Abstract
Background
The differentiation of TH17 cells, which promote pulmonary inflammation, requires the cooperation of a network of transcription factors.
Objectives
We sought to define the role of Etv5, an Ets-family transcription factor, in TH17 cell development and function.
Methods
TH17 development was examined in primary mouse T cells wherein Etv5 expression was altered by retroviral transduction, small interfering RNA targeting a specific gene, and mice with a conditional deletion of Etv5 in T cells. The direct function of Etv5 on the Il17 locus was tested with chromatin immunoprecipitation and reporter assays. The house dust mite–induced allergic inflammation model was used to test the requirement for Etv5-dependent TH17 functions in vivo.
Results
We identify Etv5 as a signal transducer and activator of transcription 3–induced positive regulator of TH17 development. Etv5 controls TH17 differentiation by directly promoting 0a and Il17f expression. Etv5 recruits histone-modifying enzymes to the Il17a–Il17f locus, resulting in increased active histone marks and decreased repressive histone marks. In a model of allergic airway inflammation, mice with Etv5-deficient T cells have reduced airway inflammation and IL-17A/F production in the lung and bronchoalveolar lavage fluid compared with wild-type mice, without changes in TH2 cytokine production.
Conclusions
These data define signal transducer and activator of transcription 3–dependent feed-forward control of TH17 cytokine production and a novel role for Etv5 in promoting T cell–dependent airway inflammation.
Keywords: TH17 cells, transcription factor, Etv5, epigenetic modifications, allergic inflammation
Helper T cells play an essential role in adaptive immunity, with specific subsets contributing effector functions to control immunity to pathogens and regulating the development of inflammatory disease. Specific cytokines in the microenvironment mediate intracellular communication that results in the activation of signal transducer and activator of transcription (STAT) proteins and the induction of a transcriptional regulatory network. IL-6, IL-21, or IL-23 can activate STAT3, which is required for TH17 cell differentiation and maintenance.1,2 TH17 cells secrete IL-17A, IL-17F, IL-21, and IL-22 and are primary mediators in controlling infection and promoting autoimmune diseases.3’4 ΓL-17–producing CD4+ T cells have been shown to be required for immunity to Candida albicans, Mycobacterium tuberculosis, Streptococcus pneumonia, and Klebsiella pneumoniae.5–8 TH17 cells also play a key role in the development of autoimmune inflammation, including experimental autoimmune encephalomyelitis, rheumatoid arthritis, and inflammatory bowel disease.9–11 STAT3 activation by IL-6, IL-21, and IL-23 results in the induction of genes encoding transcription factors, including Rorc, Maf, Irf4, and Batf, which are essential for TH17 cell differentiation.12–16 Although each of these factors are critical for TH17 cell development, 17,18 it is not clear whether additional STAT3 targets are required for TH17 cell development and function.
ETS variant 5 (Etv5) belongs to the PEA3 subfamily of ETS transcription factors that regulate gene expression by binding to a conserved GGAA/T motif.19 Etv5 has been shown to play important roles in coordinating limb development,20,21 controlling gene expression in spermatogonial stem cells22, and regulating epithelial-mesenchymal transition in many type of cancers.19 Etv5 inhibits sonic hedgehog expression in the anterior limb bud, which is essential for the anterior-posterior patterning of the vertebrate limb21. Etv5 positively regulates microRNA-21, Bcl6b, and LIM homeobox 1, which are known to control spermatogonial stem cell self-renewal.23 Chromosomal translocations resulting from the fusion between Etv5 and trans-membrane protease serine 2 correlate with prostate cancer.19
Although the role of Etv5 in developmental and cancer biology has been defined, the function of Etv5 in the immune response is still poorly understood. One report found Etv5 downstream of the IL-12–STAT4 signaling pathway, augmenting IFN-γ production in TH1 cells.24,25 The role of Etv5 in the development of other helper T cells has not been defined. In this report we show that STAT3-activating cytokines induce Etv5 expression in TH17 cells. Etv5 promotes IL-17 production in TH17 cells in vitro and in vivo and is required in T cells for the development of allergic inflammation. Mechanistically, Etv5 directly promotes Il17a and Il17f expression. Thus Etv5 is a STAT3-induced positive regulator of TH17 cell differentiation, promoting the development of allergen-induced airway inflammation.
METHODS
Mice
C57BL/6 mice were purchased from Harlan Sprague Dawley (Indianapolis, Ind). Stat3fl/fl CD4-Cre mice were previously described.26 Etv5fl/fl mice21 were crossed withCD4-Cre transgenic mice to generate Etv5 fl/fl CD4-Cre+ mice, with Cre-negative littermates as control mice. Mice were maintained under specific pathogen-free conditions. All experiments were performed with the approval of the Indiana University Institutional Animal Care and Use Committee.
In vitro T-cell differentiation
Naive CD4+CD62L+T cells were positively selected from enriched CD4+ T cells from spleens and lymph nodes by using MACS beads and columns (Miltenyi Biotec, Auburn, Calif). Naive CD4+CD62L+T cells were activated with plate-bound anti-CD3 (2 µg/mL 145-2C11; Bio-XCell, West Lebanon, NH) and soluble anti-CD28 (0.5 µg/mL; BD PharMingen, San Jose, Calif) to generate TH0 cells or with additional cytokines (all from PeproTech, Rocky Hill, NJ) and antibodies (Bio-XCell) to generate TH1 (5 ng/mL IL-12 and 10 µg/mL anti–IL-4, 11B11), TH2 (10 ng/mL IL-4 and 10 µg/mL anti–IFN-γ, XMG), TH9 (20 ng/mL IL-4, 2 ng/mL TGF-β, and 10 µg/mL anti–IFN-γ; XMG), TH17 (100 ng/mL IL-6, 10 ng/mL IL-23, 10 ng/mL IL-1β, 2 ng/mL TGF-β, and 10 µg/mL anti–IL-4, 11B11, and 10 µg/mL anti–IFN-γ, XMG), or inducible regulatory T (iTreg; 2 ng/mL TGF-β and 10 µg/mL anti–IL-4, 11B11) cells in culture conditions. Cells were expanded after 3 days with a half concentration of the original cytokines in fresh medium. Cells were harvested on day 5 for analysis.
House dust mice–induced allergic airway inflammation
Wild-type and Etv5 mutant mice were sensitized by means of intranasal injection of 40 µg of house dust mite (HDM; Greer Laboratories, Lenoir, NC) in PBS each day for 3 consecutive days over 5 weeks. In experiments as indicated, mice were treated with 200 µg of anti–IL-17A (17F3) or IgG1 (MOPC-21) antibodies (Bio-XCell) for 3 consecutive days over 5 weeks or 4 µg of IL-17A/F cytokine (PeproTech) on weeks 3, 4, and 5. Mice were killed 24 hours after the final intranasal challenge at week 5. The trachea was cannulated, and lungs were lavaged 3 times with 1 mL of PBS to collect bronchoalveolar lavage (BAL) cells. BAL cells and the single-cell suspension from lungs generated by using the lung dissociation kit from Miltenyi Biotec were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 2 hours followed by monensin for a total of 6 hours for cytokine analysis by means of intracellular staining. Single-cell suspension from the lungs was used for gene expression analysis with quantitative reverse transcriptase PCR (qRT-PCR). Cells from mediastinal lymph nodes were stimulated with HDM for 5 days, and cytokine production was analyzed by using ELISA. Lung tissue was analyzed after paraffin embedding and staining with hematoxylin and eosin for evaluation of the infiltration of inflammatory cells, and periodic acid–Schiff was used for evaluation of mucus production. Eosinophils, neutrophils, T cells, B cells, and mononuclear cells in the BAL fluid and lungs were characterized by cell size and expression of CD3, B220, CCR3, CD11c, and MHC class II by using flow cytometric analysis, as described previously.27
Retroviral expression vectors and retroviral transduction
Etv5 (Open Biosystems, Thermo Scientific, Waltham, Mass) cDNA was digested and subcloned into MSCV-YFP. Bicistronic retrovirus expressing YFP preparation of retroviral stocks was previously described.28 CD4+ T cells were transduced on day 2 with control or retrovirus vector expressing the gene of interest by using centrifugation at 2000 rpm at 25°C for 1 hour in the presence of 8 µg/mL polybrene (Sigma-Aldrich, St Louis, Mo). Viral supernatant was replaced with the former culture supernatant supplemented with 50 U/mL human IL-2. After spin infection, cells were expanded on day 3 and analyzed on day 5.
Transfection of small interfering RNA
Control small interfering RNA (siRNA) and siRNA targeting Etv5 were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif). TH17 cells were transfected with control or Etv5 siRNA on day 2 with the Amaxa Nucleofector kit (Lonza, Basel, Switzerland), rested overnight with human IL-2, and restimulated with anti-CD3 for 24 hours for gene expression and cytokine production analysis.
Luciferase reporter assay
Jurkat T cells were grown in RPMI-1640 with 10% FBS and transfected with 2 to 5 µg of the IL17A/F luciferase reporter plasmids (SwitchGear Genomics, Menlo Park, Calif) and control or increasing concentration of plasmid expressing Etv5 through Fugene6 reagent (Roche, Mannheim, Germany). After 24 hours, transfected cells were stimulated with PMA and ionomycin for 6 hours before analysis with the dual luciferase system (Promega, Madison, Wis).
Analysis of gene expression, protein expression, ELISA, and flow cytometry
qRT-PCR and ELISA were performed, as previously described.29 Gene expression was normalized to housekeeping gene (β2-microglobulin) expression. The relative gene expression was calculated by using the change-in-threshold (−ΔΔCT) method. Primers are listed in Table E2 in this article’s Online Repository at www.jacionline.org. Nuclear lysates were extracted from activated CD4+ T cells and immunoblotted with Etv5 C-20 or β-Actin C4 (Santa Cruz Biotechnology) as a control. For flow cytometry, cells were stained for surface markers and fixed with 2% paraformaldehyde for 10 minutes before analysis. For cytokine staining, CD4+ T cells were stimulated with PMA and ionomycin for 2 hours followed by monesin for a total of 6 hours, fixed, permeabilized with 0.2% saponin, and stained for IL-17A–PECy7, IL-17F–phycoerythrin, or IL-13– Alexa Fluor 647 (BD PharMingen). For Etv5 intracellular staining, cells were fixed, permeabilized with 0.1% saponin, and stained for Etv5 C-20 or goat IgG (Santa Cruz Biotechnology) for 30 minutes at 4°C, followed by donkey anti-goat IgG PE (Santa Cruz Biotechnology) for an additional 30 minutes at 4°C.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay was performed, as previously described.24 In brief, resting TH17 cells were cross-linked for 10 minutes with 1% formaldehyde and lysed by means of sonication. After preclearing with salmon sperm DNA, BSA, and Protein Agarose bead slurry (50%), cell extracts were incubated with either rabbit polyclonal STAT3 C-20, Etv5 H-100, p300 N-15 (Santa Cruz Biotechnology), H3K27ac, H3K27me3 (Milli-pore, Temecula, Calif), H3K4me3 (Abcam, Cambridge, United Kingdom), or normal rabbit IgG (Millipore) overnight at 4°C. The immunocomplexes were precipitated with protein Agarose beads at 4°C for 2 hours, washed, and eluted, and cross-links were reversed at 65°C overnight. DNA was purified, resuspended in H2O, and analyzed by using quantitative PCR. Primers are listed in Table E1 in this article’s Online Repository at www.jacionline.org. The percentage input was calculated by subtracting the amount of immunoprecipitated DNA from the IgG control from the amount of immunoprecipitated DNA from the specific antibody and normalized against the amount of input DNA.
Statistical analysis
A 2-tailed Student t test or 1-way ANOVA was used to generate P values for all data, with a P value of .05 or less considered significant.
RESULTS
Etv5-deficient T cells display defects in helper T-cell differentiation
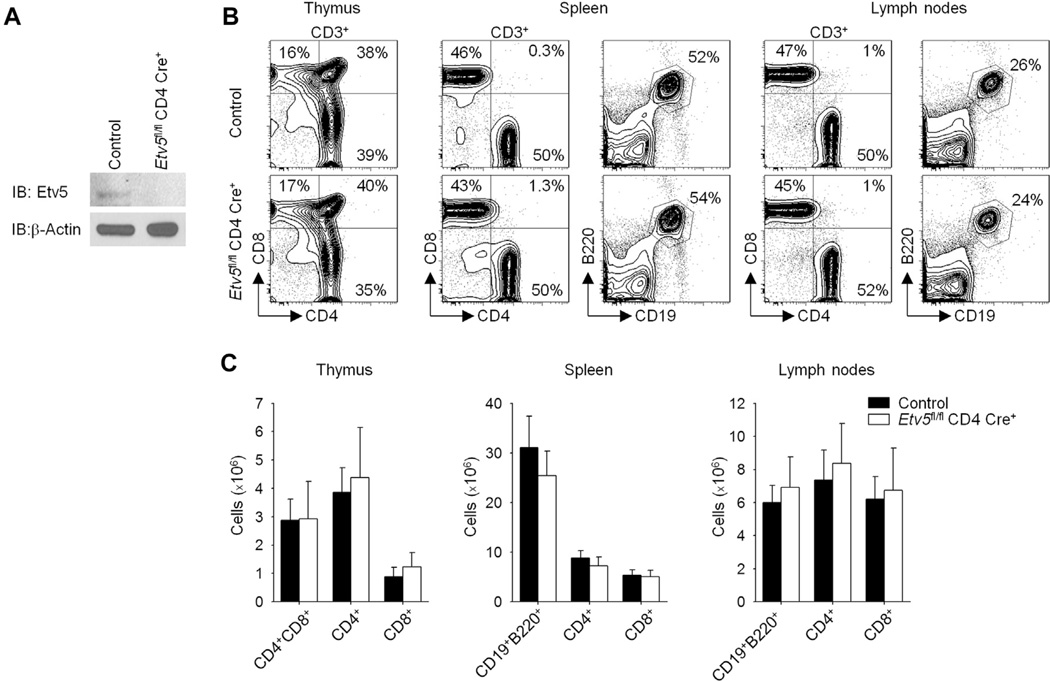
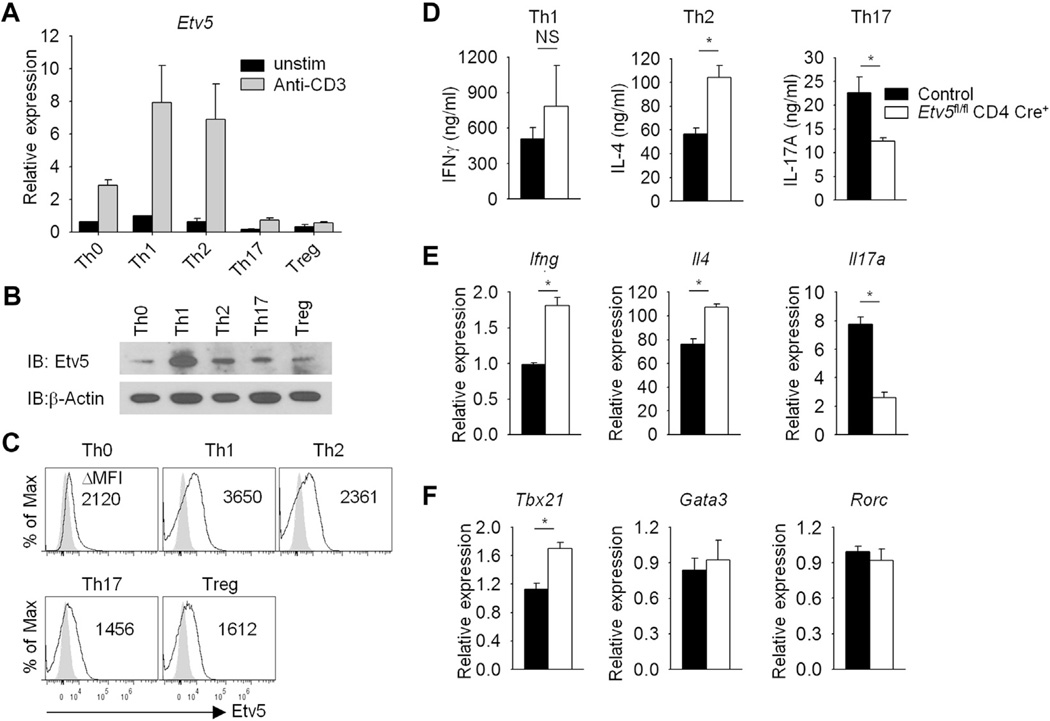
Although evidence suggests that Etv5 contributes modestly to IFN-γ production in TH1 cells,25 the role of Etv5 in helper T-cell development is still poorly understood. To define the function of Etv5 in T cells, we generated mice with Etv5 deficiency in T cells (Etv5fl/fl CD4-Cre+) by mating Etv5fl/fl mice21 with mice carrying a CD4-Cre transgene (termed Etv5 mutant mice) and confirmed the absence of Etv5 expression in Etv5-deficient T cells (Fig 1, A). Mice with Etv5-deficient T cells display T-cell populations in the thymus, spleen, and lymph nodes comparable with those seen in wild-type littermate control animals and showed normal proliferation in culture (Fig 1, B and C, and data not shown). To define the role of Etv5 in helper T-cell differentiation, we first examined Etv5 expression in helper T-cell subsets. Etv5 expression was highest in resting and activated TH1 and TH2 cells compared with other helper cell subsets at the message and protein levels (Fig 2, A-C). Naive CD4+ T cells from control and Etv5 mutant mice were used to generate TH1, TH2, and TH17 cultures, and cytokine production and gene expression were assessed. Interestingly, there were multiple defects in helper T-cell differentiation. Etv5-deficient TH2 cells produced more IL-4. In contrast, Etv5-deficient TH17 cells produced significantly less IL-17A compared with control cells (Fig 2, D). Similar results were observed at the message level in Etv5-deficient T cells compared with control cells (Fig 2, E). IFN-γ production was modestly increased in Etv5-deficient TH1 cells compared with that seen in wild-type cells (Fig 2, D and E). Examination of transcription factor expression critical for the development of each effector subset demonstrated that expression of Gata3 and Rorc was similar between control and Etv5-deficient TH2 and TH17 cells, respectively (Fig 2, F). Etv5-deficient TH1 cells had higher Tbx21 expression compared with that seen in control cells (Fig 2, F). Numbers of thymic-derived regulatory T (Treg) cells (defined as CD4+CD25+ forkhead box protein 3 [Foxp3]+ cells) in both lymph nodes and spleen and in vitro–derived Treg cells (iTreg cells) were not significantly decreased in Etv5-mutant mice compared with control mice (data not shown). Thus mice that lack Etv5 expression in T cells display altered helper T-cell differentiation in vitro.
FIG 1.
Characterization of mice with Efv5-deficient T cells. A, Naive CD4+CD62+ T cells from control and Etv5fl/fl CD4-Cre+ mice were activated with anti-CD3 and anti-CD28 for 5 days. Nuclear lysates were extracted and immunoblotted for Etv5 and β-actin as a control. B and C, Total cells were isolated from the thymus, spleen, and lymph nodes of control and Etv5 mutant mice and stained for cell-surface markers, with the percentage of positive cells (Fig 1, B) and cell numbers (Fig 1, C) shown. Data are representative of 2 independent experiments with similar results (Fig 1, A and B) or are means ± SEMs of 3 mice per group and representative of 2 independent experiments with similar results (Fig 1, C).
FIG 2.
Helper T-cell differentiation in the absence of Etv5 in T cells. Naive control and Etv5-deficient CD4+CD62L+ T cells were activated (TH0) or cultured under TH1, TH2, TH17, and Treg cell polarizing conditions. Etv5 expression was measured in helper T-cell subsets by using qRT-PCR before and after 6 hours of anti-CD3 stimulation (A), immunoblotting (B), or intracellular staining (C) before anti-CD3 stimulation. Δ Mean fluorescence intensity was calculated by subtracting the background from the signal of Etv5 antibody. TH1, TH2, and TH17 cells were used for assessing cytokine production by means of ELISA after 24 hours of anti-CD3 stimulation (D) and gene expression analysis after (Ifng, Il4, and Il17a; E) or before (Tbx21, Gata3, and Rorc; F) 6 hours of anti-CD3 stimulation by means of qRT-PCR. Data are means ± SEMs of 4 independent experiments (Fig 2, D-F) or means ± SDs of replicate samples (Fig 2, A) and representative of 3 independent experiments with similar results (Fig 2, A-C). *P < .05. NS, Not significant.
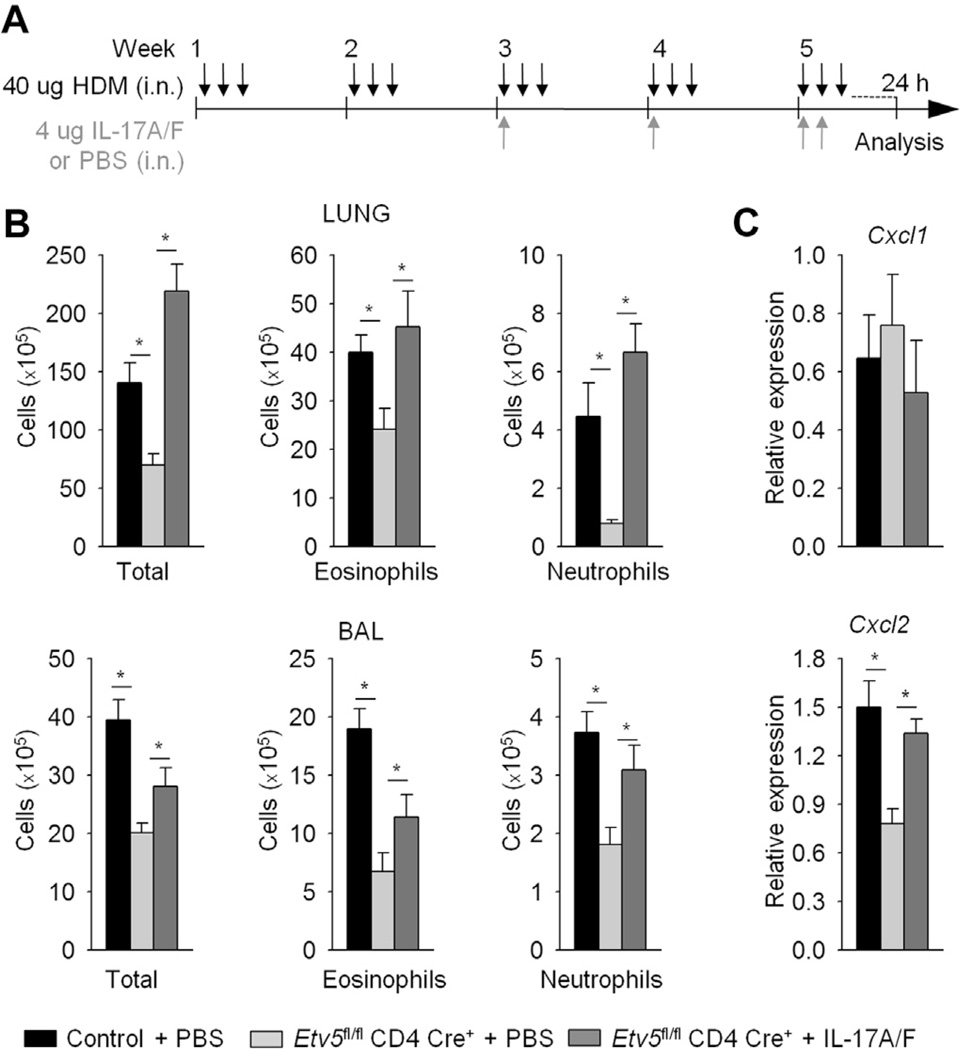
Mice with Etv5-deficient T cells had reduced allergic inflammation
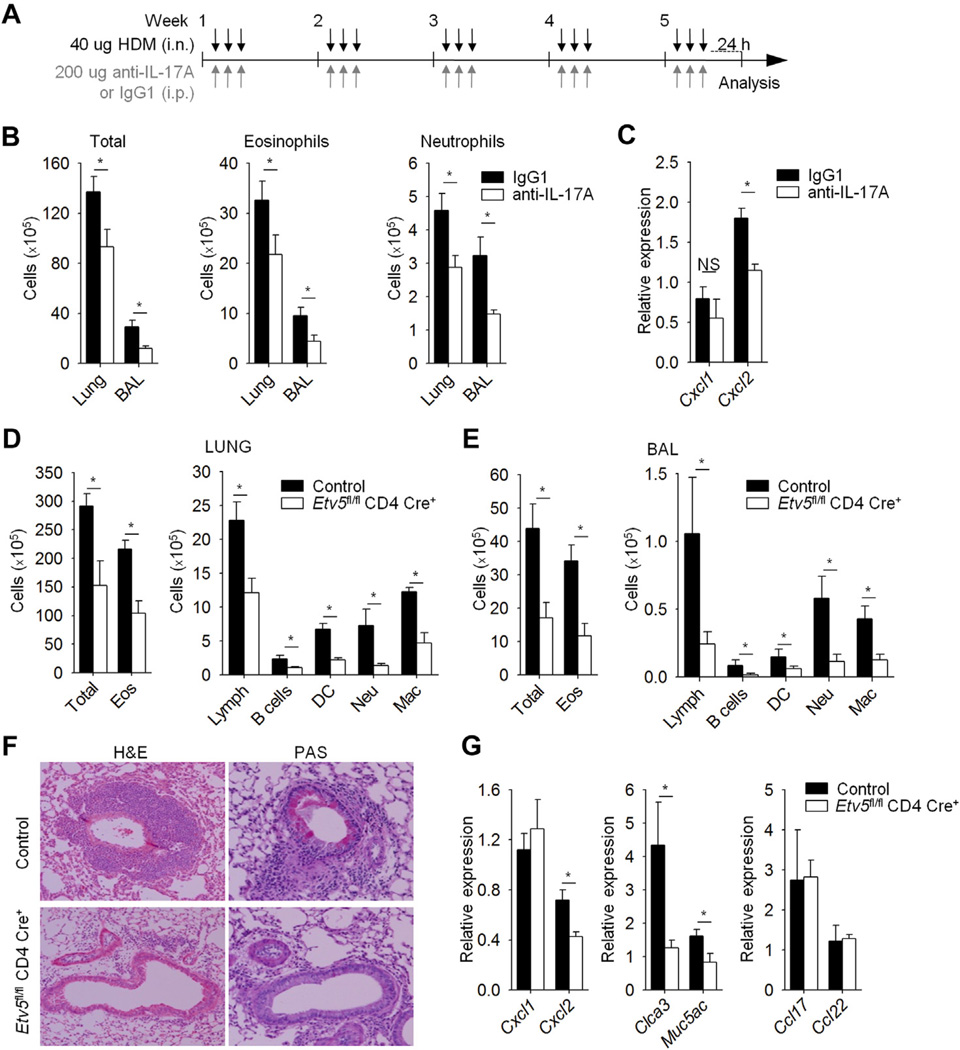
Our data suggested that Etv5 plays a repressive role in TH1 and TH2 cell differentiation while promoting TH17 cell differentiation (Fig 2). HDM-induced allergic airway inflammation is TH2 and TH17 dependent,30–33 and in exploring Etv5-dependent immunity, we first wanted to confirm the role of IL-17A in this model. We sensitized and challenged mice with intranasal HDM antigen and anti–IL-17A or IgG1 antibodies for 5 weeks and assessed pulmonary inflammation (Fig 3, A). Mice treated with anti–IL-17A antibody had significantly lower numbers of total cells, eosinophils, and neutrophils in lung tissue and BAL fluid compared with control mice (Fig 3, B). Consistent with the decrease in neutrophil numbers, expression of Cxcl2, an IL-17– induced gene, but not Cxcl1, was reduced in lung tissue from mice treated with anti–IL-17A antibody compared with control lung tissue (Fig 3, C). Our results demonstrated that IL-17A is important in mediating allergic airway inflammation on HDM allergen exposure.
FIG 3.
Etv5 mutant mice have reduced HDM-induced allergic airway inflammation. A, Wild-type mice were immunized (intranasally [i.n.]) with HDM for 5 weeks to induced allergic inflammation and treated with anti–IL-17A or IgG1 antibodies (intraperitoneally [i.p.]). B, Total and inflammatory cell counts in the lungs and BAL fluid of HDM-induced airway inflammation in wild-type mice. C, Total cells from lungs of wild-type mice were used for gene expression analysis by means of qRT-PCR. D-G, Control and Etv5 mutant mice were sensitized and challenged (intranasally) with HDM for 5 weeks to induce allergic inflammation. Inflammatory cells in the lung tissue and BAL fluid of control and Etv5 mutant mice were as follows: DC, dendritic cells; Eos, eosinophils; Lymph, lymphocytes; Mac, macrophages; Neu, neutrophils. Fig 3, F, Cell infiltration in the lungs and mucus in the airways of control and Etv5 mutant mice were evaluated by means of hematoxylin and eosin (H&E) and periodic acid–Schiff (PAS) staining. Fig 3, G, Total cells from lungs of control and Etv5 mutant mice were used for gene expression analysis by means of qRT-PCR. Data are means ± SEMs of 5 to 6 mice per group (Fig 3, A-G) and representative of 2 independent experiments with similar results. *P < .05. NS, Not significant.
We next wanted to test the role of Etv5 in vivo by inducing allergic airway inflammation in control and Etv5 mutant mice. Etv5 mutant mice had significantly decreased numbers of total cells and specific cell populations in lung tissue compared with control mice (Fig 3, D). Analysis of the cellular composition after BAL showed similar results (Fig 3, E). Histologic examination also demonstrated diminished inflammatory cell infiltrates in the lungs and decreased mucus production in the airways of Etv5 mutant mice compared with control mice (Fig 3, F). Consistent with histologic analysis, the expression of genes involved in goblet cell metaplasia (Muc5ac and Clca3) was reduced in lungs of Etv5 mutant mice compared with that seen in control mice (Fig 3, G). Given that we observed the greatest fold reduction (>5-fold) in infiltrating neutrophils in Etv5 mutant mice compared with control mice (Fig 3, D and E), we wanted to examine the expression of neutrophil-attractant chemokines in total lung RNA. Consistent with the decrease in neutrophil numbers, expression of Cxcl2, an IL-17–induced gene, was reduced in lung tissue from Etv5 mutant mice compared with that seen in control lung tissue (Fig 3, G). In contrast, there was no difference in Cxcl1 expression or in expression of the TH2-induced chemokines Ccl17 and Ccl22 (Fig 3, G).
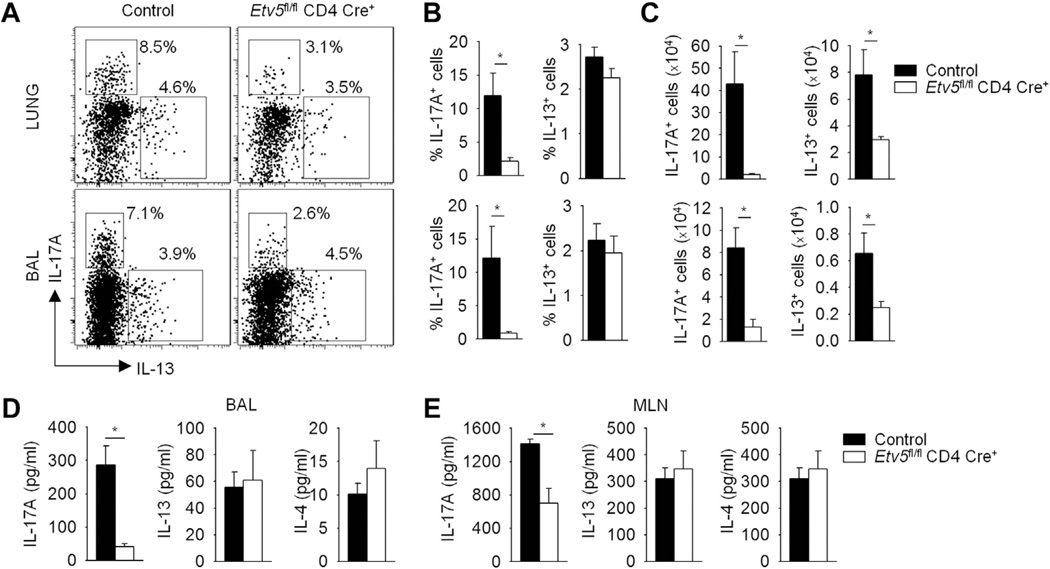
We next assessed cytokine production in the lung tissue and BAL fluid of control and Etv5 mutant mice with HDM-induced allergic inflammation. Consistent with in vitro analyses, there was a significantly decreased frequency and number of IL-17A–producing CD4+ T cells from the lung and BAL cells of Etv5 mutant mice compared with that seen in control mice (Fig 4, A-C, and data not shown). No change was observed in the frequency of IL-13–producing CD4+ T cells from the lung and BAL cells, although cell numbers were decreased in Etv5 mutant mice compared with those seen in control mice, which is concomitant with the decrease in overall inflammation in the lung (Fig 4, A-C). We further confirmed that the concentration of IL-17A, but not IL-13 and IL-4, was significantly reduced in the BAL fluid of Etv5 mutant mice compared with that seen in control mice (Fig 4, D). The frequencies of Treg cells (identified as CD4+ Foxp3+ ) in the lung and BAL cells were similar between control and Etv5 mutant mice (data not shown). We next assessed the generation of helper T-cell responses in the periphery by stimulating mediastinal lymph node cells with HDM extract. IL-17A production was significantly reduced in Etv5 mutant mice compared with that seen in control mice, although there was no difference in the amount of IL-13 or IL-4 (Fig 4, E).
FIG 4.
Etv5 regulates TH17 cells in HDM-induced allergic airway inflammation. A-C, Lung and BAL cells from HDM-induced allergic airway inflammation in control and Etv5 mutant mice (from Fig 3) were stimulated with PMA and ionomycin for 6 hours to assess cytokine production (Fig 4, A-C) by using intracellular staining, with the average percentage of positive cells (Fig 4, B) and cell numbers (Fig 4, C) shown. D and E, Cells from mediastinal lymph nodes (MLN) were stimulated with HDM for 5 days. BAL fluid (Fig 4, D) and cell-free supernatant (Fig 4, E) were used to assess cytokine production by using ELISA. Data are means ± SEMs of 6 mice per group and representative of 2 independent experiments with similar results. *P < .05.
To determine whether loss of IL-17 is the critical defect in Etv5 mutant mice, we treated HDM-sensitized Etv5 mutant mice with either PBS or IL-17A and IL-17F together and assessed pulmonary inflammation (Fig 5, A). IL-17A/F–treated Etv5 mutant mice had increased numbers total cell, eosinophil, and neutrophil numbers in lung tissue and BAL fluid compared with those seen in the PBS group (Fig 5, B). In addition, Cxcl2, but not Cxcl1, gene expression was increased when Etv5 mutant mice were treated with IL-17A/F cytokines compared with values seen in control mice (Fig 5, C). Collectively, these results suggested that the critical role of Etv5 in vivo in response to an allergen in experimental airway inflammation is the regulation of IL-17–secreting T cells.
FIG 5.
IL-17 cytokine contributes to HDM-induced allergic airway inflammation. Control and Etv5 mutant mice were immunized (intranasally [i.n.]) with HDM for 5 weeks to induce allergic inflammation. A, Mice were treated with IL-17A/F cytokine or PBS on weeks 3, 4, and 5. B, Total and inflammatory cells (eosinophils and neutrophils) in the lung tissue and BAL fluid of control and Etv5 mutant mice. C, Total cells from lungs of control and Etv5 mutant mice were used for gene expression analysis by means of qRT-PCR. Data are means ± SEMs of 5 mice per group and representative of 2 independent experiments with similar results. *P < .05.
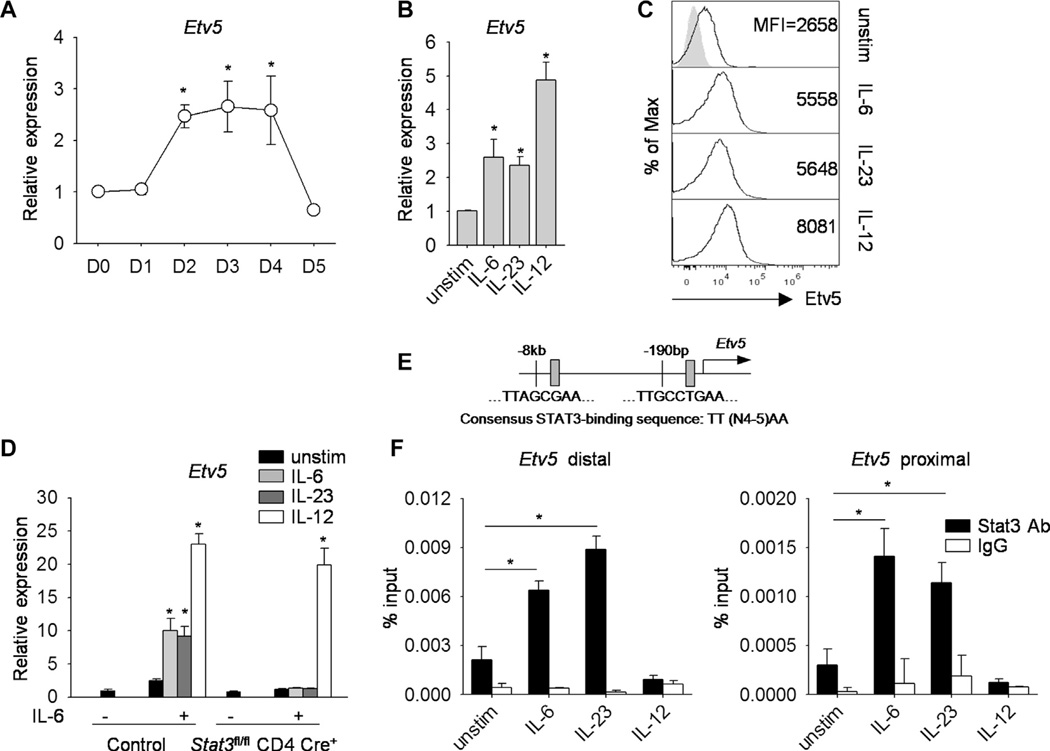
STAT3-activating cytokines induce Etv5 expression
Given that Etv5 positively regulates IL-17A in vitro and in HDM-induced allergic inflammation, we wanted to further elucidate the ability of Etv5 to promote TH17 cell differentiation. We first assessed the kinetics of Etv5 gene expression during TH17 cell differentiation. We observed an increase in Etv5 expression 48 hours after activation that gradually decreased over the subsequent 3 days of differentiation (Fig 6, A). Because STAT4 regulates Etv5 expression in TH1 cells,25 we tested whether STAT3 induced Etv5 expression in TH17 cultures. Stimulation of wild-type TH17 cells with IL-6 or IL-23 to activate STAT3 or IL-12 to activate STAT4 led to increased Etv5 mRNA and protein expression compared with that seen in unstimulated cells (Fig 6, B and C). To further confirm that Etv5 is a STAT3 target gene, we treated activated control and Stat3-deficient T cells with or without IL-6 for 48 hours to induce receptor expression. Cells were harvested, rested overnight, and restimulated with IL-6, IL-12, or IL-23 to assess Etv5 expression. As expected, IL-6, IL-23, or IL-12 induced Etv5 expression in IL-6–primed CD4+ T cells (Fig 6, D). However, IL-6– and IL-23–induced, but not IL-12–induced, expression of Etv5 was STAT3 dependent (Fig 6, D). To determine whether STAT3 could directly bind to conserved STAT-binding sites (consensus STAT3-binding sequence TTN4–5AA34) in the mouse and human Etv5 genes (Fig 6, E), differentiated wild-type TH17 cells were stimulated with STAT3-activating cytokines, and binding was examined using ChIP assay. In TH17 cells STAT3-activating cytokines, but not IL-12, resulted in STAT3 binding to the Etv5 promoter, with greater amounts at the distal than the proximal binding sites (Fig 6, F). The decreased STAT3 binding after IL-12 stimulation is likely due to displacement by STAT4 (Fig 6, F). These results suggested that STAT3-activating cytokines, including IL-6 and IL-23, promote Etv5 expression in TH17 cells.
FIG 6.
Efv5 is regulated by STAT3-activating cytokines in TH17 cells. Naive wild-type CD4+CD62L+ T cells were cultured under TH17-polarizing conditions. A, Kinetics of Etv5 gene expression during TH17 cell differentiation. B and C, TH17 cells were stimulated with IL-6, IL-23, and IL-12 for 2 hours before gene expression analysis by means of qRT-PCR (Fig 6, B) or protein expression by means of intracellular staining (Fig 6, C). D, Naive control and Sfaf3-deficient CD4+ T cells were activated with anti-CD3 and anti-CD28 in the presence or absence of IL-6 for 48 hours, rested overnight, and restimulated with IL-6, IL-12, or IL-23 for 2 hours before gene expression analysis by using qRT-PCR. E, Schematic of Etv5 promoter-containing STAT3-binding sites. F, Cells prepared as in Fig 6, B, were used for ChIP analysis with STAT3 antibody and IgG as a control. Data are means ± SDs of replicate samples and representative of 3 independent experiments with similar results (Fig 6, A-F). *P < .05.
Etv5 directly activates the Il17a-Il17f locus in TH17 cells
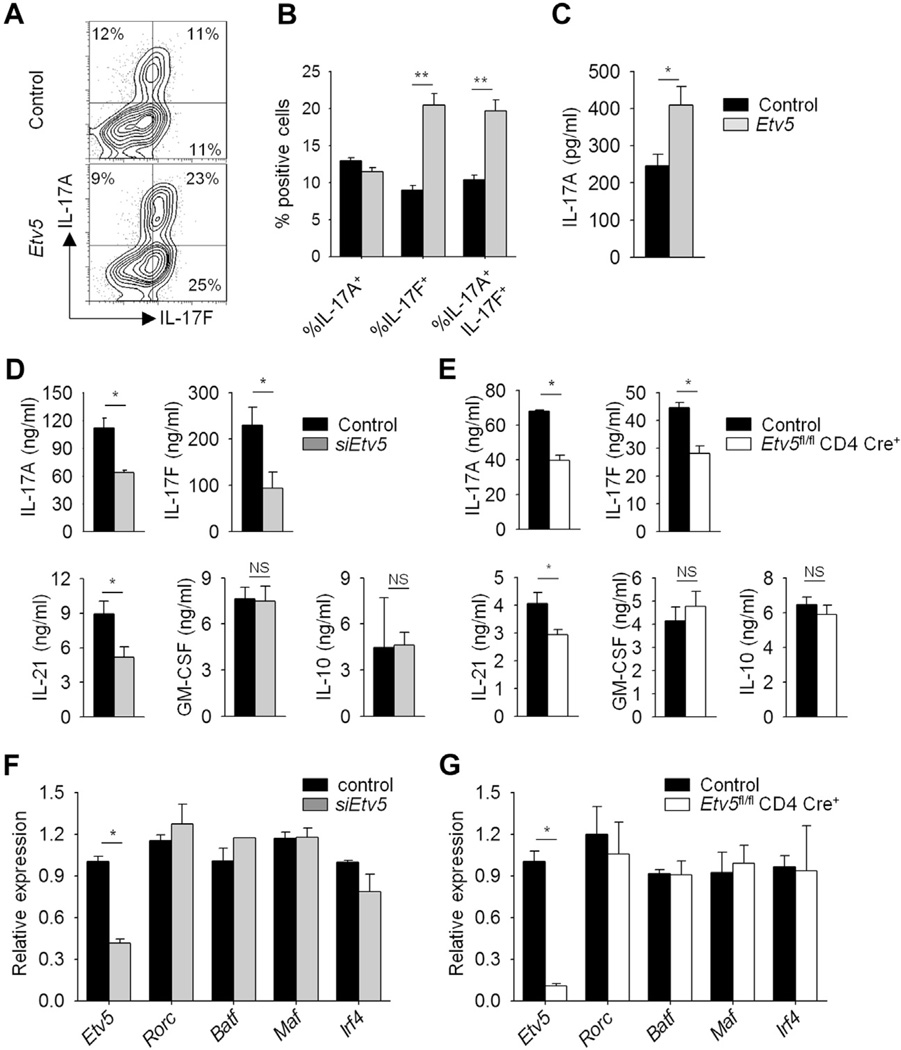
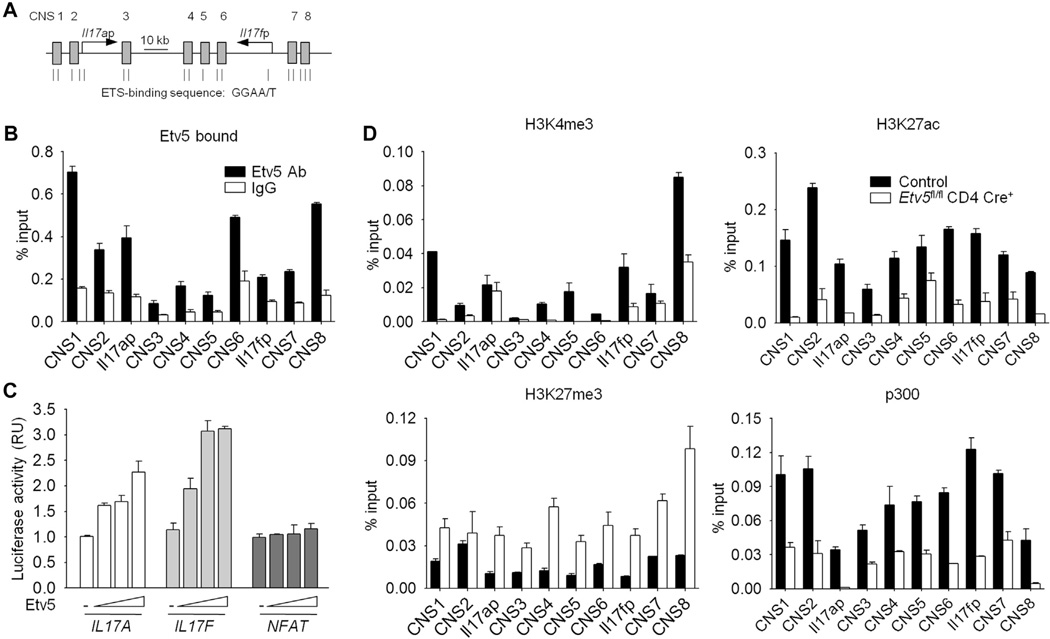
To further define the function of Etv5 in TH17 cell programming, we ectopically expressed Etv5 in TH17 cells and assessed cytokine production. Ectopic Etv5 expression in TH17 cells resulted in increased IL-17A and IL-17F production compared with that seen in control cells (Fig 7, A and B). To test whether Etv5 alone could induce IL-17 production in CD4+ T cells, Etv5 was ectopically expressed in nonpolarized CD4+ T cells (TH0 cells). Ectopic Etv5 expression in TH0 cells was able to induce IL-17A production compared with that seen in control cells (Fig 7, C). Consistent with these observations, reduced Etv5 expression in TH17 cells by transfecting T cells with siRNA targeting Etv5 (Fig 7, D) or using Etv5-deficient T cells (Fig 7, E) resulted in decreased IL-17A and IL-17F levels compared with those seen in control cells. IL-21 production was decreased in the absence of Etv5, but GM-CSF and IL-10 production was not significantly different in control cells from that seen in cells that lack Etv5 expression (Fig 7, D and E). We next examined the expression of transcription factors that are required for TH17 cell differentiation.13,15,16,35,36 Surprisingly, reduced Etv5 expression did not alter gene expression of Rorc, Batf, Maf, and Irf4 (Fig 7, F and G). Because Etv5 did not affect any of the known regulators of IL-17A–IL-17F expression, we tested whether Etv5 directly regulated expression of the Il17a-Il17f locus. The ChIP assay using differentiated TH17 cells revealed that Etv5 bound at several sites across the Il17a-Il17f locus, including the promoters and several conserved noncoding sequences (Fig 8, A and B). Jurkat T cells transfected with IL17A and IL17F luciferase reporters and a plasmid encoding Etv5 demonstrated that Etv5 promotes the transcriptional activity of the IL17A and IL17F promoters but not a nuclear factor of activated T cells promoter, in a dose-dependent manner (Fig 8, C). To define the modifications of histones correlated with active (H3K4 trimethylation and H3K27 acetylation) and repressed (H3K27 trimethylation) gene expression at the Il17a-Il17f locus, we performed ChIP assays from control and Etv5-deficient TH17 cells. We further observed decreased H3K4 methylation and H3K27 acetylation and increased H3K27 methylation across the Il17a-Il17f locus in the absence of Etv5 (Fig 8, D). In addition, the association of histone acetyltransferase p300 was decreased at the Il17a-Il17f locus in Etv5-deficient TH17 cells compared with that seen in control cells (Fig 8, D). These results suggested that Etv5 directly promotes IL-17 production, recruits histone-modifying enzyme, and mediates epigenetic changes at the Il17a–Il17f locus in TH17 cells.
FIG 7.
Etv5 promotes cytokine production in TH17 cells. A-C, Naive CD4+CD62L+ T cells were isolated from wild-type mice and differentiated under neutral conditions (TH0) or TH17 culture conditions. On day 2, cells were transduced with either control or Etv5-YFP (Etv5)–expressing retrovirus. On day 5, TH17 cells were stimulated with PMA and ionomycin for 6 hours before intracellular staining for cytokine production (Fig 7, A), with the percentage positive cells shown in Fig 7, B. Data are gated on YFP+cells. Fig 7, C, Cytokine production from sorted YFP+ TH0 cells was assessed by using ELISA after 24 hours of stimulation with anti-CD3. D and F, Wild-type TH17 cells were transfected with control or siRNA-specific Etv5, rested overnight, and restimulated with anti-CD3 to assess cytokine production by means of ELISA (Fig 7, D) and gene expression by using qRT-PCR (Fig 7, F). E and G, Control and Etv5-deficient TH17 cells were restimulated with anti-CD3 to assess cytokine production by using ELISA (Fig 7, E) and gene expression by using qRT-PCR (Fig 7, G). Data are means ± SEMs of 4 independent experiments. *P < .05 and **P < .01. NS, Not significant.
FIG 8.
Etv5 binds the Il17a-Il17f locus in TH17 cells. A, Schematic of the Il17a-Il17f locus containing ETS-binding sites. B and D, Naive CD4+CD62L+ T cells were isolated from control and Etv5 mutant mice and differentiated under TH17 culture conditions. ChIP analyses were performed by using differentiated control TH17 cells to examine transcription factor binding (Fig 8, B) or differentiated control and Etv5-deficient TH17 cells to assess histone modification and the association of histone-modifying enzyme (Fig 8, D) at the Il17a–Il17f locus. C, Luciferase activity in Jurkat T cells transfected with increased concentrations of plasmid encoding Etv5 along with IL17A, IL17F, or NFAT luciferase reporters and then activated for 6 hours with PMA and ionomycin. Data were normalized to control samples. Data are means ± SDs of replicate samples and representative of 3 to 4 independent experiments with similar results. RU, Relative unit.
DISCUSSION
The development of specialized helper T-cell subsets requires a network of transcription factors that promote the expression and production of specific cytokines. TH17 development requires the activity of STAT3-activating cytokines and the induction of STAT3 target genes, including Rorc, Batf, and Irf4, in TH17 cell differentiation. In this report we demonstrate that Etv5 is downstream of STAT3 in directly promoting Il17a/f expression and mediating changes in histone modifications at the Il17a/f locus. In the HDM-induced allergic airway inflammation model, Etv5 mutant mice had reduced lung inflammation, mucus production in the airway, and IL-17 production compared with control mice.
The requirement for IL-17 in asthma is not completely defined. Increased IL-17A and IL-17F levels are associated with severe asthma in patients.37 IL-17 produced from activated CD4+ T cells induces the expression of macrophage- and neutrophil-attracting chemokines that recruit inflammatory cells into the airways.38 However, in mouse models IL-17 has differential effects during sensitization or challenge, and the requirement for IL-17 varies among model systems.32,39–42 Importantly, the HDM adjuvant–free model is dependent on IL-17 in addition to TH2 cytokines.30–33 IL-17 production and neutrophil recruitment in the lung are increased after HDM sensitization and challenge.43 We observed a significant decrease in IL-17 production in the lung, BAL fluid, and peripheral lymphoid organs, as well as diminished neutrophil-attracting chemokine levels and neutrophil recruitment to the lung in Etv5 mutant mice compared with that seen in control mice. Although Cxcl1 and Cxcl2 are IL-17– inducible genes, only the latter was affected when IL-17 levels were reduced in Etv5-mutant mice or blocked with anti–IL-17A antibody. The result suggested Cxcl2 expression was regulated differentially compared with Cxcl1 in this model. Indeed, in a model of sterile inflammation, IL-17 controls neutrophil trafficking by preferentially regulating the expression of Cxcl2 but not Cxcl1.44 In addition, it has been demonstrated that Cxcl1 expression is associated with systemic inflammation and that Cxcl2 expression is limited to localized inflamed tissue.45
Although HDM sensitization and challenge elicits TH2 responses,30,31,43 our data demonstrated that IL-17 was the most abundant cytokine produced in the lung, suggesting IL-17 production from T cells is critical for inflammation in this model. In the absence of Etv5 in T cells, there is diminished inflammation and reduced IL-17 concentrations, without any corresponding effects on the amounts of TH2 cytokine production. In addition to decreased neutrophil recruitment to the lung, eosinophil numbers were also reduced. Because IL-17A and IL-17F are shown to have differential effects on eosinophil recruitment during antigen-induced airway inflammation,41 further studies are needed to determine whether IL-17 is responsible for the reduction in eosinophil numbers in the lung in this model. Importantly, we demonstrate that blocking IL-17A in the HDM model decreases cellular infiltrates similarly to that observed in mice lacking Etv5 in T cells. Moreover, intranasal administration of IL-17A/F to HDM-challenged Etv5 mutant mice results in a recovery of inflammation in the lung. Our results suggest that TH17 production of IL-17 provides a unique contribution to the inflammatory milieu in the development of allergic pulmonary inflammation.
Our data show that Etv5 deficiency in T cells does not alter expression of TH17 transcription factors or other TH17 cytokines, suggesting a restricted effect on expression of the Il17a-Il17f locus. This is unique among transcription factors that promote TH17 development, including retinoic acid–related orphan receptor γt, interferon regulatory factor 4, and B-cell activating transcription factor-like, which regulate multiple components of the TH17 genetic program.12,13,15,16,46 It is also important to note that Etv5 has a critical role in TH17 cytokine production despite expression of Etv5 not being specific for TH17 cells. This highlights that the amount of expression of a transcription factor is not always the most important determinant of function. Our results further document that transcription factors that are not restricted to a particular subset still play obligate roles in defining cellular phenotype in cooperation with lineage-specific factors.
These results demonstrate that Etv5 is a novel STAT3-induced transcription factor that directs expression of the Il17a-Il17f locus in TH17 cells. This is consistent with a recent report that Etv5 also contributes to IL-17 production in γδ T cells and suggests that Etv5 might have common functions among multiple IL-17– secreting T-cell populations.47 Our results further suggest that in addition to lineage regulators, such as Rorc, Irf4, and Batf, which regulate multiple genes in a subset, there are additional transcription factors that are part of the helper T-cell subset differentiation program that have a more restricted and specific function. Thus Etv5 represents a unique contributing factor that positively regulates inflammatory responses in TH17 cells and promotes allergic airway inflammation.
Supplementary Material
Key messages.
Etv5 directly promotes IL-17 production in TH17 cells.
Mice that lack Etv5 in T cells display decreased airway inflammation compared with that seen in wild-type mice.
Acknowledgments
Supported by Public Health Service grants AI045515 (to M.H.K.). D.P was supported by T32HL007910.
M. H. Kaplan has received research support from the National Institutes of Health.
We thank A. L. Dent, B. Zhou, and Kaplan laboratory members for critical reading of this manuscript.
Abbreviations used
- BAL
Bronchoalveolar lavage
- ChIP
Chromatin immunoprecipitation
- Foxp3
Forkhead box protein 3
- HDM
House dust mite
- iTreg
Inducible regulatory T
- PMA
Phorbol 12-myristate 13-acetate
- qRT-PCR
Quantitative reverse transcriptase PCR
- siRNA
Small interfering RNA
- STAT3
Signal transducer and activator of transcription 3
- Treg
Regulatory T
Footnotes
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 3.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 4.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 5.Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, et al. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422–434. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Clarke TB, Weiser JN. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest. 2009;119:1899–1909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, McAleer JP, Lin Y, Paterson DL, Zheng M, Alcorn JF, et al. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity. 2011;35:997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abraham C, Cho J. Interleukin-23/Th17 pathways and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1090–1100. doi: 10.1002/ibd.20894. [DOI] [PubMed] [Google Scholar]
- 12.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 17.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 19.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, et al. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development. 2010;137:3417–3426. doi: 10.1242/dev.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell. 2009;16:607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL. Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol Reprod. 2011;85:1114–1123. doi: 10.1095/biolreprod.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang W, Jacobson NG, Bhattacharya D, Gorham JD, Fenoglio D, Sha WC, et al. The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway. Proc Natl Acad Sci U S A. 1999;96:3888–3893. doi: 10.1073/pnas.96.7.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rijt LS, Kuipers H, Vos N, Hijdra D, Hoogsteden HC, Lambrecht BN. A rapid flow cytometric method for determining the cellular composition of bronchoalveolar lavage fluid cells in mouse models of asthma. J Immunol Methods. 2004;288:111–121. doi: 10.1016/j.jim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Pham D, Vincentz JW, Firulli AB, Kaplan MH. Twist1 regulates ifng expression in Th1 cells by interfering with runx3 function. J Immunol. 2012;189:832–840. doi: 10.4049/jimmunol.1200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathur AN, Chang HC, Zisoulis DG, Kapur R, Belladonna ML, Kansas GS, et al. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108:1595–1601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daan de Boer J, Roelofs JJ, de Vos AF, de Beer R, Schouten M, Hommes TJ, et al. Lipopolysaccharide inhibits Th2 lung inflammation induced by house dust mite allergens in mice. Am J Respir Cell Mol Biol. 2013;48:382–389. doi: 10.1165/rcmb.2012-0331OC. [DOI] [PubMed] [Google Scholar]
- 31.Gregory LG, Causton B, Murdoch JR, Mathie SA, O’Donnell V, Thomas CP, et al. Inhaled house dust mite induces pulmonary T helper 2 cytokine production. Clin Exp Allergy. 2009;39:1597–1610. doi: 10.1111/j.1365-2222.2009.03302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lajoie S, Lewkowich IP, Suzuki Y, Clark JR, Sproles AA, Dienger K, et al. Complement-mediated regulation of the IL-17A axis is a central genetic determinant of the severity of experimental allergic asthma. Nat Immunol. 2010;11:928–935. doi: 10.1038/ni.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudo M, Melton AC, Chen C, Engler MB, Huang KE, Ren X, et al. IL-17A produced by alphabeta T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18:547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci U S A. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Ramli W, Prefontaine D, Chouiali F, Martin JG, Olivenstein R, Lemiere C, et al. T(H)17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol. 2009;123:1185–1187. doi: 10.1016/j.jaci.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Laan M, Cui ZH, Hoshino H, Lotvall J, Sjostrand M, Gruenert DC, et al. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J Immunol. 1999;162:2347–2352. [PubMed] [Google Scholar]
- 39.Nakae S, Komiyama Y, Nambu A, Sudo K, Iwase M, Homma I, et al. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 40.Schnyder-Candrian S, Togbe D, Couillin I, Mercier I, Brombacher F, Quesniaux V, et al. Interleukin-17 is a negative regulator of established allergic asthma. J Exp Med. 2006;203:2715–2725. doi: 10.1084/jem.20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, et al. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira AP, Cavassani KA, Ismailoglu UB, Hullinger R, Dunleavy MP, Knight DA, et al. The protective role of TLR6 in a mouse model of asthma is mediated by IL-23 and IL-17A. J Clin Invest. 2011;121:4420–4432. doi: 10.1172/JCI44999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phipps S, Lam CE, Kaiko GE, Foo SY, Collison A, Mattes J, et al. Toll/IL-1 signaling is critical for house dust mite-specific helper T cell type 2 and type 17 [corrected] responses. Am J Respir Crit Care Med. 2009;179:883–893. doi: 10.1164/rccm.200806-974OC. [DOI] [PubMed] [Google Scholar]
- 44.Brackett CM, Muhitch JB, Evans SS, Gollnick SO. IL-17 promotes neutrophil entry into tumor-draining lymph nodes following induction of sterile inflammation. J Immunol. 2013;191:4348–4357. doi: 10.4049/jimmunol.1103621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Call DR, Nemzek JA, Ebong SJ, Bolgos GR, Newcomb DE, Wollenberg GK, et al. Differential local and systemic regulation of the murine chemokines KC and MIP2. Shock. 2001;15:278–284. doi: 10.1097/00024382-200115040-00005. [DOI] [PubMed] [Google Scholar]
- 46.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, et al. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.