sound measured ultrasonically, to yield the elastic, or Young’s, modulus (ratio of stress to strain). Bone composition was determined by the ashing technique, in which the bones were weighed, heated to 100°C for 24 hours, weighed, and heated at 600°C, before being weighed again. Bone weights were used to calculate the water, organic, and mineral fractions.
Micro–computed tomography (micro-CT)
Micro-CT analysis of the tibiae (formalin-fixed) from 10-week-old WT and CD248−/− mice was used to measure the trabecular and cortical bone parameters. Bones were scanned using a SkyScan 1072 x-ray microtomograph at 50 kV/197 μA. Images were obtained at a resolution of 5 μm with a rotation step of 0.67°. Images were reconstructed using NRecon version 1.4.4 (SkyScan), and trabecular bone parameters were analyzed using CTAn version 1.7.0.2 software (SkyScan). Cortical parameters (cortical area, second moment of area [representing resistance to bending], and periosteal and endosteal parameters) were measured in slices close to the center of each bone, using in-house scripts in ImageJ software (version 1.33u; downloaded from the National Institutes of Health web site at http://rsb.info.nih.gov/ij/).
Isolation and culture of mouse cells
Osteoblasts/preosteoblasts were isolated from newborn mouse pup calvaria (1–3 days postpartum) by serial digestion at 37°C with collagenase D (1 mg/ml in Hanks’ balanced salt solution [HBSS]; Sigma) and EDTA (4 mM in phosphate buffered saline [PBS]) as follows: 10 minutes with collagenase (discard supernatant), 30 minutes with collagenase (retain supernatant), PBS wash, 10 minutes with EDTA (retain fraction), HBSS wash, and 30 minutes with collagenase (retain supernatant). The resulting cells were cultured on tissue culture plastic in osteoblast differentiation medium (Dulbecco’s modified Eagle’s medium, 10% fetal calf serum, l-glutamine, 10 mM β-glycerophosphate, and 50 μg/ml ascorbic acid) to induce mineralization. Where indicated, culture medium was supplemented with 100 ng/ml recombinant mouse PDGF-BB or with 10 ng/ml transforming growth factor β (TGFβ) (both from eBioscience) for the duration of the experiment. These cytokines were replaced with each change of medium (every 3–4 days). Mineralized nodules were visualized by fluorescence microscopy following overnight incubation with 0.2 mM calcein. Macrophages were isolated from the bone marrow of WT C57BL/6 mice and cultured with 50 ng/ml macrophage colony-stimulating factor (M-CSF). Osteoclasts were generated by culturing bone marrow macrophages with 50 ng/ml M-CSF and 10 ng/ml RANKL (both from R&D Systems).
Isolation and culture of human cells
Osteoblasts were isolated by explant culture of diced cancellous bone from the femoral head of a patient with osteoarthritis. Peripheral blood mononuclear cells (PBMCs) were donated, with informed consent, from healthy volunteers (with approval from the North of Scotland Research Ethics Committee). PBMCs were isolated by centrifugation over Lymphoprep (Axis-Shield) and seeded into 75-cm2 flasks in culture medium supplemented with 20 ng/ml M-CSF (R&D Systems) for 7 days, to allow adherence and expansion of M-CSF–dependent monocytes. To generate osteoclasts, the M-CSF–dependent monocytes were harvested by trypsinization and gentle scraping, and then replated at a seeding density of 200,000 cells/ml in medium containing 20 ng/ml M-CSF and 100 ng/ml RANKL (both from PeproTech). The medium was refreshed every 2–3 days, and after ~5–7 days in culture with RANKL, more than 80% of the cells were determined to be αvβ3 positive, with numerous multinucleated osteoclasts.
Western blotting
Mouse and human cells were lysed in nonreducing buffer and run on a 10% polyacrylamide gel. Gel bands were transferred to a 0.45-μm PVDF membrane (Flowgen) and blots were blocked with 5% nonfat dried milk in Tris buffered saline, followed by staining with anti-human CD248 monoclonal antibody (mAb) B1/35 (generated in-house; previously described in a 2005 study by MacFadyen et al [15]), anti-mouse CD248 mAb PI3 (generated in-house; previously described in a 2007 study by MacFadyen et al [5]), anti-mouse p44/42 MAPK mAb (#4695; eBioscience), anti-mouse phospho-p44/42 MAPK mAb (#9101; eBioscience), or β-actin (eBioscience). Detection was subsequently performed with species-specific secondary antibodies conjugated to horse-radish peroxidase (Amersham). The blot was visualized by enhanced chemiluminescence (Amersham) and autoradiography (Kodak X-Omat).
Real-time reverse transcription–polymerase chain reaction (RT-PCR) from cell lysates
Cell pellets were assayed by real-time PCR for messenger RNA (mRNA) expression, using TaqMan gene expression assays (Applied Biosystems) for human CD248 (Hs.00535586), mouse CD248 (Mm.00547485), and mouse c-fos (Mm.00487425). Samples were reverse transcribed using a Cells-to-cDNA kit (Ambion) or TaqMan Reverse Transcription kit (Applied Biosystems) with PCR Master Mix and TaqMan Universal Master Mix II reagent. Assays were run on a 7900HT Real-Time PCR system (Applied Biosystems) for 40 cycles according to the following protocol: 50°C for 2 minutes, 95°C for 10 minutes, 95°C for 15 seconds, 95°C for 1 minute, and holding at 10°C. Expression values for the gene of interest were normalized to those for 18S ribosomal RNA and were transformed to assume a doubling of product with each PCR cycle, calculated using the comparative threshold cycle (Ct) method and the 2−ΔCt formula, as Ct gene of interest – Ct housekeeping gene, or expression of the gene of interest was normalized to that of the housekeeping gene, and results were expressed as the fold change relative to values in control, untreated samples.
MTT proliferation assay
Mouse and human cells were seeded at 50,000 cells per well in a 96-well plate in osteoblast differentiation medium (as described above) for 6 days. The culture medium was supplemented for the duration of the experiment with either PDGF-BB at 100 ng/ml or TGFβ1 at 10 ng/ml, and replaced at each change of medium time point (every 3–4 days). Osteoblast proliferation after stimulation was compared to that under control conditions (unsupplemented medium). An MTT assay was then performed, in which the cells were incubated with MTT solution at 37°C for 4 hours and then dried, prior to incubation with DMSO at 37°C to dissolve the formazan crystals. The amount of product produced was measured using a spectrophotometer at 540 nm.
Statistical analysis
All analyses of bone parameters were performed using Student’s 2-tailed t-test. Results of in vitro functional assays were compared by one-way analysis of variance with Tukey’s post hoc analysis (with 95% confidence intervals). All results are expressed as the mean ± SD or mean ± SEM.
RESULTS
CD248 expression on mature osteoblasts in vitro and in vivo
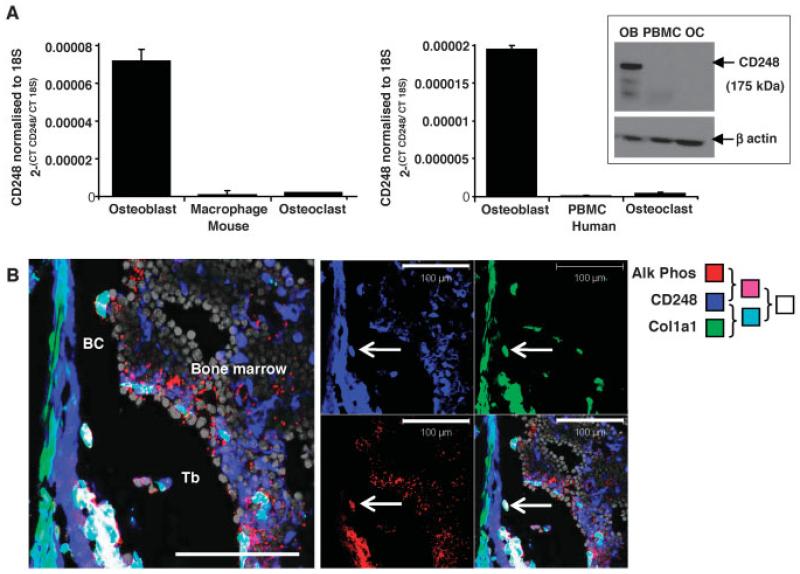
According to the gene expression data from in silico data-mining (10), we found that CD248 is highly expressed on osteoblasts and a range of other stromal cells, but not on leukocytes or osteoclasts. Analyses by real-time PCR confirmed this finding in primary murine and human osteoblasts when compared with osteoclasts and PBMCs or macrophages (Figure 1A).
Figure 1. CD248 is expressed in human and mouse osteoblasts, but not osteoclasts.
A, Real-time polymerase chain reaction was used to analyze the expression of CD248 in primary cell lysates of osteoblasts, macrophages, and osteoclasts from wild-type mice (left) or osteoblasts, peripheral blood mononuclear cells (PBMCs), and osteoclasts from human subjects (right). Bars show the mean ± SD of 3 samples per group. Inset, CD248 expression was also assessed by Western blotting of human cell lysates of osteoblasts (OB), PBMCs, and osteoclasts (OC), using anti-human CD248 (recognizing the band at 175 kd) with anti–β-actin as control (recognizing the band at 47 kd). B, Tibiae from newborn mice were assessed by confocal microscopy for CD248 (blue), Col1a1 (green fluorescent protein under the control of the Col1a1 promoter; green), and alkaline phosphatase (Alk Phos) enzymatic activity (red); nuclei were stained in grey. The merged image in the left panel shows the colocalization of Col1a1 and CD248 (cyan), colocalization of CD248 and alkaline phosphatase (purple), and colocalization of all 3 markers (white). Bar (left panel) = 100 μm. Arrows indicate examples of cells expressing all 3 markers. BC = bone collar; Tb = trabecular bone.
In addition, cell lysates from human tissue were analyzed by Western blotting (Figure 1A, inset). CD248 protein was detectable on osteoblasts, but was undetectable or barely detectable on PBMCs and osteoclasts. The presence of CD248 on osteoblasts, but not osteoclasts, is consistent with previous descriptions of the restricted expression of CD248 on mesenchyma-derived cells (5,15).
In many cases, when cells are removed from their natural environment, their phenotype and gene expression profiles are altered. We therefore checked whether CD248 could be detected on osteoblasts in situ. We assessed the expression of type I collagen and the activity of alkaline phosphatase to identify osteoblasts in sections of bone from newborn mice. Type I collagen (comprising Col1a1 and Col1a2) is the major constituent of the bone organic extracellular matrix laid down by osteoblasts, and therefore Col1a1 is a marker of mature, matrix-producing osteoblasts. Alkaline phosphatase is an enzyme produced by mature osteoblasts that facilitates precipitation of calcium crystals during mineralization of the bone extracellular matrix. As shown in Figure 1B, analyses by confocal microscopy of the long bone (tibia) of a newborn mouse revealed that mature osteoblasts coexpressed Col1a1 and alkaline phosphatase around the growth plate, bone collar, and trabecular surfaces. Many of these osteoblasts also expressed CD248. (In Figure 1B, white staining indicates coexpression of all 3 markers; individual examples are identified by arrows.)
Association of CD248 genetic deletion in mice with stronger, tougher, and stiffer bones
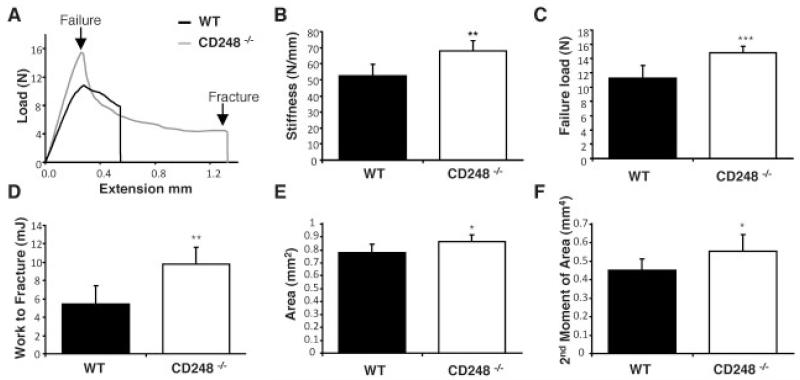
To measure the effect of deletion of CD248 on whole bone properties, the femora and tibiae of 10-week-old WT and CD248−/− C57BL/6 mice (previously described in a study by Nanda et al [8]) were tested to the point of destruction by 3-point bending (Figure 2). Only results from the tibiae are shown, as similar results were obtained in both bones.
Figure 2. CD248-deficient mice have thicker, stiffer bones that are harder to break.
A–D, Tibiae from 10-week-old wild-type (WT) and CD248−/− C57BL/6 mice were tested to the point of destruction by 3-point bending. Results shown are a representative trace of bone extension as load is applied (A) as well as bone stiffness (B), failure load (C), and work required to fracture (D) in each group. E and F, Micro–computed tomography analysis of the tibiae of 10-week-old WT and CD248−/− C57BL/6 mice was performed to measure cortical bone parameters of cortical area (E) and second moment of area (F). Bars show the mean ± SD of 6 samples per group. * = P < 0.05; ** = P < 0.01; *** = P < 0.001 versus WT, by Student’s t-test.
Findings from the bending test showed that the tibiae from CD248−/− mice had superior mechanical properties. The load required for failure of the tibia was compared between WT and CD248−/− mice (Figure 2A), from which the area under the curve (equating to the work required to fracture) was calculated. The stiffness, failure load (strength), and work to fracture (toughness) values (Figures 2B-D) were all significantly increased in the CD248−/− mouse bones compared to those of WT controls. CD248−/− mouse tibiae also had significantly greater cortical area (P = 0.041) and second moment of area (P = 0.042) than did WT controls, as measured by micro-CT (Figures 2E and F).
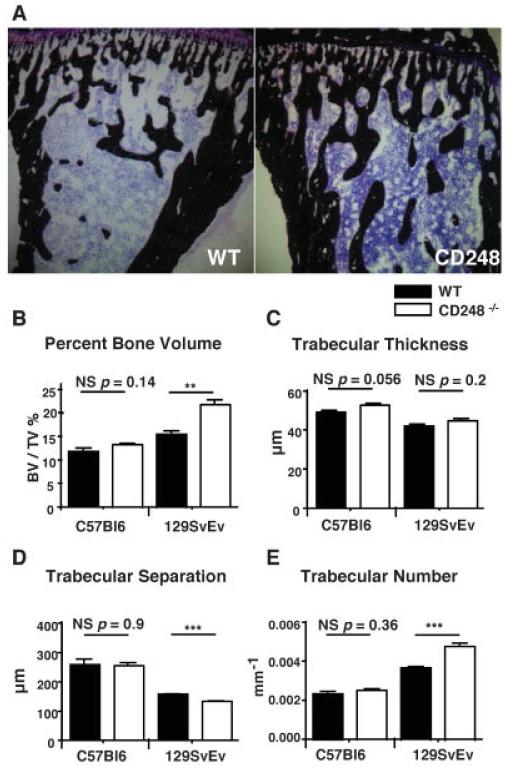
Von Kossa’s staining of sections of the mouse tibia revealed increased trabecular bone in CD248−/− mice compared to WT control mice, as shown in Figure 3A. This phenotype on 2 genetic backgrounds, C57BL/6 and 129SvEv was investigated by micro-CT. Increased bone volume/tissue volume was seen in CD248−/− mice on the 129SvEv strain (Figure 3B). The cause of the increased bone volume in these mice could be attributed to a significant increase in the number of trabeculae (Figure 3E), a trend toward increased trabecular thickness (Figure 3C), and a corresponding reduction in trabecular separation (Figure 3D). In mice on the C57BL/6 background, a strain recognized as having lower peak bone mass (16), these differences failed to reach statistical significance.
Figure 3. CD248-deficient mice have increased trabecular bone volume.
A, The tibiae of wild-type (WT) and CD248−/− (129SvEv strain) mice were stained with von Kossa’s stain to detect trabecular bone formation (dark brown; counterstained with hematoxylin [purple]). B–E, Micro–computed tomography was performed to measure the trabecular bone structure in WT and CD248−/− mice on the C57BL/6 and 129SvEv genetic backgrounds, assessed as the percentage of bone volume/tissue volume (BV/TV) (B), trabecular thickness (C), trabecular separation (D), and trabecular number (E). Bars show the mean ± SD of 6 samples per group. ** = P < 0.01; *** = P < 0.001, by Student’s t-test. NS = not significant.
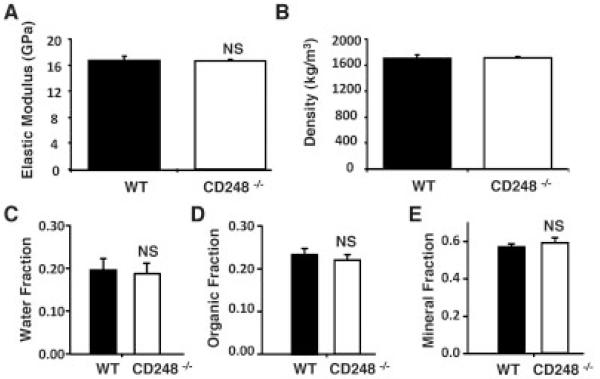
Deletion of CD248 had no effect on the material properties or composition of the cortical bone in either strain of mice. The elastic modulus and cortical density identified by 3-point bending, as well as the mineral, organic, and water fractions analyzed by ashing, were the same in WT and CD248−/− mouse tibiae (Figures 4A-E), indicating that no change in bone composition occurs with deletion of CD248. Only results from mice on the C57BL/6 background are shown, as the same results were obtained in both strains.
Figure 4. CD248-deficient mice have normal bone composition.
The parameters of bone composition included the elastic modulus (A) and bone density (B), measured in wild-type (WT) and CD248−/− mice on the C57BL/6 background, as well as the water (C), organic matter (D), and mineral (E) fractions, determined using ashing in the C57BL/6 and 129SvEv strains of WT and CD248−/− mice. Bars show the mean ± SD of 6 samples per group. Student’s t-test was used to assess differences between the 2 genotypes. NS = not significant.
Increased bone mineralization capability of osteoblasts from CD248-deficient mice
To determine whether osteoblasts were the primary cells responsible for the bone phenotype, we isolated osteoblasts from the calvaria of newborn WT and CD248−/− mice and cultured them in osteogenic medium for 21 days. Bone formation was visualized using a calcein incorporation assay, followed by fluorescence microscopy (Figure 5A). A 10-fold increase in bone nodules was seen in the CD248−/− mouse osteoblast cultures after 21 days.
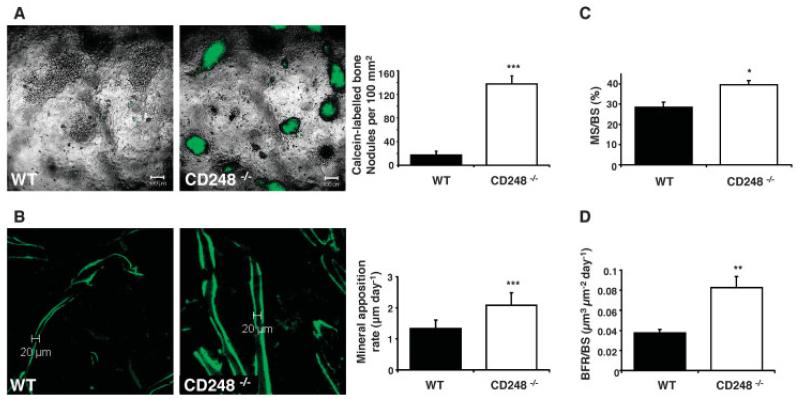
Figure 5. Increased osteoblast activity is the cause of increased bone formation in CD248-deficient mice.
A, Cultures of osteoblasts from wild-type (WT) and CD248−/− mice were stained with calcein to label mineralized bone nodules (left), and the bone nodules were quantified in the osteoblast cultures from both groups (right). The experiment was repeated twice; representative results are shown. B, Ten-week-old WT and CD248−/− C57BL/6 mice were given 2 injections of calcein 7 days apart to label mineralizing bone surfaces (left). The distance between these labels, measured in the trabecular bone at the epiphysis of tibia sections, was determined as the mineral apposition rate (right). C and D, The percentage of bone surface that was actively mineralized over the course of the 7-day period of the experiment (mineralized surface/bone surface [MS/BS]) (C) and the bone formation rate per bone surface (BFR/BS) per day (D) were determined in each group. All analyses were performed by investigators blinded with regard to genotype. Bars in A, C, and D show the mean ± SD of 3 samples per group. Bars in B show the mean ± SD of 6 samples per group. * = P < 0.05; ** = P < 0.01; *** = P < 0.001 versus WT, by Student’s t-test.
To confirm that this increase in osteoblast activity was also present in vivo, calcein incorporation assays were used to assess the rate of mineral apposition. Bone sections from 10-week-old C57BL/6 mice injected with calcein twice, 7 days apart, showed fluorescently labeled zones representing mineralizing surfaces at the 2 injection time points (Figure 5B, left). The distance between these 2 labels establishes the amount of mineral laid down at a given location over the 7-day period, which was converted into the MAR (expressed in μm per day) (Figure 5B, right). CD248−/− mice exhibited a significantly increased MAR compared to WT controls.
In addition, 2 other measures of bone formation, the percentage of mineralized surface (MS/BS) and the rate of bone formation (BFR/BS; in μm3 per μm2 per day), were increased in the CD248−/− mice compared to WT control mice (Figures 5C and D). The MS/BS indicates the amount of the trabecular bone surface area that has been mineralized over the 7-day period of the experiment. The BFR/BS is a composite measure that takes into account the percentage of surface that is being remodeled during the experimental period, as well as the depth of bone formed along that surface, to give the area of new bone formed per day.
Role of CD248 in PDGF signaling in osteoblasts
PDGF is a growth factor and a potent mitogen for cells of mesenchymal origin. PDGF is a glycoprotein consisting of either a homodimer of the α- or β-chains (PDGF-AA or PDGF-BB, respectively) or a heterodimer of the 2 chains (PDGF-AB). PDGF functions by signaling through the receptor tyrosine kinase PDGFRα or PDGFRβ. PDGFRα can bind to all forms of the PDGF dimer, whereas PDGFRβ can bind to only the BB and AB forms. Binding of the ligand to its receptor results in PDGFR phosphorylation, followed by phosphorylation of the MAPK ERK-1/2 and, finally, up-regulated transcription of target genes, including c-fos (17).
Given that a role of CD248 in PDGF signaling has been reported (17), we hypothesized that the increase in osteoblast differentiation and activity in CD248-deficient mouse cells was due to the role of CD248 in the PDGF signaling cascade. Microarray analysis comparing global gene expression in WT and CD248−/− mouse embryonic fibroblasts confirmed the previous findings in the study by Tomkowicz et al (17), that mRNA expression of c-fos, the downstream target of PDGF signaling, was reduced in CD248-deficient mouse cells, to levels that were one-half those observed in WT mouse cells, whereas there was no reduction in PDGFRα or PDGFRβ expression (results not shown).
PDGFR expression on osteoblasts has been widely reported (18,19), and we confirmed these findings with the use of alkaline phosphatase as an osteoblast marker. As shown in Figure 6A, CD248 and PDGFRα colocalized on the osteoblast cell surface. (In Figure 6A, purple staining indicates dual labeling for CD248 and PDGFRα, green indicates staining for alkaline phosphatase, and white indicates colocalization of all 3 proteins.)
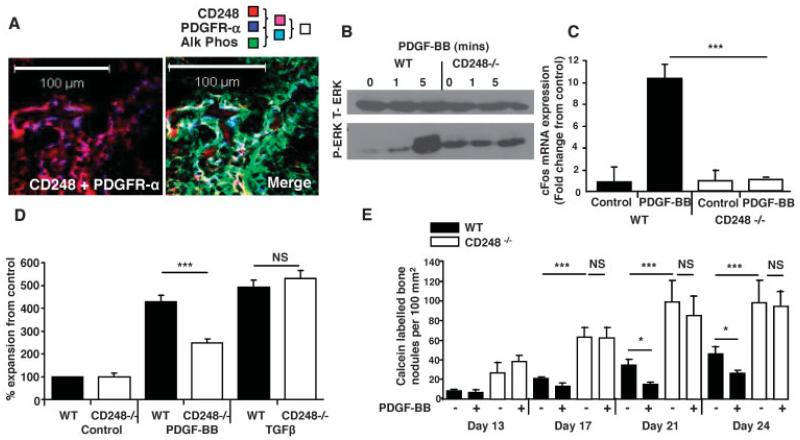
Figure 6. CD248 signals via the platelet-derived growth factor receptor (PDGFR) to maintain osteoblasts in an immature state.
A, Confocal microscopy images of newborn mouse tibiae show the colocalization of CD248 and PDGFRα (left), and the colocalization of CD248, PDGFRα, and alkaline phosphatase (Alk Phos) (merged image; right). B, Total ERK (t-ERK) and phosphorylated ERK (p-ERK) were assessed by Western blotting in osteoblasts from WT and CD248−/− mouse tibiae exposed to PDGF-BB for 0, 1, or 5 minutes. C, Real-time polymerase chain reaction was performed to analyze c-fos expression, normalized to the values for GAPDH, in osteoblasts from WT and CD248−/− mice after stimulation with PDGF-BB. Results are the mean ± SD fold change in 3 samples per group, relative to that in control, untreated cultures (set at 1). D, Proliferation of WT and CD248−/− mouse osteoblasts was assessed by MTT proliferation assay after 6 days of stimulation with PDGF-BB or transforming growth factor β (TGFβ). Results, at an absorbance at 550 nm, are the mean ± SEM of 3 samples per group, normalized to the control, untreated cell response. E, Results of in vitro mineralization assays, with or without PDGF-BB, show that bone nodule formation was inhibited at all time points after stimulation with PDGF-BB in WT mouse osteoblasts, but not in CD248−/− mouse osteoblasts. Bars show the mean ± SEM of 6 samples per group. Experiments were repeated twice; representative results are shown. * = P < 0.05; *** = P < 0.001, by Student’s t-test. NS = not significant.
To test whether PDGF signaling was perturbed in CD248−/− mouse osteoblasts, the cells were stimulated with PDGF-BB for 0, 1, or 5 minutes prior to Western blot analysis for phosphorylation of ERK, which is downstream of PDGFR. WT mouse cells showed robust ERK phosphorylation after 5 minutes of stimulation with PDGF-BB, whereas no effect of PDGF stimulation was seen in CD248−/− mouse cells (Figure 6B).
Since PDGF signaling induces c-fos transcription, we also compared the c-fos expression levels in osteoblasts from WT and CD248−/− mice. We found that WT mouse cells showed a 12-fold up-regulation of c-fos mRNA following PDGF-BB stimulation; in contrast, CD248−/− mouse cells showed no such response (Figure 6C). These results match the findings in the study by Tomkowicz et al (17), in which the presence of CD248 was found to be required for PDGF signaling in pericytes.
Given this required presence of CD248 for PDGF signaling, we investigated the functional effects of CD248 knockout in mouse osteoblasts. Using MTT proliferation assays, we found that proliferation of PDGF-stimulated CD248−/− mouse osteoblasts was decreased, similar to that previously reported with regard to CD248−/− mouse pericytes (17). As shown in Figure 6D, CD248−/− mouse cells, in comparison to WT control cells, had attenuated proliferation in response to PDGF-BB. In contrast, there was no defect in the ability of CD248−/− mouse cells to proliferate in response to TGFβ stimulation, indicating that CD248−/− cells do not have an intrinsic proliferation defect, but rather may undergo a specific failure of PDGF signal transduction.
To establish whether the interaction of CD248 and PDGF signaling plays a role in osteogenesis, we differentiated primary WT and CD248−/− mouse osteoblasts in the presence or absence of PDGF-BB in vitro (Figure 6E). CD248−/− mouse osteoblasts produced more bone nodules throughout the duration of the experiment. From day 21 onward, it was apparent that PDGF inhibited the production of bone nodules in WT mouse osteoblasts (P < 0.05 versus unsupplemented medium), whereas this effect was not evident in the CD248−/− mouse osteoblasts (P not significant versus unsupplemented medium). These results showed that the presence of CD248 is required to elicit PDGF-dependent proliferation of osteoblasts and regulation of bone mineralization.
DISCUSSION
During bone development, osteoblast progenitor cells that are involved in tissue remodeling and repair (preosteoblasts) are mobilized to the site of bone formation (20,21). In preosteoblasts, PDGF signaling via ERK acts as a break on cell maturation, maintaining the cells in a proliferative and migratory state (22,23). Other investigators have shown that overexpression of c-fos (the major product of PDGF signaling) in osteoblasts causes accelerated cell cycle progression, further implicating PDGF signaling in the control of osteoblast activity (24). Once precursor cells have differentiated into mature osteoblasts, they become nonproliferative and produce type I collagen (Col1a1) (21,25). Imatinib mesylate, a potent inhibitor of PDGFRβ signaling, has been found to promote osteoblast differentiation (26) and to suppress proliferation of osteoblast precursors (27) in vitro. In a mouse model in which PDGFRβ was deleted (in mesenchymal stromal cells, using an inducible Cre-Lox system), accelerated osteoblast maturation and an increase in woven bone were observed during fracture repair (28).
Recently, Tomkowicz et al (17) identified a mechanism that may explain the multiple functions of CD248. Their findings showed that CD248 is required for the phosphorylation of ERK and subsequent c-fos expression following PDGF stimulation in murine pericytes. Given that the signaling defect is downstream of PDGFR, but upstream of ERK, it has been proposed that CD248 directly interacts with phosphatidylinositol 3-kinase (PI3K), or an unknown cofactor, to regulate cell proliferation and migration. These observations support our findings that CD248 acts within the PDGF signaling cascade to regulate osteoblast function. The CD248−/− mouse has increased bone mass and its osteoblasts have greater mineralization capacity in vitro. We propose that this phenotype is caused by a defect in PDGF signal transduction that allows osteoblasts to mature more rapidly, resulting in increased bone formation, but with no change in bone composition.
It has been previously suggested that targeting the PDGF signaling pathway could be a useful approach in tissue regeneration and repair (28). PDGF and PDGFR are widely expressed, and their signaling affects functions in many cell types. In contrast, CD248 has more restricted expression, and is seen in adult human subjects predominantly at sites of inflammation, fibrosis, remodeling, or metastasis. This cell-specific and temporal regulation of CD248 expression in mesenchymal cells, in comparison to other members of the PDGF/PI3K cascade, and its presence at the cell surface make CD248 amenable to antibody or small molecule–mediated blockade. Our data suggest that targeting CD248 could enable a more discriminative approach to targeting the PDGF signaling pathway in stromal cells in general, and osteoblasts in particular.
Maia et al (6) have recently shown that antibody-induced arthritis is ameliorated in CD248-deficient mice. They suggested that CD248 blockade might therefore be therapeutically beneficial in rheumatoid arthritis. Given their findings and the results described herein, inhibition of CD248 function may be an ideal strategy with which to suppress inflammatory arthritis and reverse inflammation-induced osteoporosis.
ACKNOWLEDGMENTS
We are grateful to Gillian C. Milne and Kevin S. MacKenzie at the Histology and Microscopy Facility, Musculoskeletal Programme, University of Aberdeen for assistance and expertise in histologic processing and micro-CT analysis.
Supported by a grant from Arthritis Research UK and funding from the European Union (AutoCure Consortium). Dr. Naylor’s work was supported by a Foundation Fellowship award from Arthritis Research UK.
Footnotes
Dr. Duffield has received consulting fees, speaking fees, and/or honoraria from Amira Pharmaceuticals, Gilead, Takeda, Boehringer Ingelheim, Regulus Therapeutics, and Promedior (less than $10,000 each) and owns stock or stock options in Promedior.
REFERENCES
- 1.Tomkowicz B, Rybinski K, Foley B, Ebel W, Kline B, Routhier E, et al. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc Natl Acad Sci U S A. 2007;104:17965–70. doi: 10.1073/pnas.0705647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lax S, Hardie D, Wilson A, Douglas M, Anderson G, Huso D, et al. The pericyte and stromal marker CD248 (endosialin) is required for efficient lymph node expansion. Eur J Immunol. 2010;40:1884–9. doi: 10.1002/eji.200939877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S, et al. Endosialin/TEM1/CD248 is a pericyte marker of embryonic and tumor neovascularisation. Microvasc Res. 2008;76:180–8. doi: 10.1016/j.mvr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Smith SW, Eardley KS, Croft A, Nwosu J, Howie AJ, Cockwell P, et al. CD248+ stromal cells are associated with progressive chronic kidney disease. Kidney Int. 2011;80:199–207. doi: 10.1038/ki.2011.103. [DOI] [PubMed] [Google Scholar]
- 5.MacFadyen J, Savage K, Wienke D, Isacke CM. Endosialin is expressed on stromal fibroblasts and CNS pericytes in mouse embryos and is downregulated during development. Gene Expr Patterns. 2007;7:363–9. doi: 10.1016/j.modgep.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Maia M, de Vriese A, Janssens T, Moons M, van Landuyt K, Tavernier J, et al. CD248 and its cytoplasmic domain: a therapeutic target for arthritis. Arthritis Rheum. 2010;62:3595–606. doi: 10.1002/art.27701. [DOI] [PubMed] [Google Scholar]
- 7.Bagley RG, Weber W, Rouleau C, Yao M, Honma N, Kataoka S, et al. Human mesenchymal stem cells from bone marrow express tumor endothelial and stromal markers. Int J Oncol. 2009;34:619–27. doi: 10.3892/ijo_00000187. [DOI] [PubMed] [Google Scholar]
- 8.Nanda A, Karim B, Peng Z, Liu G, Qiu W, Gan C, et al. Tumor endothelial marker 1 (Tem1) functions in the growth and progression of abdominal tumors. Proc Natl Acad Sci U S A. 2006;103:3351–6. doi: 10.1073/pnas.0511306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marty C, Langer-Machova Z, Sigrist S, Schott H, Schwendener RA, Ballmer-Hofer K. Isolation and characterization of a scFv antibody specific for tumor endothelial marker 1 (TEM1), a new reagent for targeted tumor therapy. Cancer Lett. 2006;235:298–308. doi: 10.1016/j.canlet.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Su AL, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krempen K, Grotkopp D, Hall K, Bache A, Gillian A, Rippe RA, et al. Far upstream regulatory elements enhance position-independent and uterus-specific expression of the murine α1(I) collagen promoter in transgenic mice. Gene Exp. 1999;8:151–63. [PMC free article] [PubMed] [Google Scholar]
- 12.Yata Y, Scanga A, Gillian A, Yang L, Reif S, Breindl M, et al. DNase 1-hypersensitive sites enhance α1(I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37:267–76. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 13.Huesa C, Yadav MC, Finnila MA, Goodyear SR, Robins SP, Tanner KE, et al. PHOSPHO1 is essential for mechanically competent mineralization and the avoidance of spontaneous fractures. Bone. 2011;48:1066–74. doi: 10.1016/j.bone.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somerville JM, Aspden RM, Armour KE, Armour KJ, Reid DM. Growth of C57Bl/6 mice and the material and mechanical properties of cortical bone from the tibia. Calcif Tissue Int. 2004;74:469–75. doi: 10.1007/s00223-003-0101-x. [DOI] [PubMed] [Google Scholar]
- 15.MacFadyen JR, Haworth O, Roberston D, Hardie D, Webster MT, Morris HR, et al. Endosialin (TEM1, CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett. 2005;579:2569–75. doi: 10.1016/j.febslet.2005.03.071. [DOI] [PubMed] [Google Scholar]
- 16.Beamer WG, Donahue LR, Rosen CJ, Baylink DJ. Genetic variability in adult bone density among inbred strains of mice. Bone. 1996;18:397–403. doi: 10.1016/8756-3282(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 17.Tomkowicz B, Rybinski K, Sebeck D, Sass P, Nicolaides NC, Grasso L, et al. Endosialin/TEM-1/CD248 regulates pericyte proliferation through PDGF receptor signaling. Cancer Biol Ther. 2010;9:908–15. doi: 10.4161/cbt.9.11.11731. [DOI] [PubMed] [Google Scholar]
- 18.Kose KN, Xie JF, Carnes DL, Graves DT. Pro-inflammatory cytokines downregulate platelet derived growth factor-α receptor gene expression in human osteoblastic cells. J Cell Physiol. 1996;166:188–97. doi: 10.1002/(SICI)1097-4652(199601)166:1<188::AID-JCP20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Horner A, Bord S, Kemp P, Grainger D, Compston JE. Distribution of platelet-derived growth factor (PDGF) A chain mRNA, protein, and PDGF-α receptor in rapidly forming human bone. Bone. 1996;19:353–62. doi: 10.1016/s8756-3282(96)00217-7. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–31. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 21.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–44. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Fernandez MA, Gallois A, Riedl T, Jurdic P, Hoflack B. Osteoclasts control osteoblast chemotaxis via PDGF-BB/PDGF receptor β signaling. PLOS One. 2008;3:e3537. doi: 10.1371/journal.pone.0003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber R, Karreth F, Kandler B, Fuerst G, Rot A, Fischer MB, et al. Platelet-released supernatants increase migration and proliferation, and decrease osteogenic differentiation of bone marrow-derived mesenchymal progenitor cells under in vitro conditions. Platelets. 2004;15:29–35. doi: 10.1080/09537100310001643999. [DOI] [PubMed] [Google Scholar]
- 24.Sunters A, Thomas DP, Yeudall WA, Grigoriadis AE. Accelerated cell cycle progression in osteoblasts overexpressing the c-fos protooncogene. J Biol Chem. 2004;279:9882–91. doi: 10.1074/jbc.M310184200. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Malaval L, Aubin JE. The mature osteoblast phenotype is characterized by extensive plasticity. Exp Cell Res. 1997;232:97–105. doi: 10.1006/excr.1997.3501. [DOI] [PubMed] [Google Scholar]
- 26.O’Sullivan S, Noat D, Callon K, Porteous F, Horne A, Wattie D, et al. Imatinib promotes osteoblast differentiation by inhibiting PDGFR signaling and inhibits osteoclastogenesis by both direct and stromal cell-dependent mechanisms. J Bone Miner Res. 2007;22:1679–89. doi: 10.1359/jbmr.070719. [DOI] [PubMed] [Google Scholar]
- 27.Fierro F, Illmer T, Jing D, Schleyer E, Ehninger G, Boxberger S, et al. Inhibition of platelet-derived growth factor receptor β by imatinib mesylate suppresses proliferation and alters differentiation of human mesenchymal stem cells in vitro. Cell Prolif. 2007;40:355–66. doi: 10.1111/j.1365-2184.2007.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokunaga A, Oya T, Ishii Y, Motomura H, Nakamura C, Ishizawa S, et al. PDGF receptor β is a potent regulator of mesenchymal stromal cell function. J Bone Miner Res. 2008;23:1519–27. doi: 10.1359/jbmr.080409. [DOI] [PubMed] [Google Scholar]