The development of light-regulated processes represents a noninvasive means to exert a high level of spatial and temporal control over a chemical or biological system.[1] The ability to light regulate the activity of biologically relevant molecules by the installation of photolabile protecting groups that are removed upon irradiation with ultraviolet light has been demonstrated numerous times in this context. Examples of this approach include the regulation of gene expression,[2] protein production,[3] protein function,[4] as well as antisense,[5] DNA,[6] and RNA function.[7] Other caged biologically relevant small molecules include ATP, Ca2+, theophylline, and fatty acids.[8] However, the decaging process only allows for the irreversible activation or deactivation of the system under study, thus limiting the scope of its utility.
Many important processes, for example, information storage, signal transduction, signal processing, and gene expression all need to be regulated in a reversible fashion. An important example is the regulation of hox genes, which have been found to be turned on, off, and on again in model organisms during development.[9] The reversible regulation of these genes facilitates proliferation, differentiation, and morphogenesis of bone tissue and other tissue involved in digitation. Possessing a means of externally regulating these (or similar) events with Nature’s precision has the potential to aid in the understanding of fundamental developmental processes.[9]
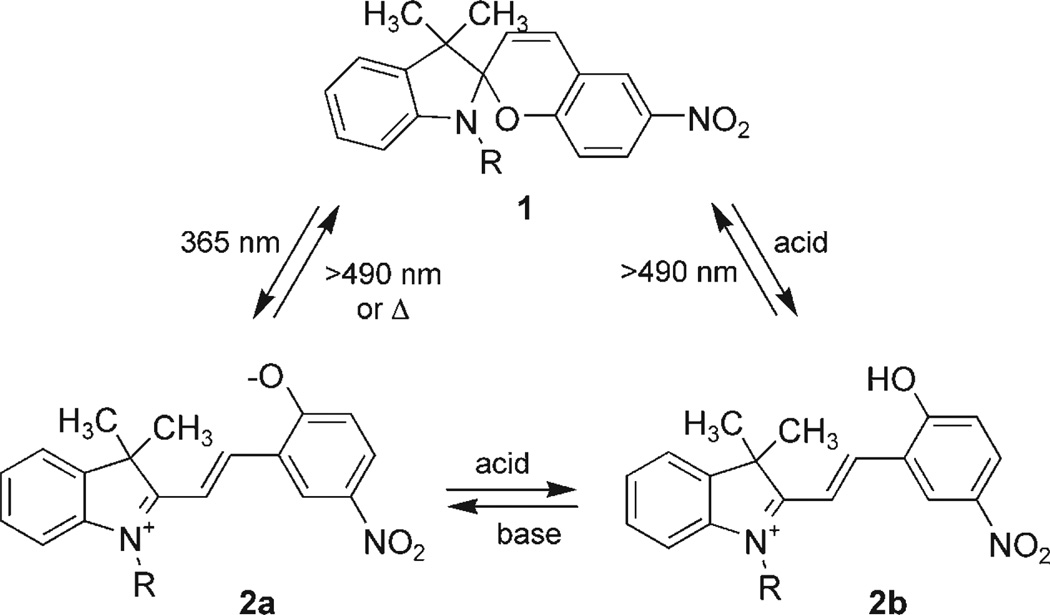
Obviously, Nature has evolved her own highly sophisticated mechanism to reversibly sense light using the photochemical cis→trans isomerization of retinal. This configurational change is detected by opsin proteins, which then trigger a signal transduction cascade.[10] Synthetic molecules that are isomerizable in a reversible fashion under light irradiation of different wavelengths have also been developed, including diazobenzenes, dihydropyrenes, spirooxazines, anthracenes, fulgides, and spiropyrans.[10–12] These molecules have been used in the investigation of transport channels, CAP binding affinity, papain activity, DNAzyme cleavage, and various other applications.[13] However, no synthetic reversible photochemical genetic switch has been demonstrated to date. We envisioned the design of such a switch by mimicking Nature, substituting the small molecule–protein interaction with a small molecule–RNA interaction. Here, we report the first step towards the goal of assembling a reversible, light-switchable gene control element at the RNA level by developing a light-switchable small-molecule–RNA aptamer pair.[14] A spiropyran (1) has been selected as the light-responsive molecule since it displays unique chemical properties.[10] Spiropyrans are both light and pH sensitive, exhibit distinctive chromophores in different forms (colorless, purple, and yellow) enabling a visual detection of the switching event (Scheme 1), and undergo rapid isomerization between the spiropyran (1) and the merocyanine form (2a and 2b) upon irradiation with UV light (or pH changes).[12, 15] These molecules have been extensively employed in the construction of nanoscale switches and photoisomerizable polymers for materials applications.[16, 17] As the second component of the switch, an RNA oligonucleotide has been found to adopt sequence specific tertiary structures capable of binding small molecules. These oligonucleotides are known as RNA aptamers.[18] By using the SELEX (systematic evolution of ligands by exponential enrichment) process, RNA aptamers have been engineered to selectively bind to a variety of different small molecules.[19]
Scheme 1.
Spiropyran 1 (colorless) and its merocyanine forms 2a (purple) and 2b (yellow).
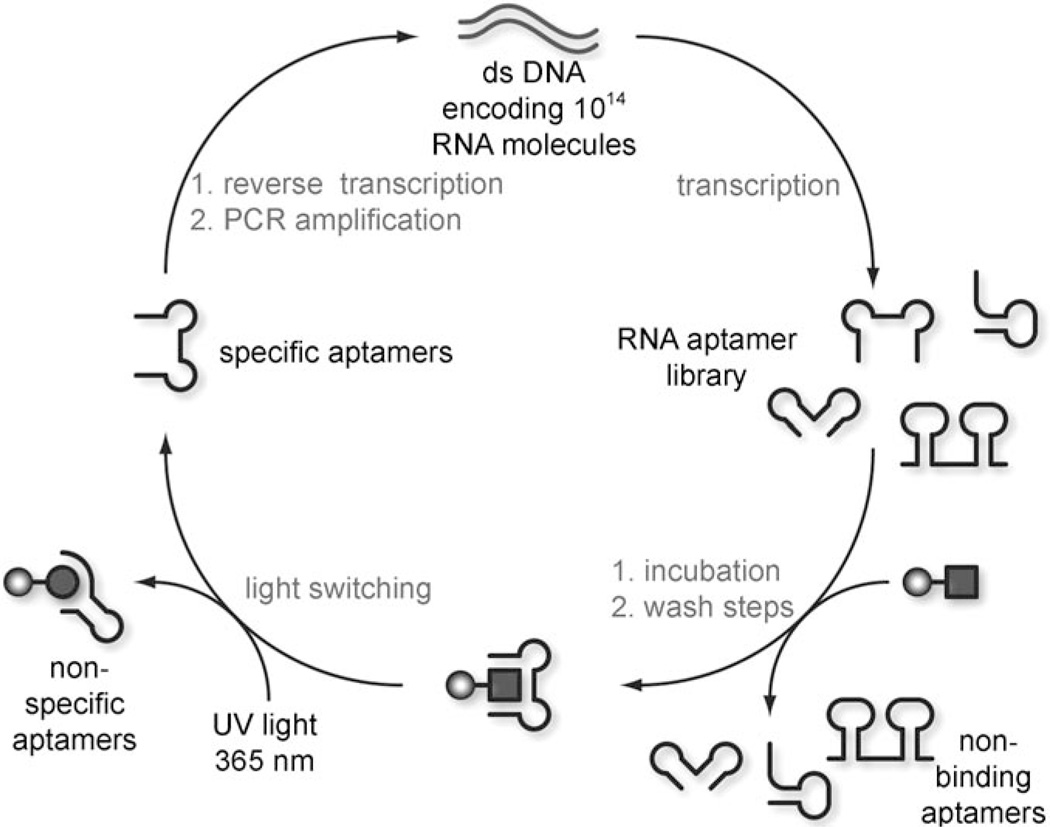
We applied this in vitro selection process in the evolution of an RNA aptamer capable of selectively recognizing only one photochemical isomer of a spiropyran molecule. The spiropyran 1 (R=(CH2)2O2C(CH2)2NH2) was immobilized on a sepharose resin, and subjected to ten rounds of affinity selection (Scheme 2). The selection was designed to employ the photoisomerization as part of the partitioning process.
Scheme 2.
Photochemical in vitro selection. An RNA library is incubated with a resin containing the spiropyran 1 (square). RNA aptamers incapable of binding are washed away and the resin is switched to the merocyanine form 2a (circle) with UV light of 365 nm. RNAs which are nonspecifically bound, or recognize both isomers are retained, whereas specific binders are eluted and collected. This enriched pool of RNA aptamers is then reverse transcribed, PCR amplified, and transcribed back to RNA to continue the cycle. The selection cycle was performed ten times until significant enrichment of the RNA pool was detected.
A 32P-labeled RNA library was incubated with resin carrying the spiropyran in its closed form 1, and nonbinding members were removed through subsequent washing of the resin. The resin was then subjected to irradiation with UV light at 365 nm for 1 h (25 W, hand-held UV lamp), resulting in a visible isomerization to the purple merocyanine 2a (such prolonged irradiation is not necessary with the nonimmobilized spiropyran as complete switching is observed within 2 min). The RNA aptamers that selectively bind only the closed isomer 1 dissociate from the resin and are collected, whereas nonspecific binders remain bound to the resin and are discarded. This feature of the selection obviates the need for the negative selection step traditionally required in in vitro evolution strategies. The collected aptamers were then reverse transcribed, amplified by PCR, and transcribed into RNA again to be taken through another round of selection. After ten rounds, an enrichment of ~13% of sequences carried through the selection was noted (as detected by radioactive quantitation by liquid scintillation counting), and the resulting aptamer pool was cloned and sequenced (see the Supporting Information). Sequencing results revealed five aptamer families with a certain degree of sequence homology, thus suggesting convergence of the selection. Several RNA aptamers were analyzed for their binding affinity and selectivity for the desired spiropyran isomer 1. Initial titration experiments were performed to assess the binding of individual sequences to the spiropyran. Whereas several of the isolated aptamers exhibited micromolar binding to the resin, to our surprise very few exhibited preferential binding to 1 over 2a (Supporting Information). Radioactively labeled RNA aptamers were incubated with resin carrying 1, which was then washed until no radioactivity was detected in the eluent. The resin was then suspended in binding buffer, and, after 1 h, the amount of RNA aptamer present in solution was determined on a scintillation counter. The suspended resin was then switched to 2a by irradiation at 365 nm (25 W, hand-held UV lamp), and after 1 h the solution was assayed for radioactivity. A randomly selected control sequence (CNTL) was determined to have no binding to either the spiropryan (in any form) or the sepharose resin, thus showing comparable levels of RNA present before and after irradiation. The other aptamers (SP3, SP4, SP5, and SP18) were found to bind immobilized 1. Of all the aptamers assayed, SP3 exhibited binding with the highest level of selectivity, as a significant portion of the RNA was released from the resin into solution upon switching from 1 to 2a. SP5 was capable of binding both photochemical states, 1 and 2a, of the spiropyran, but did not exhibit specificity. SP4 and SP18 exhibited some specificity for 1, however it was minimal compared to SP3.
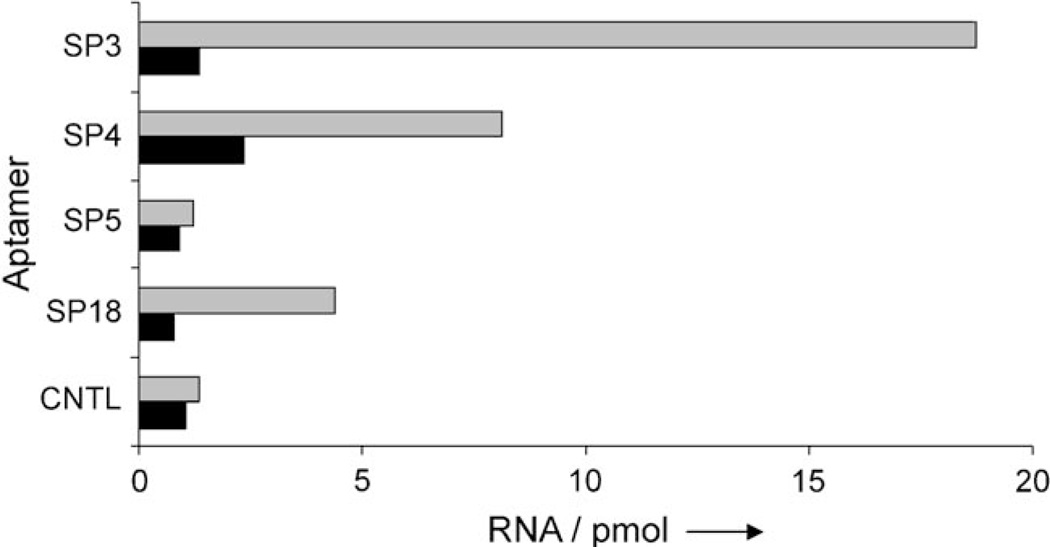
These experiments indicate that the SP3 aptamer (5’-GGAUUAACUCCUAUCCGAUUUGAAGCAGUACCCUAUUCCA-3’) is a selective binder for the closed spiropyran form 1 and does not bind to the merocyanine form 2a, as indicated by a marked 14-fold increase of radioactivity after resin switching (Figure 1). As SP3 demonstrated the best selectivity, its binding constant was determined to be 19 µm by both radioactive titration assays and equilibrium dialysis. We are aware that 1 is present as a racemate; this results in a binding constant of 10 µm in the case of enantioselective aptamer recognition.[20]
Figure 1.
Specificity assay for isolated aptamers. Isotopically labeled RNA was incubated with the spiropyran resin 1, then washed, resuspended in fresh buffer, and radioactivity in the supernatant was quantitated (dark bars), the resin was then switched to 2a through irradiation at 365 nm, and the supernatant was quantitated again (light bars). The RNA aptamer SP3 demonstrated the greatest specificity for spiropyran 1 over the merocyanine 2a. RNA=aptamer concentration.
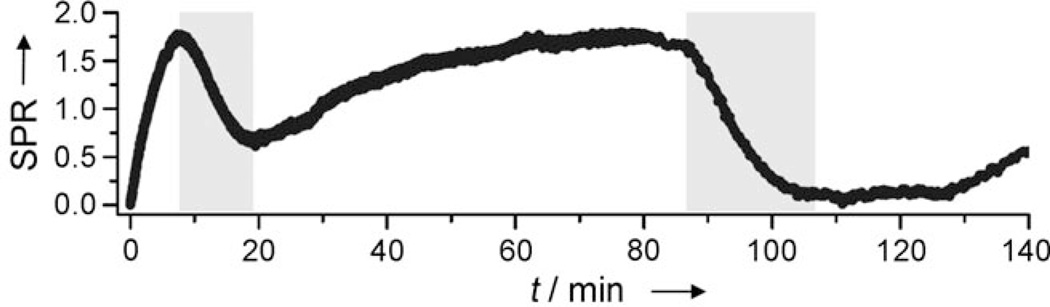
Following the identification of an aptamer sequence, the reversibility of the developed photochemical switch was demonstrated by surface plasmon resonance (SPR) studies. The spiropyran molecule 1 was modified with a poly(ethylene glycol) spacer and a thiol group (R=(CH2)2NHCO(CH2)2(OCH2)8-CH2)2SH, Supporting Information) and immobilized on a gold slide along with a mercaptohexanol control. The surface was then incubated with the SP3 aptamer (90 µm); this resulted in an increase of the SPR signal (the mercaptohexanol control did not display any signal change; data not shown). Subsequent irradiation with UV light of 365 nm for 10 min by using fiberoptics connected to a UV LED system (Prizmatix BLCC-02) led to a decrease in the SPR signal indicating a dissociation of the aptamer from the surface. This correlates to the isomerization event as the spiropyran is switched from closed form 1 to the open form 2a. After irradiation, the spiropyran moiety thermally reverts back to the closed form 1 in a gradual process, and rebinding of the aptamer is observed (Figure 2). This process was then repeated with a second 20 min irradiation event. Thermal, instead of photochemical switching of 2a to 1 was conducted because of an interference of white light irradiation and the SPR camera.
Figure 2.
Surface plasmon resonance experiment demonstrating the reversibility of the light-regulated SP3 aptamer binding. The spiropyran 1 was immobilized on a gold surface, incubated with the SP3 aptamer, and irradiated at 365 nm. SPR=change in percent reflectivity.
In summary, the engineered RNA aptamer SP3 is specific for one geometrical state of the spiropyran 1, and the binding event can be reversibly switched by using light irradiation, thus representing a unique tool in the development of biological switches. Over the course of our studies, two other photo-switchable molecule–aptamer pairs from a diazobenzene[21] and a dihydropyrene[22] have been reported, and effectively complement our switch. The advantages of spiropyran 1 over other reversible systems lie in its photochemical switching being virtually complete (>95% of 2a after UV irradiation at 365 nm) because of the distinctively different absorption maxima of 1 (350 nm) and 2a (563 nm),[23] unlike diazobenzenes, which reach a photostationary state of 70–90% cis when exposed to UV light of 365 nm.[24] The developed light-switchable small-molecule–aptamer pair has the potential to be developed into a photochemical riboswitch that can then be used to spatially and temporally regulate gene function in a reversible fashion. This switch also has potential in materials applications, including nanodevices and data-processing circuits and storage devices.[17, 25]
Experimental Section
SPR Protocol
The spiropyran 1 (1 µm, 3 × 1.5 µL) and a mercaptohexanol control (1 µm, 3 × 1.5 µL) were incubated on a gold surface (SPRchip; GWC Technologies) for 2 h to afford efficient immobilization. The surface was then washed with water (3 × 1 mL), and placed into the SPR imager (SPRimager II, GWC Technologies). The RNA aptamer was transcribed, purified, resuspended in binding buffer, and quantitated on a nanodrop spectrophotometer. Aptamer solution (ca. 100 µL, 30–96 µm) was injected into the SPR chamber and allowed to bind to the immobilized aptamer for 30 min. After binding had been detected, the gold slide was irradiated with UV light of 365 nm for approximately 10 min, dissociation and adsorption were monitored in real time, and the switching was repeated to demonstrate the reversibility of the aptamer binding.
Acknowledgements
D.D.Y. acknowledges a graduate research fellowship from the ACS Medicinal Chemistry Division. A.D. is a Beckman Young Investigator and a Cottrell Scholar. This research was supported in part by research grant no. 5-FY05-1215 from the March of Dimes Birth Defects Foundation and the NIGMS (1R01M079114). We gratefully acknowledge Dr. Lin He for assistance with the surface plasmon resonance measurements.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a) Young DD, Deiters A. Org. Biomol. Chem. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]; b) Dmochowski I, Tang XJ. Mol. Biosyst. 2007;3:B15. [Google Scholar]; c) Mayer G, Heckel A. Angew. Chem. 2006;118:5020–5042. [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]; d) Curley K, Lawrence DS. Curr. Opin. Chem. Biol. 1999;3:84–88. doi: 10.1016/s1367-5931(99)80015-5. [DOI] [PubMed] [Google Scholar]; e) Adams SR, Tsien RY. Annu. Rev. Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 2.a) Young DD, Deiters A. Angew. Chem. 2007;119:4368–4370. [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:4290–4292. doi: 10.1002/anie.200700057. [DOI] [PubMed] [Google Scholar]; b) Link KH, Shi YH, Koh JT. J. Am. Chem. Soc. 2005;127:13088–13089. doi: 10.1021/ja0531226. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Harada M, Sisido M, Hirose J, Nakanishi M. FEBS Lett. 1991;286:6–8. doi: 10.1016/0014-5793(91)80928-v. [DOI] [PubMed] [Google Scholar]; d) Lin WY, Albanese C, Pestell RG, Lawrence DS. Chem. Biol. 2002;9:1347–1353. doi: 10.1016/s1074-5521(02)00288-0. [DOI] [PubMed] [Google Scholar]; e) Cambridge SB, Geissler D, Keller S, Curten B. Angew. Chem. 2006;118:2287–2289. doi: 10.1002/anie.200503339. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:2229–2231. doi: 10.1002/anie.200503339. [DOI] [PubMed] [Google Scholar]
- 3.Goard M, Aakalu G, Fedoryak OD, Quinonez C, St Julien J, Poteet SJ, Schuman EM, Dore TM. Chem. Biol. 2005;12:685–693. doi: 10.1016/j.chembiol.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 4.a) Wu N, Deiters A, Cropp TA, King D, Schultz PG. J. Am. Chem. Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]; b) Deiters A, Groff D, Ryu YH, Xie JM, Schultz PG. Angew. Chem. 2006;118:2794–2797. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:2728–2731. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]; c) Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat. Chem. Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 5.a) Shestopalov IA, Sinha S, Chen JK. Nat. Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]; b) Tang XJ, Maegawa S, Weinberg ES, Dmochowski IJ. J. Am. Chem. Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]; c) Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Lusic H, Young DD, Lively MO, Deiters A. Org. Lett. 2007;9:1903–1906. doi: 10.1021/ol070455u. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heckel A, Mayer G. J. Am. Chem. Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]; c) Tang X, Dmochowski IJ. Org. Lett. 2005;7:279–282. doi: 10.1021/ol047729n. [DOI] [PubMed] [Google Scholar]
- 7.a) Mikat V, Heckel A. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shah S, Rangarajan S, Friedman SH. Angew. Chem. 2005;117:1352–1356. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]; c) Ando H, Furuta T, Tsien RY, Okamoto H. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 8.a) Young DD, Deiters A. Bioorg. Med. Chem. Lett. 2006;16:2658–2661. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]; b) Xia J, Huang XP, Sreekumar R, Walker JW. Bioorg. Med. Chem. Lett. 1997;7:1243–1248. [Google Scholar]; c) Berger CL, Svensson EC, Thomas DD. Proc. Natl. Acad. Sci. USA. 1989;86:8753–8757. doi: 10.1073/pnas.86.22.8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salser SJ, Kenyon C. Development. 1996;122:1651–1661. doi: 10.1242/dev.122.5.1651. [DOI] [PubMed] [Google Scholar]
- 10.Dugave C, Demange L. Chem. Rev. 2003;103:2475–2532. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- 11.a) Pieroni O, Fissi A, Angelini N, Lenci F. Acc. Chem. Res. 2001;34:9–17. doi: 10.1021/ar990141+. [DOI] [PubMed] [Google Scholar]; b) Willner I. Acc. Chem. Res. 1997;30:347–356. [Google Scholar]; c) Kumar GS, Neckers DC. Chem. Rev. 1989;89:1915–1925. [Google Scholar]
- 12.Berkovic G, Krongauz V, Weiss V. Chem. Rev. 2000;100:1741–1753. doi: 10.1021/cr9800715. [DOI] [PubMed] [Google Scholar]
- 13.a) Bose M, Groff D, Xie JM, Brustad E, Schultz PG. J. Am. Chem. Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]; b) Liu Y, Sen D. J. Mol. Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]; c) Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Borisenko V, Burns DC, Zhang ZH, Woolley GA. J. Am. Chem. Soc. 2000;122:6364–6370. [Google Scholar]; e) Willner I, Rubin S, Riklin A. J. Am. Chem. Soc. 1991;113:3321–3325. [Google Scholar]
- 14.a) Bauer G, Suess B. J. Biotechnol. 2006;124:4–11. doi: 10.1016/j.jbiotec.2005.12.006. [DOI] [PubMed] [Google Scholar]; b) Mandal M, Breaker RR. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]; c) Winkler WC, Breaker RR. ChemBioChem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 15.a) Sakata T, Yan YL, Marriott G. J. Org. Chem. 2005;70:2009–2013. doi: 10.1021/jo048207o. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Giordani S, Cejas MA, Raymo FM. Tetrahedron. 2004;60:10973–10981. [Google Scholar]; c) Asanuma H, Shirasuka K, Yoshida T, Takarada T, Liang XG, Komiyama M. Chem. Lett. 2001:108–109. [Google Scholar]
- 16.Raymo FM. Adv. Mater. 2002;14:401. [Google Scholar]
- 17.Raymo FM, Giordani S. J. Am. Chem. Soc. 2001;123:4651–4652. doi: 10.1021/ja005699n. [DOI] [PubMed] [Google Scholar]
- 18.a) Isaacs FJ, Dwyer DJ, Collins JJ. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]; b) Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]; c) Famulok M. Curr. Opin. Struct. Biol. 1999;9:324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 19.a) Osborne SE, Ellington AD. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]; b) Gold L, Brown D, He YY, Shtatland T, Singer BS, Wu Y. Proc. Natl. Acad. Sci. USA. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 20.Eulberg D, Klussmann S. ChemBioChem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi G, Hagihara M, Dohno C, Nakatani K. J. Am. Chem. Soc. 2007;129:8678. doi: 10.1021/ja071298x. [DOI] [PubMed] [Google Scholar]
- 22.Lee HW, Robinson SG, Bandyopadhyay S, Mitchel RH, Sen D. J. Mol. Biol. 2007;371:1163–1173. doi: 10.1016/j.jmb.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 23.Raymo FM, Giordani S, White AJ, Williams DJ. J. Org. Chem. 2003;68:4158–4169. doi: 10.1021/jo0340455. [DOI] [PubMed] [Google Scholar]
- 24.Fischer E, Frankel M, Wolovsky R. J. Chem. Phys. 1955;23:1367–1367. [Google Scholar]
- 25.Mir M, Katakis I. Mol. BioSyst. 2007;3:620–622. doi: 10.1039/b708858b. [DOI] [PubMed] [Google Scholar]