Abstract
A key event in the domestication and breeding of the oil palm, Elaeis guineensis, was loss of the thick coconut-like shell surrounding the kernel. Modern E. guineensis has three fruit forms, dura (thick-shelled), pisifera (shell-less) and tenera (thin-shelled), a hybrid between dura and pisifera1–4. The pisifera palm is usually female-sterile but the tenera yields far more oil than dura, and is the basis for commercial palm oil production in all of Southeast Asia5. Here, we describe the mapping and identification of the Shell gene responsible for the different fruit forms. Using homozygosity mapping by sequencing we found two independent mutations in the DNA binding domain of a homologue of the MADS-box gene SEEDSTICK (STK) which controls ovule identity and seed development in Arabidopsis. The Shell gene is responsible for the tenera phenotype in both cultivated and wild palms from sub-Saharan Africa, and our findings provide a genetic explanation for the single gene heterosis attributed to Shell, via heterodimerization. This gene mutation explains the single most important economic trait in oil palm, and has implications for the competing interests of global edible oil production, biofuels and rainforest conservation6.
Oil palm fruits (drupes) are derived from three fused carpels and consist of epicarp, mesocarp and endocarp tissues surrounding one or more kernels. Hybrids (tenera) between dura and pisifera (Fig. 1) have a distinct fiber ring, derived from the endocarp, that surrounds the coconut-like shell of the oil palm seed7. The Shell gene responsible for this phenotype has co-dominant monogenic inheritance, first reported in the Belgian Congo in the 1940’s7. However, tenera fruit forms were recognized and exploited in Africa well before then2,8. Given the central role played by the Shell gene, oil palm breeding utilizes reciprocal recurrent selection of maternal (dura) and paternal (pisifera) pools4. The Deli dura population, direct descendants of the four original African palms planted in Bogor Botanical Garden, Indonesia (1848), has excellent combining ability with the AVROS (Algemene Vereniging van Rubberplanters ter Oostkust van Sumatra) and other pisifera parental palms. AVROS pisifera palms were derived from the famous “Djongo” palm from Congo, but more recently several different accessions of dura and tenera have also been sourced from Africa4. Tenera palms are thought to have been selected by pre-colonial cultures in West Africa due to their higher oil yields, and are the basis for modern oil palm breeding4.
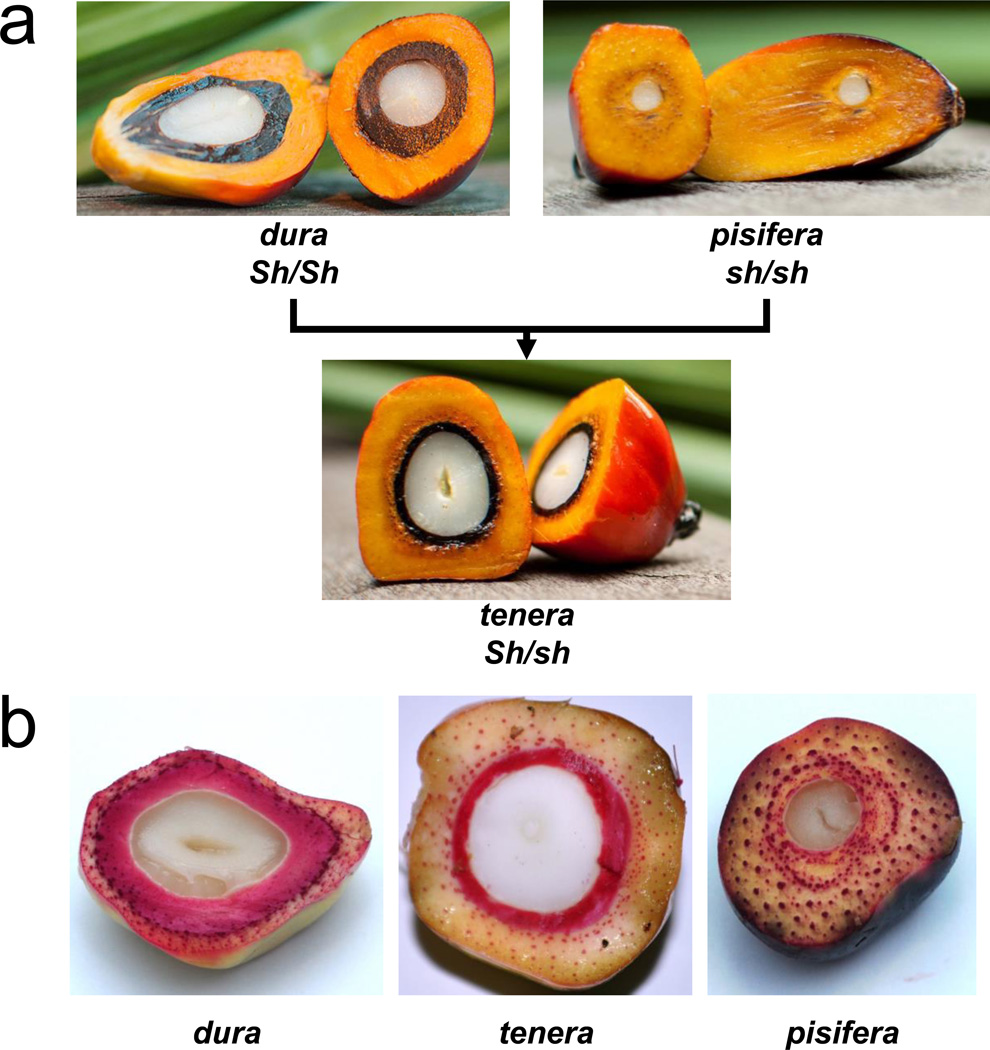
Figure 1. Fruit forms of the African oil palm Elaeis guineensis.
a, Dura (Sh/Sh) fruit forms have a thick lignified shell surrounding the kernel, which is absent in pisifera (sh/sh). F1 hybrid palms (Sh/sh) have an intermediate fruit form (tenera) which is much higher yielding than either parent in terms of mesocarp oil. b, Dura, tenera and pisifera fruit stained for lignin (red) with phloroglucinol.
The Shell gene lies 4.7 cM and 9.8 cM away from the closest molecular markers9–11 but has proven extremely challenging to identify given the large genome, long generation times and difficulty of phenotyping in experimental populations of oil palm, which are widely distributed among different plantations (Methods). We employed a two-tiered approach to identify the Shell gene, taking advantage of the recently completed oil palm genome sequence12. First, 240 F1 progeny, derived by controlled self-pollination of the Nigerian tenera accession T128 and grown over two decades in plantations throughout Malaysia, were scored for fruit form phenotype13. In addition to 200 RFLP and SSR markers, the progeny were genotyped for 4,451 SNP markers derived from the oil palm genome sequence12 by the Infinium iSelect® Assay (Illumina). A genetic map with 16 linkage groups was constructed, and the Shell gene locus was placed in T128 Linkage Group 7 (Supplementary Fig. 1), consistent with prior studies (Methods), and mapped by sequence similarity to a 3.4 Mb assembly Scaffold 43 (p3-sc00043) at the end of Chromosome 212. A tiling path of BAC contigs corresponding to Scaffold 43 was selected from a high-information content physical map of pisifera and sequenced12. Further SNP assays were designed from an improved assembly, and additional genotypes incorporated into the map. In a review of single marker data, recombinant breakpoints were identified indicating the gene lay in a 450kb interval (Methods).
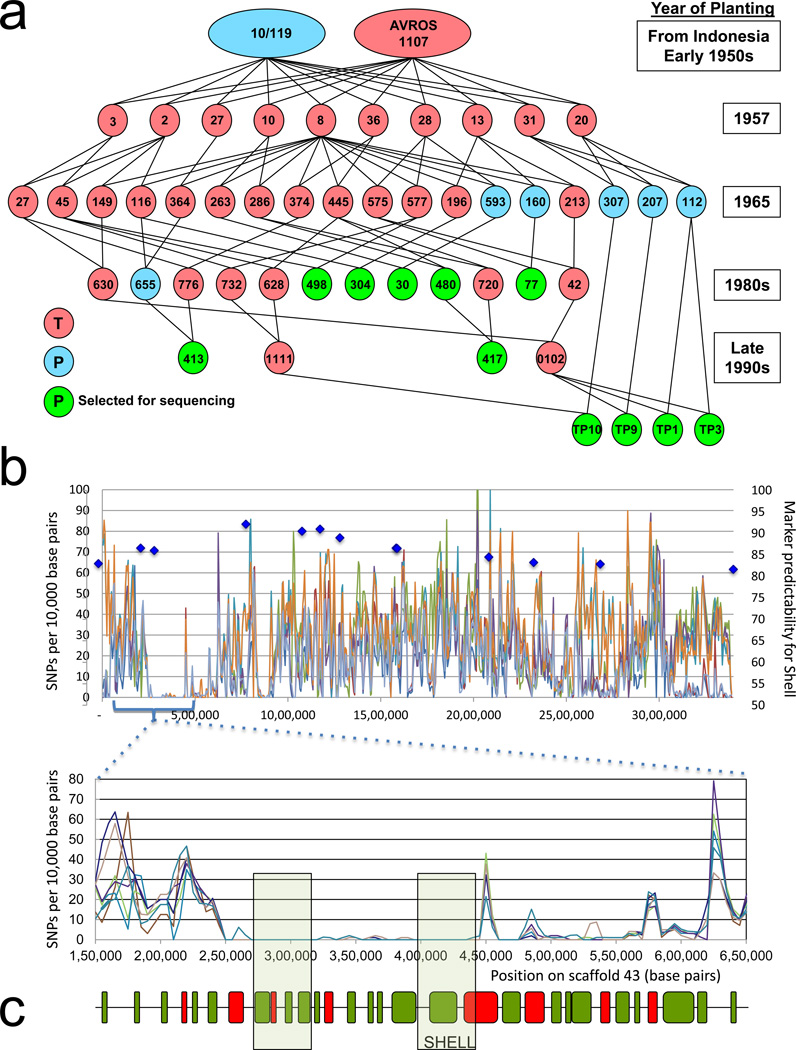
Next, we employed homozygosity mapping using the AVROS pedigree (Fig. 2a) and whole genome re-sequencing (Methods Summary). In this technique, candidate genes appear as regions with low diversity in homozygous inbred individuals14,15. Fourteen individual pisifera palm genomes were sequenced at 20× genomic coverage and 29 additional pisifera palms were sequenced as a pool (pool 1) at 40× coverage (Illumina HISEQ 2000). SNPs were called throughout the genome, and those in Scaffold 43 were scored for homozygosity. The resulting homozygosity plot had a local minimum of 200 Kb (centered on 400,000 bp in p3-sc00043, Fig. 2b). This 200 Kb region contained about 30 annotated genes, only five of which were fully homozygous, and only one of which lay in the genetic interval containing Shell (Fig. 2c). This gene encodes a homologue of SEEDSTICK (STK), which is responsible for ovule and seed development in Arabidopsis16,17 (Fig. 2c; Supplementary Fig. 2).
Figure 2. Homozygosity mapping of the shell trait.
a, AVROS pedigree palms heterozygous (tenera, pink) and homozygous (pisifera, blue and green) for Shell are indicated, linked to parents and progeny planted over 5 decades. Individually sequenced palms are indicated (green). Three additional pisifera palms from this extended pedigree were also individually sequenced (not shown). The remaining pisifera palms were sequenced as a pool. b, The Shell locus was mapped onto scaffold p3-sc00043 (3.4Mb) and SNP density was plotted along the scaffold (upper panel). Markers surrounding Shell are plotted against the % of pisifera palms with the SNP haplotype in the Nigerian T128 F1 mapping population. Shell was mapped between flanking markers, denoted by blue diamonds, at 400 Kb and 1 Mb. A local diversity minimum of 200 Kb was found within this interval (inset). c, Genes (green) and transposons (red) are indicated as boxes. The local diversity minimum contained 4 homozygous genes (boxed), only one of which (Shell) was located between the flanking markers in panel b.
PCR amplicon sequencing (Methods) identified allelic differences between Shell in Deli dura (ShDeliDura), and in the AVROS (shAVROS) and T128 (shMPOB) pisifera haplotypes derived from Congo and Nigeria, respectively (Fig. 3). A nucleotide substitution in shMPOB results in a leucine to proline amino acid change in the conserved DNA binding and dimerization domain, while a substitution in shAVROS results in a lysine to asparagine amino acid change only two amino acids removed (Fig. 3). In related proteins, this highly conserved lysine residue is involved in nuclear localization, and in direct DNA binding18,19, while the substitution by a proline only two amino acid residues N-terminal to this position would disrupt the alpha helix that is involved in MADS dimerization and DNA binding19. Analysis of an additional 336 palms was used to validate these alleles within established phenotyping norms (Methods). These included four pisifera palms from introgression trials of shMPOB into tenera carrying the shAVROS allele. Sequencing confirmed that these four palms were heteroallelic, indicating that the two alleles failed to complement, and confirming the identity of the gene (Methods). To further explore segregation in E. guineensis populations, Shell Exon 1 sequence was generated from a diversity panel of 379 palms representing nine distinct wild oil palm populations collected from Angola4, Madagascar4, Nigeria3 and Tanzania4, and a subset of a 110,000-accession seed bank collected over the past five decades4 (Methods). We found that all palms evaluated carried either the ShDeliDura, shAVROS or shMPOB alleles in exon 1 (Fig. 3, Supplementary Fig. 3).
Figure 3. Sequence diversity of Shell.
Multiple sequence alignment of Arabidopsis STK (AEE82818.1) and SHP1 (P29381.1) proteins, rice protein OsMADS13 (AAF13594.1), poplar predicted protein (XP_002327282.1), grape MADS-box protein 5 (XP_002281890.1), tomato TAGL1 (AAM33101.2) and peach SHATTERPROOF-like (ABG75908.1) (Out Groups), E. oleifera, E. guineensis and exemplars of Deli dura (Oil Palm – Dura Allele) and pisifera from T128 – Nigerian and AVROS – Congo (Oil Palm – Pisifera Allele). The pisifera fruit form is caused by two disruptive SNPs that affect a highly conserved amino acid motif in the MADS-box DNA binding and dimerization domain. Gene names (out group examples) or sample identifiers (oil palm examples) are in parentheses.
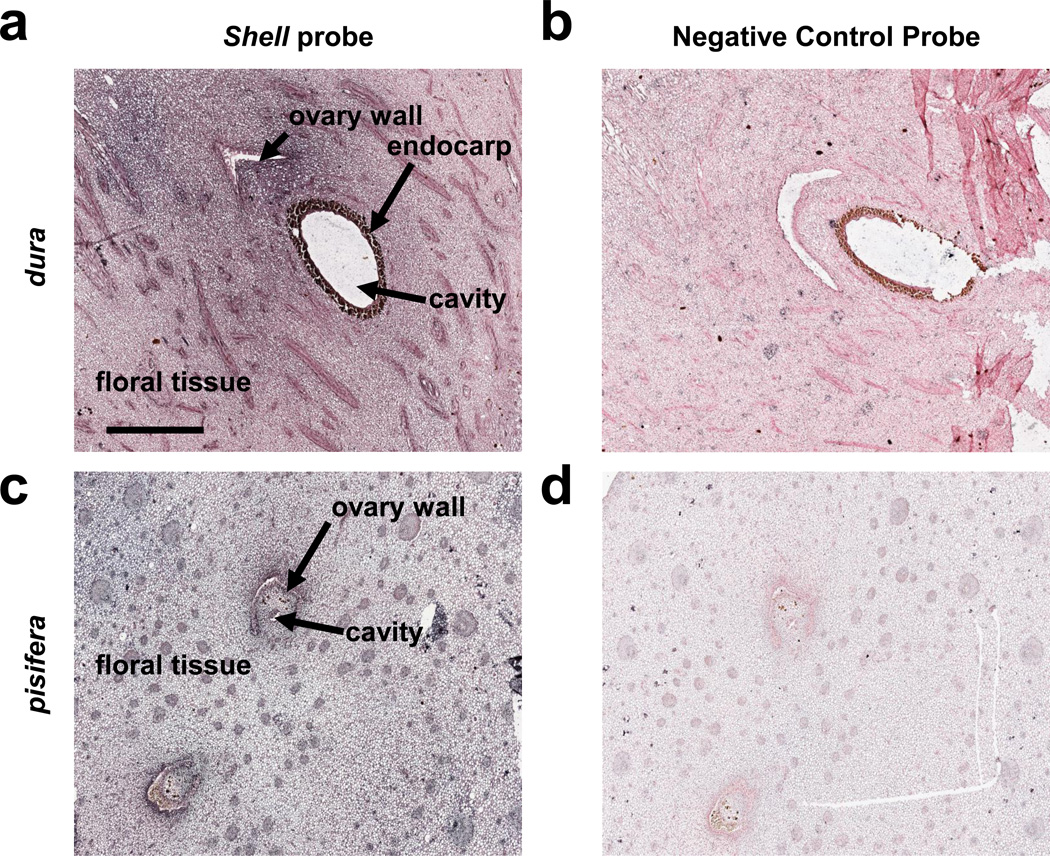
Analysis of 10,916,126 RNA-seq reads from 22 different libraries12 revealed only 159 reads matching Shell, all of which were found in just four libraries: from whole florets one day after anthesis, kernels 10 and 15 weeks, and mesocarp 15 weeks after anthesis (WAA). In situ hybridization was performed on fruits between 1 and 5 WAA (Fig. 4), at the earliest stages of shell formation3,20. Uniform but weak hybridization signals were detected in the mesocarp of both the thick-shelled (dura) and shell-less (pisifera) fruit forms (Fig. 4a & c), but very strong signals were detected in the outer layers of the developing kernel in only the dura type (Fig. 4a), consistent with the function of Shell. The oil palm shell is heavily lignified (Fig. 1b), derived from the endocarp, and surrounds the kernel (or pit). It is not found in pisifera palms which are often female sterile.
Figure 4. Expression of Shell in early oil palm fruit.
Longitudinal sections of 1 WAA Dura fruit (a, b) and cross sections of 5 WAA pisifera fruit (c, d) were hybridized to a pool of two unique Shell locked nucleic acid (LNA) probes (a, c) or to a scrambled sequence LNA negative control probe (b, d). Colorimetric in situ hybridization probe detection is blue and all sections were counterstained red. The scale bar (0.8 mm) shown in panel b applies to all four panels. Structural hallmarks of fruits are indicated by labeled arrows in panels a and c.
The function and expression of the Shell gene is conserved in higher plants16. In Arabidopsis, SHATTERPROOF (SHP) and STK are Type II MADS-box proteins of the C and D class, respectively, and form a network of transcription factors that control differentiation of the ovule, seed and lignified endocarp21. In tomato, SHP homologues control fleshy fruit expansion in the endocarp22 while in peaches, which are also drupes, homologues of both genes have been implicated in lignified split-pit formation23,24. In rice, the Shell orthologue is OsMADS13 (Supplementary Fig. 4), a homologue of STK25 and SHP26 that controls ovule identity, so that mutants are female sterile27. STK and SHP bind to DNA as heteromultimers with SEPALLATA (SEP) MADS box proteins, including OsMADS24 in rice16, and the highly conserved MADS domain is involved in both DNA binding and in dimerization18,19. We postulated that the mutations we detected in Shell could account for the remarkable single gene heterosis exhibited in tenera palms7 if they disrupted heterodimerization, as well as DNA binding16,18. In yeast 2 hybrid assays, we found that only the dura allelic form of Shell, but neither of the pisifera alleles, interacted as a heterodimer with OsMADS24. Furthermore, the dura form of Shell homodimerized with itself and with the pisifera forms but the pisifera forms did not homodimerize with each other (Table 1, Supplementary Figure 5 and Supplementary Table 1). If productive heterodimers compete with non-productive homodimers in heterozygotes, this would neatly account for hybrid vigor according to theoretical models28. Overdominance at a single locus accounts for similarly remarkable increases in hybrid yield in tomato29.
Table 1.
Pisifera fruit form mutations disrupt the interaction of SHELL and OsMADS24
| Binding Domain Fusion | Activation Domain Fusion | Interaction |
|---|---|---|
| ShDeliDura | ShDeliDura | + |
| ShMPOB | ShDeliDura | + |
| ShAVROS | ShDeliDura | + |
| OsMADS24 | OsMADS24 | − |
| OsMADS24 | ShDeliDura | + |
| OsMADS24 | ShMPOB | − |
| OsMADS24 | ShAVROS | − |
Symbols: +, dimerization detected in yeast two-hybrid assay. −, no interaction detected. Experiments were performed as described (Methods). Complete datasets are provided in Supplementary Figure 5 and Supplementary Table 1.
The unraveling of the genetic basis for the shell-less phenotype paves the way for designing molecular strategies for genotyping trees that breed true for the phenotype, and modulating Shell activity for desired fruit forms. A marker for Shell could be used by seed producers to reduce or eliminate dura contamination, and to distinguish the dura, tenera and pisifera plants in the nursery long before they are field planted – the advantage here being that they could be planted separately based on the shell trait. This is useful as the pisifera palms have vigorous vegetative growth, and planting them in high density encourages male inflorescence development and pollen production4,30. As foreseen by the breeder A. Devuyst in the Belgian Congo (in the same year that the structure of DNA was solved)2, accurate genotyping for enhanced oil yields will optimize and ultimately reduce the acreage devoted to oil palm plantations, providing an opportunity for conservation and restoration of dwindling rainforest reserves6.
Methods
Plant Materials and Germplasm Collection
Oil palm germplasm materials used in this study were collected through bilateral agreements between Malaysia and the countries of origin and were in accordance with the Convention on Biological Diversity (CBD) 1992. The reference Deli dura palm was MPOB 0.212/70, the reference pisifera palm was MPOB 0.182/77, and the reference E.oleifera palm was MPOB 0.211/2460. The mapping family used for generating the genetic linkage map was derived from controlled self pollination of the high iodine value (IV, a measure of level of unsaturation) virescens tenera palm, T128 (Accession number: MPOB 371), from MPOB's Nigerian germplasm collection13. A total of 241 palms was originally planted from 1993 to 1997 at several locations in Malaysia, namely the MPOB-UKM Research Station Bangi at Selangor, MPOB Ulu Paka Research Station at Terengganu, MPOB Keratong Research Station at Johore, MPOB Lahad Datu Research Station at Sabah, United Plantations at Perak and FELDA Research Station at Pahang. Of the 241 palms, 240 were still available for marker analysis. The palm T128 is relevant to the breeding programme as it has outstanding attributes in terms of oil yield, fatty acid composition and low height increment. It has been crossed with at least six dura, eight tenera and three pisifera palms for progeny testing and widening the genetic base. A collection of 379 palms (287 dura, 86 tenera and 6 pisifera scored fruit types) from Angola, Congo, Madagascar, Nigeria and Tanzania were taken from the diversity panel maintained by MPOB, and screened for new alleles as described. Unopened leaf samples (spear leaves) were collected from individual palms and immediately frozen under liquid nitrogen and then stored at −80°C until DNA preparation. DNA was extracted and purified from the leaf samples as previously described31.
Whole genome sequencing and assembly
An AVROS pisifera palm was sequenced to high coverage12 on the 454 XL next generation sequencing platform (454/ Roche). Sequence reads were generated from DNA fragment libraries and from a complex series of linker libraries where read ends span fragment sizes ranging from 0.75 Kb to >30 Kb. Sequence reads were assembled with the Newbler assembler (Roche 454, Bradford, CT) producing a reference sequence of the oil palm genome12. Scaffolds from the reference assembly containing markers genetically mapped in the shell interval were identified. A BAC physical map was constructed from a 10-fold BAC library constructed from the same AVROS pisifera used to generate the reference sequence. BAC end sequences were also generated from each BAC in the library using standard Sanger sequencing on the 3730 sequencing platform (Life Technologies). BAC end sequences were assembled into the reference genome with the Newbler assembler. A minimum tiling path of BAC clones spanning the SHELL interval was selected, and BAC clones in the tiling path were sequenced in pools to high coverage with 454 XL technology. The BAC pool sequences were assembled and merged with the original 454 whole genome shotgun sequence data and all BAC end sequences. Improved scaffold coverage and scaffold length spanning the shell interval was produced.
Genetic mapping
241 progeny palms were derived from self-pollination of the Nigerian tenera palm T1284, of which 240 palms were available throughout the study. Two palms could not be phenotyped accurately while 124 tenera, 46 pisifera and 68 dura, palms were phenotyped with high confidence at the time of genetic map construction. All 240 progeny were scored for 4451 SNP markers using the Infinium iSelect® Assay (Illumina), as well as three RFLP markers32 four SSR markers3 and 193 additional SSR markers developed from the oil palm genome sequence12. The linkage map was then constructed using JoinMap v433. The genotype data was formatted as required for mapping according to an F2 population. Markers showing a segregation profile of 1:2:1 for the phenotypes were used in the map construction.
Two sets of the genotype data were then created, whereby one set is the converse of the other to account for phase differences in the T128 selfed F2 population. Markers that exhibited severe distortion (p < 0.0001) and markers having more than 10% missing data were excluded. Both sets of genotype data were then grouped at a recombination frequency of ≤ 0.2. Eighteen nodes were selected to create 18 initial groups for calculating the linkage groups. The linkages were calculated and loci ordered based on the maximum likelihood algorithm. None of the markers showed severe distortion (p < 0.0001). The few distorted markers observed were significant at p < 0.05 – 0.1. These markers were removed from further analysis when necessary. Markers exhibiting nearest neighbour stress (N.N. Stress) value > 2 (cM) were identified and excluded from the analysis. Markers contributing to insufficient linkages were also determined and removed. The T128 co-dominant map constructed consisted of 16 groups9, and shell was placed on linkage group 7, within the expected region (Supplementary Figure 1).
The SNP markers surrounding shell were subsequently mapped by sequence similarity to a 3.4Mb assembly Scaffold 43 (p3-sc00043). A tiling path of BAC contigs corresponding to Scaffold 43 was selected from a high-information content physical map of pisifera and sequenced12. An additional 50 SNP assays were designed from an improved assembly corresponding to Scaffold 43 (p5-Scaffold 60). Thirty additional SNP markers were also designed from the scaffolds p3-sc00191, p3-sc00203 and p3-sc02216, which were also associated with markers on linkage Group 7, and reassembled as p5-sc00263. These 80 SNP markers (designated as SNPE) were genotyped in the T128 selfed population using the Sequenom MassArray® iPlex platform and 63 were polymorphic giving a final co-dominant genetic linkage map consisting of 818 markers [719 SNP (inclusive of SNPE), 96 SSR, 2 RFLP markers and shell] in 16 linkage groups. Shell remained in linkage group 7, together with the 63 SNPE markers developed from the selected scaffolds. The final size of linkage group 7 is ~182 cM with an average of 1.2 cM between two adjacent markers (Supplementary Figure 1).
Single marker mapping was employed to determine the recombination breakpoints surrounding shell in each individual palm. To obtain a genetic interval containing shell, palms with multiple breakpoints in the region and/or inconsistent haplotypes were discarded in case of mis-genotyping. Markers flanking shell were selected using these conservative criteria and found to lie 450 Kb apart on scaffold p3-Sc0043 (Fig. 2b & c).
Fruit form phenotyping
Oil palm trees were grown to maturity in open plantations, making accurate phenotyping of fruit form for some samples difficult due to variation in fertility, yield and environment. Several fruits were harvested from each palm, and shell thickness and fruit form was determined using established criteria3. To assess the accuracy of the fruit form phenotypic data used in the study, we reviewed 460 phenotype calls which were made between 2003 and 2012. In this period, up to three independent attempts were made to visually determine the fruit form phenotypes of 340 palms, 241 of which were from the T-128 selfed population used to map shell, and 99 from a different population for which re-phenotyping data were available. In the data, ambiguous calls were made 26 times (or 5.7% of total phenotype determinations) where breeders were unsure of the fruit form phenotype.
Homozygosity mapping
A total of 43 individual pisifera palms from the AVROS pedigree (originating from Congo) were sourced from MPOB, Sime Darby and Kulim plantations. 14 of the AVROS pisifera palms were independently sequenced while the DNA from the remaining 29 palms were pooled for sequencing (Pool 1). Whole genome shotgun sequence data was generated on the HISEQ 2000 (Illumina). Individual trees and pools of trees were sequenced to 20 and 40 fold raw sequence coverage, respectively. Individual reads were mapped to sequence scaffolds from the pisifera reference genome assembly, and highest probability SNPs were located on scaffold 43 of the pisifera Build 3 (p3_sc00043). In each of the 14 genome sequences, SNPs were summed over 10kb windows along the scaffold and plotted against map location. This scaffold was computationally annotated for genes by comparing to public databases for the A. thaliana and rice genomes using the NCBI BLAST similarity searching tool. The genes with the highest homozygosity within the predicted interval from the genetic map were screened for putative function and for nucleic and amino acid changes between the pisifera and dura lines, as well as for amino acid differences in other species.
Validation and genotyping
DNA sequencing of the eight exons of the SHP1 gene was carried out for all palms of the mapping family as well as the AVROS pisifera palms used in homozygosity mapping. One of the 14 sequenced pisifera palms (TP10) proved to be heterozygous for the shAVROS and shMPOB haplotypes, due to contamination in a breeding trial, and was not used for the homozygosity analysis. PCR primers were designed based on the reference pisifera genome sequence to amplify the entirety of SHP1 exon 1. SHP1-specific primer sequences were TTGCTTTTAATTTTGCTTGAATACC (forward primer upstream of exon 1) and TTTGGATCAGGGATAAAAGGGAAGC (reverse primer downstream of exon 1). Primer sequences were confirmed to be unique in the reference pisifera genome, and to avoid any identified polymorphic nucleotides. A 5’ M13 forward sequence tag (GTTTTCCCAGTCACGACGTTGTA) was added to the Exon 1 forward PCR primer. A 5’ M13 reverse sequence tag (AGGAAACAGCTATGACCAT) was added to the Exon 1 reverse primer. SHP1 exon 1 was amplified from genomic DNA and PCR amplification was performed using 20 ng of purified genomic DNA under standard PCR amplification conditions. Amplicons were treated with exonuclease I and shrimp alkaline phosphatase to remove unincorporated primers and deoxynucleotides. An aliquot of each amplicon was sequenced using M13 forward as primer on an ABI 3730 instrument using standard conditions. Each amplicon was sequenced twice in the forward direction. An aliquot of each amplicon was additionally sequenced using M13 reverse as primer. Each amplicon was sequenced twice in the reverse direction. All sequencing data was aligned to the reference pisifera genome sequence. Data were analyzed to determine the genotype at each of the two SNP positions identified to be associated with the pisifera fruit form. We sequenced SHP Exon 1 from 336 individual palms from the T-128 mapping population, the samples used to construct homozygosity maps, and a collection of palms in crosses with advanced lines (100 pisifera, 148 tenera, 86 dura and 2 with ambiguous phenotype). 323 (96.7%) had SHP genotypes concordant with their phenotype, and 11 (3.3%) had discordant phenotypes, reflecting the accuracy of phenotyping in the plantation (see above). SHP exon 1 was also sequenced from all 4 pisifera palms derived from TxT crosses between shMPOB and shAVROS (Felda AA and MPOB 0.305), and proved to be heteroallelic as predicted. An additional 3 pisifera palms were also found to be heteroallelic, including TP10 which was sequenced completely and proved to be a contaminant in the AVROS pedigree (see above). The other 2 heteroallelic palms were likely similarly contaminated.
A second attempt was made to phenotype and re-sequence the 11 apparently discordant trees, enabling the re-evaluation of 9 trees for fruit type. The second phenotype call of 1 palm was concordant with the genotype prediction, while the second phenotype calls of 7 palms were ambiguous, and 1 palm retained the original phenotype. This palm was re-genotyped and proved to have a consistent genotype. It is plausible that the 9 palms remaining (or 2.7% of the genotyped population) had been misphenotyped originally, given that fruit form phenotyping error is believed to be in excess of 5% (see above), highlighting the need for a molecular assay which more accurately predicts fruit form. This assumption was confirmed in 6 of the 9 palms for which haplotypes were available, as haplotypes were consistent with genotype not phenotype, ruling out recombination as an explanation for discrepancy. The map expansion immediately around the shell gene is similarly explained by mis-phenotyped palms. That is, the 4 SNP markers closest to shell surround the SHP gene, but all have 9/238 “recombinants” including 6 of the 9 mis-phenotyped palms.
In situ Hybridization. 5 micron (µm) sections of tissues were mounted on Superfrost slides, dried overnight, and baked at 60°C. Sections were deparaffinized in xylene, immersed in 100% ethanol and dried. The sections received light treatment with protease. Two locked nucleic acid (LNA) probes to the target mRNA were designed and supplied by Exiqon (5’-DIG-ATTAACAAGCAGCGACATACTT and 5’-DIG-TTGATGGTGTGAATAGTGTTGT). A Scramble-miR negative control LNA probe was also provided by Exiqon (5’-DIG-GTGTAACACGTCTATACGCCCA). Optimized probe cocktail solution in Exiqon hybridization buffer was placed on the tissue sections. The sections were covered with polypropylene coverslips and heated to 60°C for 5 minutes followed by hybridization at 37°C overnight. Sections were washed in high stringency solution (0.2× SSC with 2% bovine serum albumin) at 60°C for 10 minutes. The LNA probes were detected using alkaline phosphatase conjugate (NBT/BCIP, blue precipitate). Sections were counterstained with Nuclear Fast Red. Sections were rinsed and mounted with coverslips. Aperio scans of the slides were made and images were extracted with ImageScope software.
Yeast two-hybrid assays
The coding sequences for oil palm ShDeliDura, ShMPOB, ShAVROS and rice OsMADS24 were synthesized as two ~300 bp gBlocks each that overlapped by 30 bp (Integrated DNA Technologies). Gibson assembly of the two fragments was performed using kit manufacturer’s protocols (NEB). EcoRI and BamHI sites were added to the gBlock sequences for simple ligation into MatchMaker Gold Yeast Two-Hybrid vectors. Each sequence was cloned into both the binding domain vector, pGBKT7, and the activation domain vector, pGADT7. Shell sequences encoded amino acids 2 to 175, including the entire MADS-box, I and K domains. The C domain was excluded from yeast two-hybrid constructs to avoid auto-activation of selection genes in the yeast two-hybrid system. The ShDeliDura peptide sequence encoded by the vectors was: GRGKIEIKRIENTTSRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSIRSTIDRYKKACANSSNSGATIEINSQQYYQQESAKLRHQIQILQNANRHLMGEALSTLTVKELKQLENRLERGITRIRSKKHELLFAEIEYMQKREVELQNDNMYLRAKIAEN. The ShMPOB peptide sequence encoded by the vectors was identical to the above sequence, with the exception that the underlined leucine residue (L) was converted to proline (P). The ShAVROS peptide sequence encoded by the vectors was identical to the above sequence, with the exception that the underlined lysine residue (K) was converted to asparagine (N). OsMADS24 sequences encoded amino acids 2 to 177, including the entire MADS-box, I and K domains, but excluding the C domain. The OsMADS24 sequence encoded by the vectors was: GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSGQSMTRTLERYQKFSYGGPDTAIQNKENELVQSSRNEYLKLKARVENLQRTQRNLLGEDLGTLGIKELEQLEKQLDSSLRHIRSTRTQHMLDQLTDLQRREQMLCEANKCLRRKLEES. Auto-activation control tests were performed by transforming each fusion vector into yeast alone, and each vector showed no auto-activation of selection reporter genes. Co-transformations were performed for all 16 pairwise combinations of BD and AD vectors and scored for growth on SD-Leu-Trp, SD-Lue-Trp-His, SD-Leu-Trp-His-Ade and X-gal media plates. Positive interactions were scored as blue co-tranformants (on X-gal plate) that were able to grow on SD-Lue-Trp-His and SD-Leu-Trp-His-Ade selection plates (Supplementary Fig. 5).
Supplementary Material
Acknowledgements
We thank Johannes Jansen (PRI, Wageningen, Netherlands) for assistance with JoinMap, and Beijing Genome Institute, The Genome Institute at Washington University, Tufts University Core Facility and the Arizona Genome Institute for mapping and genome sequencing. We thank Kulim, Sime Darby, FELDA Agricultural Services and Applied Agricultural Resources for providing materials and phenotype information of individual palms. Phylogeny, Inc. (Powell, OH, USA) performed in situ hybridization services. Creative Biolabs (Shirley, NY, USA) performed yeast two-hybrid co-transformations and interaction assays. We thank Jim Birchler and Zach Lippman for useful discussions on hybrid vigor. We appreciate the unflagging support from Datuk Dr. Choo Yuen May, Director General of MPOB as well as the Ministry of Plantation Industries and Commodities, Malaysia. The project was endorsed by the Cabinet Committee on the Competitiveness of the Palm Oil Industry (CCPO) and funded by MPOB. R.A.M. is supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation, and by a grant from NSF 0421604 "Genomics of Comparative Seed Plant Evolution".
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. R.S. initiated work on the shell marker/gene. R.S., E-T.L.L., M.O.A. and R.S.M. conceptualized the research programme. R.S.,E-T.L.L., M.O.A., R.N., M.A.A.M., N.L., S.W.S., J.M.O., R.S.M. and R.A.M. developed the overall strategy, designed experiments and coordinated the project. R.A.M. conceptualized the homozygosity mapping strategy. R.A.M., R.S. and R.N. identified samples for homozygosity mapping. R.N. and M.D.A. developed and maintained the mapping population and assisted in phenotyping. R.S., M.O.A., L.C.L.O, T.N.C., J.N., N.L., M.A.B., B.B., A.V.B., C.W., J.M.O. and R.S.M. conducted laboratory experiments and data analyses. R.S. and L.C.L.O. constructed the genetic map. E-T.L.L., R.R., K.L.C., M.A.H., N.A., S.W.S., M.H., C.W. and A.V.B. performed bioinformatics analyses. C.W., A.V.B, N.L., J.M.O, R.A.M., R.S. resequenced the candidate gene and characterized the mutations. R.S., E-T.L.L., M.O.A., R.N., N.L., S.W.S., J.M.O., R.S.M. and R.A.M. prepared and revised the manuscript.
E. guineensis and E. oleifera genome sequences have been deposited at DDBJ/EMBL/GenBank under the accessions ASJS00000000 and ASIR00000000, respectively. Gene sets available at genomsawit.mpob.gov.my.
R.A.M. is a consultant for Orion Genomics, LLC.
References
- 1.Janssens P. Le palmier à huile au Congo Portugais et dans l’enclave de Cabinda. Descriptions de principales Variétés de Palmier (Elaeis guineensis) Bull. Agric. Congo Belge. 1927;18:29–92. [Google Scholar]
- 2.Devuyst A. Selection of the oil palm (Elaeis guineensis) in Africa. Nature. 1953;172:685–686. doi: 10.1038/172685a0. [DOI] [PubMed] [Google Scholar]
- 3.Hartley C. In: The Oil Palm. Hartley C, editor. Longman; 1988. pp. 47–94. [Google Scholar]
- 4.Rajanaidu N, et al. In: Advances in Oil Palm Research. Basiron Y, Jalani BS, Chan KW, editors. Bangi, Selangor: Malaysian Palm Oil Board (MPOB); 2000. pp. 171–237. [Google Scholar]
- 5.Corley RHV, Tinker PB. The Oil Palm. Oxford: Blackwell Science; 2003. pp. 1–26. [Google Scholar]
- 6.Danielsen F, et al. Biofuel plantations on forested lands: double jeopardy for biodiversity and climate. Conserv Biol. 2009;23:348–358. doi: 10.1111/j.1523-1739.2008.01096.x. [DOI] [PubMed] [Google Scholar]
- 7.Beirnaert A, Vanderweyen R. Contribution a l’etude genetique et biometrique des varieties d’Elaeis guineensis Jacq. Publs. Inst. Nat. Etude agron. Congo Belge, Ser. Sci. 1941;27:1–101. [Google Scholar]
- 8.Godding R. Observation de la production de palmiers selectionnes a Mongana (Equateur) Bull Arig. Congo belge. 1930;21:1263. [Google Scholar]
- 9.Billotte N, et al. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.) Theor Appl Genet. 2005;110:754–765. doi: 10.1007/s00122-004-1901-8. [DOI] [PubMed] [Google Scholar]
- 10.Mayes S, Jack PL, Corley RH, Marshall DF. Construction of a RFLP genetic linkage map for oil palm (Elaeis guineensis Jacq.) Genome. 1997;40:116–122. doi: 10.1139/g97-016. [DOI] [PubMed] [Google Scholar]
- 11.Moretzsohn MC, Nunes CDM, Ferreira ME, Grattapaglia D. RAPD linkage mapping of the shell thickness locus in oil palm (Elaeis guineensis Jacq.) Theoretical and Applied Genetics. 2000;100:63–70. [Google Scholar]
- 12.Singh R, et al. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. doi: 10.1038/nature12309. (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajanaidu N, Rao V, Abdul Halim H, ASH O. Genetic resources: New developments in Oil Palm breeding. Elaeis. 1989;1:1–10. [Google Scholar]
- 14.Gschwend M, et al. A locus for Fanconi anemia on 16q determined by homozygosity mapping. Am J Hum Genet. 1996;59:377–384. [PMC free article] [PubMed] [Google Scholar]
- 15.Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 16.Favaro R, et al. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell. 2003;15:2603–2611. doi: 10.1105/tpc.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinyopich A, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, et al. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell. 1996;8:81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Immink RG, Kaufmann K, Angenent GC. The 'ABC' of MADS domain protein behaviour and interactions. Semin Cell Dev Biol. 2010;21:87–93. doi: 10.1016/j.semcdb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Bhasker S, Mohankumar C. Association of lignifying enzymes in shell synthesis of oil palm fruit (Elaeis guineensis--dura variety) Indian J Exp Biol. 2001;39:160–164. [PubMed] [Google Scholar]
- 21.Dinneny JR, Yanofsky MF. Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. Bioessays. 2005;27:42–49. doi: 10.1002/bies.20165. [DOI] [PubMed] [Google Scholar]
- 22.Vrebalov J, et al. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tani E, Polidoros AN, Tsaftaris AS. Characterization and expression analysis of FRUITFULL- and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiol. 2007;27:649–659. doi: 10.1093/treephys/27.5.649. [DOI] [PubMed] [Google Scholar]
- 24.Tani E, et al. Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol Biochem. 2009;47:690–700. doi: 10.1016/j.plaphy.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahn LM, et al. Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol Dev. 2006;8:30–45. doi: 10.1111/j.1525-142X.2006.05073.x. [DOI] [PubMed] [Google Scholar]
- 27.Dreni L, et al. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007;52:690–699. doi: 10.1111/j.1365-313X.2007.03272.x. [DOI] [PubMed] [Google Scholar]
- 28.Veitia RA, Vaiman D. Exploring the mechanistic bases of heterosis from the perspective of macromolecular complexes. FASEB J. 2011;25:476–482. doi: 10.1096/fj.10-170639. [DOI] [PubMed] [Google Scholar]
- 29.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 30.Jacquemard JC, Ahizi P. Induction of male inflorescences in pisifera palms. Oleagineaux. 1981;36:51–58. [Google Scholar]
Methods References
- 31.Nam J, et al. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA. 2004;101:1910–1915. doi: 10.1073/pnas.0308430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh R, et al. Mapping quantitative trait loci (QTLs) for fatty acid composition in an interspecific cross of oil palm. BMC Plant Biol. 2009;9:114. doi: 10.1186/1471-2229-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Ooigen JW. In: JoinMap®4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, editor. Netherlands: Wageningen; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.