Highlights
-
•
We report the rational design of structure-switching DNA aptamers for potassium.
-
•
The shift between non-binding and binding-competent states was determined experimentally.
-
•
The stability of the non-binding state was estimated computationally.
-
•
A linear free energy relationship between these values was established.
Abbreviations: CD, circular dichroism; GQS, G-quartet sequence; TBA, thrombin binding aptamer
Keywords: Structure-switching aptamer, Thrombin-binding aptamer, G-quartet, Population-shift mechanism, mFold
Abstract
Structure-switching molecules provide a unique means for analyte detection, generating a response to analyte concentration through a binding-specific conformational change between non-binding and binding-competent states. While most ligand-binding molecules are not structure switching by default, many can be engineered to be so through the introduction of an alternative non-binding (and thus non-signalling) conformation. This population-shift mechanism is particularly effective with oligonucleotides and has led to the creation of structure-switching aptamers for many target ligands. Here, we report the rational design of structure-switching DNA aptamers, based on the thrombin binding aptamer (TBA), that bind potassium with affinities that bridge the gap between previously reported weak-binding and strong-binding aptamers. We also demonstrate a correlation between the free energy of the experimentally determined binding affinity for potassium and the computationally estimated free energy of the alternative (non-binding) structure.
1. Introduction
Structure-switching molecules generate a signal in response to a binding-specific conformational change, Fig. 1A. In the absence of target ligand, the molecule adopts a molecular shape that is not competent for ligand-binding and is thus non-signalling. The switch between this non-binding state and a binding-competent state is controlled by a switching equilibrium constant, KS. The binding-competent state has an intrinsic affinity for ligand, defined by the dissociation constant, . Stabilization of this binding-competent state by ligand thus serves to quantitatively signal its presence. This population-shift mechanism has been well described for previous systems in vitro [1–4] and riboswitches, characterized only in the past decade [5,6], represent an ideal example of the power of the population-shift mechanism in nature.
Fig. 1.
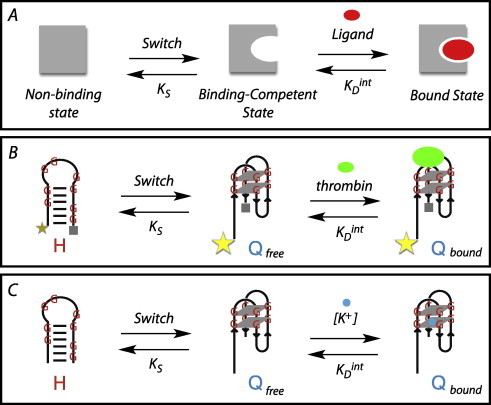
Structure-switching molecules operate via a population-shift mechanism. (A) Structure-switching molecules can exist in two conformations – a non-binding (and thus non-signalling) conformation, and a binding-competent state. The partitioning between these two states is dictated by a switching equilibrium constant, KS, which reflects the inherent stability of the non-binding state, as well as the intrinsic affinity for the target ligand, as defined by the dissociation constant, . In the absence of ligand, partitioning is dominated by KS, maintaining the molecule in a non-binding conformation. In the presence of ligand, becomes an important contributor to partitioning between non-binding and bound states. The observed binding affinity for a structure-switching molecule, , reflects contributions from both KS and . In this way, ligand presence facilitates the capture of molecules in the binding-competent state, transitioning the structure-switching molecule to a bound (signalling) state in a concentration-dependent fashion. (B) Ellington and co-workers have modified the thrombin binding aptamer (TBA) into a structure-switching DNA aptamer (19), which partitions between a non-binding hairpin conformation (H) and the binding-competent state (Qfree), which is captured by thrombin protein as a G-quadruplex motif (Qbound). Sequences have affinities for thrombin that reflect the inherent stability of hairpin conformations. Formation of the G-quadruplex motif separates the fluorophore (star) from the quencher (square), which is detectable by fluorescence spectroscopy. (C) By way of its G-quartet sequence, TBA is also an aptamer for potassium, and is similarly partitioned as in (B), and captured as a G-quadruplex motif (Qbound) by potassium ions.
The observed binding affinity for a structure-switching molecule, , reflects contributions from both KS and . Greater stability of the non-binding state (i.e., larger KS values) is reflected in larger values, requiring a larger concentration of ligand to populate the signalling state. This molecular switching can be coupled to a number of specific optical, electrochemical, and biochemical outputs, including FRET or fluorescence emission, making it an extremely versatile tool in the laboratory (reviewed in [3]).
The optimal sensitivity (change in signal/change in target concentration) of a structure-switching molecule can be achieved by optimizing the thermodynamics of switching between a binding-competent and alternatively folded structure. While this “tuning” of the observed binding affinity can be done by identifying aptamers with inherently different values [7] an alternative approach is to manipulate KS [8]. Specifically stabilizing or destabilizing either the binding-competent state or the non-binding conformation can modulate KS. By manipulating KS in this way, Plaxco and co-workers have expanded, narrowed, and edited the dynamic ranges of several structure-switching molecules [9–12].
Aptamers are nucleic acid sequences that have been selected for their ability to bind specific molecular targets, which can range from small organic molecules to large proteins [13]. Their binding affinities (KD values) range from 1 pm to several μM, but generally fall in the nanomolar range. In this way, nucleic acid aptamers have the ability to bind analytes with specificities previously reserved for proteins, but come with the added advantage of in vitro engineering, ease of chemical synthesis, long-term storage viability, and limited immunogenicity for therapeutic applications [14–16].
Previous work by Toole and co-workers has identified a 15-nucleotide DNA aptamer capable of binding the protein thrombin with high affinity (i.e., is small; 25–200 nM) [17]. A closer examination of the structure of this thrombin binding aptamer (TBA) revealed a G-quartet sequence (GQS). A G-quartet (a.k.a. quadruplex) is a structure formed when four-strands of DNA (or RNA) associate to form stacks of G-quartets (platforms) (Fig. 1B and Q) [18].
Most aptamers are not inherently structure-switching – that is they do not produce a significant conformational change, and thus a readily measurable signal, upon target binding. Thus, an inherent problem in designing structure-switching molecules is the creation of an alternative non-binding conformation. In later work by Ellington and co-workers, the thrombin binding aptamer was redesigned as a structure-switching molecule capable of signalling the presence of thrombin using fluorescence spectroscopy [19], Fig. 1B. This was done by the addition of nucleotides on the 5′-end that were complementary to the 3′-end of the TBA sequence, forcing the sequence into a stem–loop structure in the absence of the thrombin ligand (H). The addition of thrombin ligand allowed for a conformational change into the binding-competent state (Q), which was signalled by a change in fluorescence intensity from a fluorophore attached to the 5′-end.
Significant structural stability of quadruplexes comes from base-stacking, hydrogen bonding interactions, and coordination with metal cations, and there have been extensive structural studies on this thrombin binding aptamer [20–25]. For many guanine quadruplexes, including TBA and these structure-switching molecules derived from it, the cation of choice is potassium. Thus, a G-quartet is inherently an aptamer for K+.
While progress has been made on developing DNA quadruplex-forming sequences into potassium sensors [26–33], little work has been done ‘fine-tuning’ these sensors to have differing sensitivities (i.e., different values). In the present work, we examine the ability of previously studied thrombin binding aptamers to serve as a structure-switching aptamers for potassium (Fig. 1C). The 15-nucleotide TBA sequence, containing no additional nucleotides to afford an alternative non-binding conformation should represent the lower limit of potassium binding (e.g., in Fig. 1C). Work by Gross and co-workers have determined the KD of this strong-binding sequence to be 5 μM [34]. Two structure-switching thrombin aptamers examined by Ellington and co-workers [19] would be predicted to exist primarily in a non-binding conformation in the absence of potassium (H, in Fig. 1C). As such, these weak-binding aptamers would be predicted to require significantly greater potassium concentrations to populate the bound state (Q). While these structure-switching aptamers have been studied for their ability to bind the protein thrombin, potassium-binding studies have not been reported in the literature.
Here, we use computationally estimated free energies of the non-binding hairpin states to design three new G-quadruplex conformational switches with binding affinities to predictably fill in the gap between the previously reported strong- and weak-binding potassium-binding DNA aptamers. This project aims to correlate the experimentally determined for potassium-induced G-quartet formation with the computationally estimated stability associated with an alternative hairpin structure [35,36]. Correlation of these experimental and computational parameters will allow for the design of a G-quartet sequence with a predictable potassium-binding affinity.
2. Materials and methods
2.1. DNA preparation
DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. Sequences and their abbreviations are provided in Table 1 whereby the first number indicates the total number of nucleotides, the second indicates the number of nucleotides in the loop, and the last indicates the number of base pairs in the stem. For example, 20-10-5 is a 20-nucleotide hairpin loop with 10 nucleotides in the loop and 5 base pairs in the stem. These six sequences represent three newly designed oligos which were based on previous research on the thrombin binding aptamer (TBA) [34] and two previously studied structure-switching analogs (referred to herein as 20-10-5 and 19-11-4) [19]. Oligonucleotides were prepared by resuspending the lyophilized DNA in distilled and autoclaved water and quantitated using standard procedures.
Table 1.
Sequence information for structure-switching DNA aptamers for potassium.
| Oligonucleotide | Sequence a | ε260, L (mol strand)−1 cm−1b |
|---|---|---|
| 20-10-5 | 5′-ccaacGGTTGGTGTGGTTGG-3′ | 188,600 |
| 19-11-4 | 5′-ccaaGGTTGGTGTGGTTGG-3′ | 181,800 |
| 20-11-4-5′A | 5′-accaaGGTTGGTGTGGTTGG-3′ | 195,600 |
| 20-11-4-5′C | 5′-cccaaGGTTGGTGTGGTTGG-3′ | 189,000 |
| 18-11-3 | 5′-caaGGTTGGTGTGGTTGG-3′ | 174,600 |
| TBA | 5′-GGTTGGTGTGGTTGG-3′ | 143,300 |
Quartet-forming nucleotides are shown in uppercase, and flanking sequence is shown in lowercase.
Provided by Integrated DNA Technology.
2.2. Determination of hairpin stability using mFold
Stabilities of hairpins were computationally estimated using the DNA folding form within the program mFold [35] and the following parameters: linear DNA, 22 °C folding temperature, 10 mM Na+, and no changes to the remaining default parameters. These parameters were chosen to most closely match the conditions at the start of CD titration experiments (22 °C, 10 mM Li+). In situations where more than one stable fold was predicted (as was the case for five of the six oligos studied), the free energy value for the most stable fold was used. For all cases, as there was at least 1 kcal/mol difference between the free energy value reported and that for the nearest less stable fold (representing at least a 10-fold difference in contribution of this fold to the overall population), this approach is justified. While a more rigorous treatment of the thermodynamic data was undertaken using a binding polynomial approach (data not shown), the results obtained were not different than those determined using the more simplified method.
2.3. Circular dichroism (CD)
CD spectroscopy was performed using a Jasco CD J810 Spectropolarimeter, and data were analyzed with Jasco Spectra Manager Suite software [37]. DNA oligonucleotides were prepared to a concentration of either ∼5 or 15 μM in 10 mM LiCacodylate or 20 mM Tris (pH 7.0) buffer. DNA was renatured at 90 °C for 5 min and allowed to cool at room temperature. Spectra were acquired at 22 °C every nm from 210 to 310 nm with a bandwidth of 1 nm and a response time of 1 s/nm. Data were buffer subtracted and normalized to provide molar residue ellipticity values.
Titrations were performed with KCl to determine the amount of K+ necessary for G-quadruplex formation. To determine values, ellipticity data at λmax for a particular sequence were fit with KaleidaGraph v. 4.5.2 (Synergy software) according to the two-state Hill equation:
| (1) |
where εQ is the normalized CD signal corresponding to the fully folded GQS; εH is signal for the hairpin state; [K+] is the potassium ion concentration, is the potassium ion concentration needed to fold half the DNA, and n is the Hill coefficient. In addition, some data were plotted as fraction folded plots, calculating fraction folded as follows:
| (2) |
and were fit using the following equation:
| (3) |
the standard free energy (ΔG) of the folding transition of a given GQS can be calculated from the relationship:
| (4) |
where R is the gas constant and T is the temperature in K, and [Q]/[H] represent the concentrations of quartet and hairpin states respectively. This expression can be evaluated at any given potassium ion concentration using the and Hill coefficient (n) to obtain ΔGobs, the observed free energy of the folding transition [38]:
| (5) |
this equation corresponds to an observed free energy in K+ concentration, which couples folding and ion binding free energies. Data shown in Table 2 were obtained from fitting using Eq. (1) and calculation of ΔGobs using Eq. (5); the errors listed are those reported from the curve fitting.
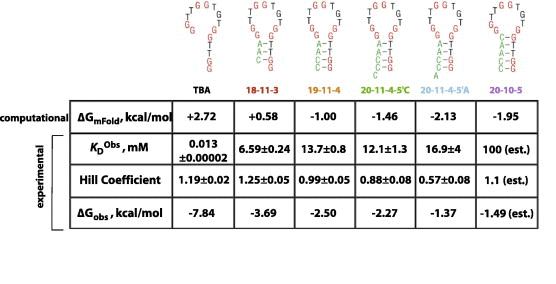
Table 2.
Computationally estimated and experimental data for structure-switching DNA aptamers for potassium. Computational data were obtained for all six sequences using the program mFold. ΔGmFold are the free energy values reported for the folded structures provided by the program and using parameters that most closely resemble those experienced during experimental conditions. Experimental values were determined by monitoring changes in molar ellipticity at 295 nm by CD potassium titrations. These data were fit to a two-state Hill equation to obtain and Hill coefficient (n) values. These two values were then used to calculate ΔGobs for all six sequences. Errors reported are those from data fitting. Data for 20-10-5 were estimated based on CD data as there was an insufficient upper baseline for data fitting.
 |
3. Results
3.1. Design of structure-switching DNA aptamers for potassium
In the present work, a systematic approach was taken to rationally design three new structure-switching DNA aptamers with smaller or larger KS values (i.e., more stable or weaker alternative structures) than three previously studied aptamers, Fig. 2. For all sequences, quartet-forming sequence is shown in red and black, and structure-switching sequence is in green. The 15-nucleotide thrombin binding sequence (TBA) is predicted to have the strongest binding to potassium ions [34], lacking any sequence capable of forming an alternative structure. The structure-switching DNA aptamers (20-10-5, and 19-11-4) [19], are predicted to have larger KS values, binding potassium only at high concentrations due to the population of a non-binding hairpin conformation. Nucleotides were added or removed to strengthen (larger KS) or weaken (smaller KS) the hairpin folded state, respectively, creating the three new aptamers. It should be noted that for any constructs involving a redesign of the stem structure, modifications were only made to the portion of the stem that is not the potassium-binding motif. That is, the quartet-forming sequence remains identical in all six structures. Sequences other than the minimal thrombin binding aptamer (TBA) sequence are identified by a numerical code indicating the total number of bases, bases in the loop domain, and bases in the hairpin stem. For example, 20-11-4-5′A is a 20 nucleotide stem–loop containing 11 nucleotides in the loop domain and closed by a 4 base-pair stem with an unpaired adenosine on the 5′-end (a dangling A).
Fig. 2.
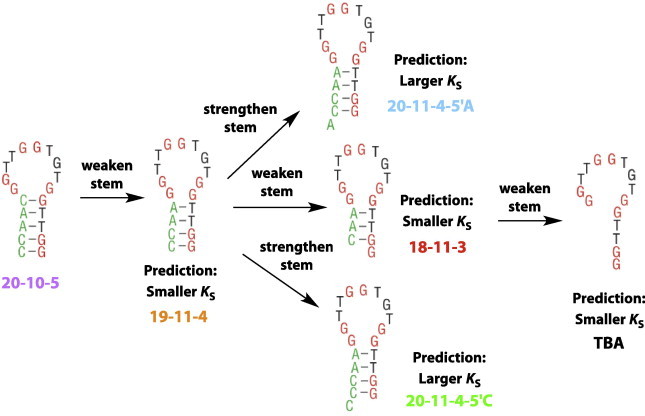
Structure-switching DNA aptamers for potassium. Beginning with three previously studied aptamers that represent the predicted lower (TBA) and upper (20-10-5 and 19-11-3) limits of potassium binding, three additional sequences were created by either weakening the stem (predicted to decrease KS) or strengthening the stem (predicted to increase KS). These sequences are named using a code indicating the total number of nucleotides, nucleotides in the loop domain, and number of base pairs in the stem. For example 20-11-4-5′C contains 20 total nucleotides, 11 nucleotides in the loop domain, 4 base-pairs in the stem, and an unpaired cytosine residue on the 5′-end. Each sequence name is also color-coded to correlate with data presented throughout the paper.
3.2. Computational estimation of ΔGmFold (KS values) for structure-switching DNA aptamers for potassium
Thermodynamic parameters (ΔGmFold, kcal/mol) were computationally estimated for the six sequences in Fig. 2 using the program mFold [35] and parameters most comparable to conditions used experimentally (10 mM monovalent ion, 22 °C). As this program is only capable of examining secondary structure formation, values reported are only for possible stem–loop structures and not for any structures arising from quadruplex formation. These values are reported in Table 2. The expected trend is observed. TBA, containing few switch-forming nucleotides, reported the least stable alternative structure (i.e., smallest KS) with sequences containing 3, 4, or 5 base-pair stems report more stable alternative structures (i.e., larger KS values).
It is worth noting that the predicted structures in Fig. 2 are based on possible base-pairing options. While these proposed two-dimensional foldings are based on mFold possible structures, the actual structures may be more complex [39].
3.3. Experimental determination of ΔGobs (via and Hill coefficient values) for structure-switching DNA aptamers for potassium
Potassium-induced structure switching between hairpin and quartet states was examined using circular dichroism spectroscopy, monitoring the change in molar ellipticity as a function of potassium concentration between 210 and 310 nm. There is a clear distinction between CD signals for the hairpin and quartet states, with the greatest signal changes occurring at 240, 270, and 295 nm, Fig. 3A.
Fig. 3.
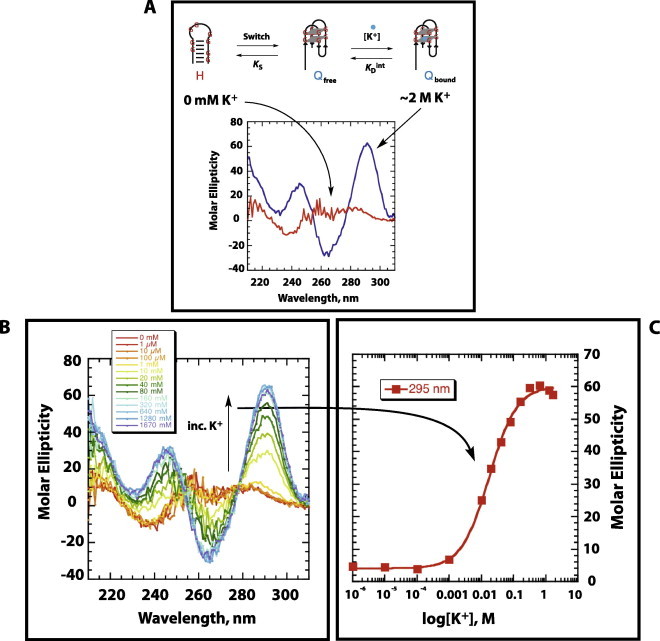
Determination of and Hill coefficient (n) values using circular dichroism (CD) spectroscopy. (A) Circular dichroism spectroscopy was used to “signal” the presence of a non-binding hairpin state (H) or the G-quadruplex state (Q). In the absence of potassium, conformational switching favors the hairpin state, generating a specific CD signal (red). Addition of saturating levels of potassium ion allow for the conformational switch to the quartet state (blue). (B) Monitoring potassium titrations by CD show changes in molar ellipticity at three wavelengths; 240, 270, and 295 nm. (C) The increase in molar ellipticity at 295 nm is evaluated as a function of potassium concentration and fit to a two-state Hill equation to determine and Hill coefficient (n) values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
This change in CD signal was monitored over several additions of potassium ion, Fig. 3B. The change in molar ellipticity at 295 nm as a function of potassium concentration was plotted and fit to a two-state hill equation, Fig. 3C, to determine and Hill coefficient values, which were then used to calculate ΔGobs. While changes in hyperchromicity for hairpin to quartet transitions at wavelengths of 240, 270, and 295 nm have been well documented [19,40], for our data, the signal change of greatest magnitude and with the least amount of signal noise occurred at 295 nm. Indeed, while all three wavelengths provided experimental KD and Hill coefficient values that were nearly identical within the error of the data fitting (data not shown), the magnitude of the error associated with the values obtained from the 295 nm data were 5–10-fold smaller than that from the 240 or 270 nm data. Thus, it is reasonable to assume that the fitting from the 295 nm data most accurately reflects the experimental binding constants for these aptamers.
Potassium titrations were performed for all six sequences, and the CD traces and curve fits are shown in Fig. 4. , Hill coefficient, and ΔGobs values are reported in Table 2. A summary plot showing fraction folded curves (which normalizes data to allow for direct comparison of all six sequences) is shown in Fig. 5. Again, the expected trend was observed. TBA, containing few switch-forming nucleotides and the smallest computationally estimated KS value reported the smallest and ΔGobs values. The potassium binding constant for this strongest binder ( = 13 μM) is consistent with previously reported values of 5 μM [34]. Sequences containing 3, 4, or 5 base-pair stems reported more stable alternative structures (i.e., larger and more negative ΔGobs values).
Fig. 4.
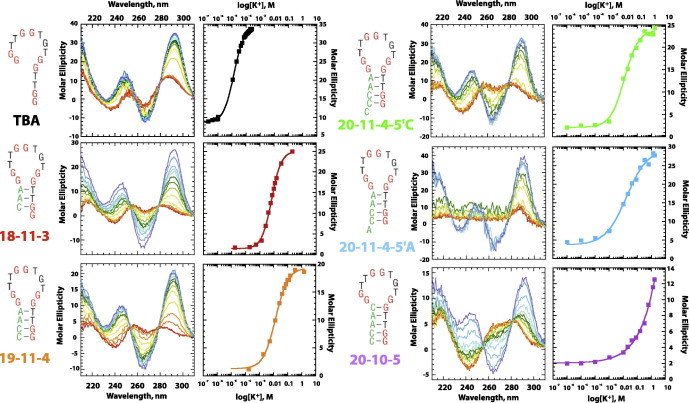
Determination of and Hill coefficient (n) values for structure-switching DNA aptamers for potassium. As per Fig. 3, for each sequence studied, potassium titrations were monitored by CD. and Hill coefficients (n) were obtained by plotting the change in molar ellipticity at 295 nm as a function of potassium concentration, and fitting the data to a two-state hill equation.
Fig. 5.
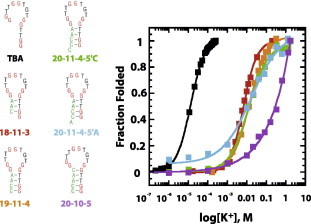
Fraction folded plots for structure-switching DNA aptamers for potassium. Data from Fig. 4 were used to generate fraction folded plots for all six sequences.
3.4. Correlation of computationally estimated (ΔGmFold) and experimentally determined (ΔGobs) thermodynamic parameters
To determine the extent of correlation between computationally estimated (ΔGmFold) and experimentally determined (ΔGobs) parameters, the observed free energy of potassium binding (as determined by circular dichroism) was plotted as a function of the free energy of folding for each of the six sequences, Fig. 6. Consistent with both computational and experimental data following expected trends, a linear free energy relationship is evidenced by the high degree of correlation between these values (R2 = 0.9789), establishing the validity of this approach.
Fig. 6.
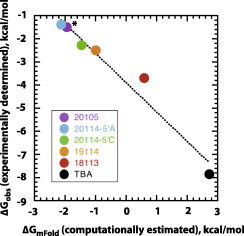
Linear free energy relationship between experimental (ΔGobs) and computationally estimated (ΔGmFold) data. Free energy data reported in Table 2 were plotted to obtain a linear correlation (y = −3.88x − 1.27, R2 = 0.9789).
4. Discussion
Structure-switching molecules, which couple a binding-specific conformational change to a reportable signal (Fig. 1A) present a new quantitative and versatile method of analyte detection. Work by Plaxco and co-workers has demonstrated the ability to rationally fine-tune aptamer binding properties by optimizing the thermodynamics of switching between binding-competent and alternatively folded states. This optimization has generally resulted from changes that modulate KS, the switching equilibrium constant (Fig. 1A), and has resulted in structure-switching aptamers with altered dynamic ranges for substrate binding [1,3,8–12].
The concept of a structure-switching aptamer was utilized by Ellington and co-workers to transform the 15-nucleotide DNA aptamer for the protein thrombin into two structure-switching DNA aptamers, capable of detection of their target protein ligand [19] (Fig. 1, and 20-10-5 and 19-11-4 in Fig. 2). As the central structural motif in these structure-switching aptamers is a G-quadruplex sequence (GQS), which also has an affinity for potassium ions, these DNA sequences are also structure-switching aptamers for potassium ion (Fig. 1C).
The 15-nucleotide thrombin binding aptamer (TBA) sequence is predicted to have a strong affinity for potassium, as the lack of any flanking sequence prevents the population of a non-binding conformation. Consistent with this, Gross and workers have reported the of this strong binder to be 5 μM [34]. Structure-switching TBA aptamers, populating a non-binding state in the absence of potassium ions, are predicted to have a weaker affinity for potassium ions, although these data have not been previously reported in the literature. The creation of a more stable alternative structure (H in Fig. 1C) increases KS, decreasing the population of aptamer in a binding-competent state (Qfree in Fig. 1C), requiring a higher ligand concentration for signal change. Thus, while the intrinsic affinity () of the binding-competent state has not necessarily changed from that of the unextended TBA sequence, what has changed by modulating Ks is the fraction of molecules in this state. Much like is the case for competitive inhibition with enzymes, an unfavorable off-pathway equilibrium (e.g., formation of an alternative structure) can be countered by an increase in ligand concentration. Thus, the observed affinity of the aptamer for its ligand () will change with changes in KS. Work by Plaxco and co-workers has shown a correlation between these two parameters, determining KS by urea melting and by fluorescence intensity increases [8].
As is demonstrated in the present work, structure-switching nucleic acid aptamers are particularly amenable to rational design using computational methods to predict the thermodynamics of switching between a binding-competent and alternatively folded structure. This has great value in creating an aptamer that operates within a given range of analyte concentration. That is, the sensitivity (change in signal/change in target concentration) can be tuned to the desired range of analyte concentration. The dynamic range for potassium ion detection by thrombin binding aptamer sequences was established by previously studied sequences. The lower limit (strong-binding) was set at 5 μM by the 15-nucleotide TBA sequence [34] while the upper limit (weak-binding) was set by the structure-switching aptamers used by Ellington and co-workers [19], although potassium binding constants for these sequences have not been reported in the literature. In the present work, the gap between these upper and lower limits was filled using structure-switching DNA aptamers with potassium binding constants that were predicted based on the stabilities of their non-binding alternative conformations.
Using computationally estimated free energies of the non-binding hairpin states, we have designed three new structure-switching DNA aptamers with different stabilities of off-pathway folds (i.e., different KS values, Fig. 2). The observed affinity for potassium ion () was evaluated experimentally by CD titration experiments, Figs. 3–5. Computationally estimated and experimentally determined free energy data is shown in Table 2 and followed the predicted trends. The linear free energy relationship between these data (Fig. 6) was substantiated by the near-unity slope of the plot, which establishes the validity of this experimental approach. To our knowledge, this study represents the first demonstration of the correlation between computationally estimated switching equilibrium constants (KS) and experimentally determined binding affinities () of structure-switching aptamers. This has many important implications as the relationship between stability of a non-binding state and the ligand concentration required to “switch states” can allow for the prediction of the necessary KS value to obtain a structure-switching aptamer that would operate in the desired range.
The introduction of additional sequence in nucleic acids to facilitate the design of structure-switching aptamers has the potential to also generate unexpected structures, which could have significant sensitivity and/or selectivity issues. For structure-switching aptamers, the formation of a specific binding-competent structure is generally associated with the presence of ligand. In the case of TBA, this is the formation of an antiparallel G-quartet in the presence of potassium ions. Thus, to ensure the selectivity of this binding event, it is essential that no other ligands are capable of generating the same specific structure, which would result in the generation of a false positive. In a recent study by Buess-Herman and co-workers, extended G-quartet-based aptamer sequences were shown to adopt both parallel and antiparallel structures by CD and UV spectroscopy, depending on the nature of the cation [41]. While all structures (including TBA) adopted the standard anti-parallel conformation in the presence of potassium (which is the ion used in the present work), the presence of different cations was shown to change the conformation of the quadruplex present in extended TBA sequences. This has significant relevance in the use of structure-switching molecules based on TBA as biosensors as these conformational shifts could give rise to false positives. Thus, control experiments examining issues of selectivity, such as those used in the recent work by Chen and co-workers [42] will be essential for any application of a TBA-based aptamer as a biosensor.
Acknowledgements
The authors thank Dr. Philip C. Bevilacqua and the Pennsylvania State University for circular dichroism equipment, and Dr. Martin Serra for help on analysis of thermodynamic data. This work was supported by research awards to A.D.P. from Mercyhurst University and through a Research Development Grant to A.D.P. from Penn State Altoona Division of Mathematics and Natural Sciences. Funding for open access charge: Mercyhurst University. A.D.P. conceived and designed the project, A.D.P., A.T.C., and S.N.S. acquired the data, all authors analysed and interpreted the data, A.D.P. and G.A.R.-W. wrote the paper.
Appendix A. Supplementary data
This document contains Supplementary Fig. 1.
This document contains Supplementary Fig. 2.
This document contains Supplementary Fig. 3.
This document contains Supplementary Fig. 4.
References
- 1.Ricci F., Vallée-Bélisle A., Porchetta A., Plaxco K.W. Rational design of allosteric inhibitors and activators using the population-shift model. In vitro validation and application to an artificial biosensor. J. Am. Chem. Soc. 2012;134:15177–15180. doi: 10.1021/ja304672h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plaxco K.W., Soh H.T. Switch-based biosensors: a new approach towards real-time, in vivo molecular detection. Trends Biotechnol. 2011;29:1–5. doi: 10.1016/j.tibtech.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallée-Bélisle A., Plaxco K.W. Structure-switching biosensors: inspired by nature. Curr. Opin. Struct. Biol. 2010;20:518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Idili A., Plaxco K.W., Vallée-Bélisle A., Ricci F. Thermodynamic basis for engineering high-affinity, high-specificity binding-induced DNA clamp nanoswitches. ACS Nano. 2013;7:10863–10869. doi: 10.1021/nn404305e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman S.K. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA. 2003;9:377–383. doi: 10.1261/rna.2200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth A., Breaker R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drabovich A.P., Okhonin V., Berezovski M., Krylov S.N. Smart aptamers facilitate multi-probe affinity analysis of proteins with ultra-wide dynamic range of measured concentrations. J. Am. Chem. Soc. 2007;129:7260–7261. doi: 10.1021/ja072269p. [DOI] [PubMed] [Google Scholar]
- 8.Vallée-Bélisle A., Ricci F., Plaxco K.W. Thermodynamic basis for the optimization of binding-induced biomolecular switches and structure-switching biosensors. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porchetta A., Vallée-Bélisle A., Plaxco K.W., Ricci F. Allosterically tunable, DNA-based switches triggered by heavy metals. J. Am. Chem. Soc. 2013;135:13238–13241. doi: 10.1021/ja404653q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallée-Bélisle A., Ricci F., Plaxco K.W. Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range. J. Am. Chem. Soc. 2012;134:2876–2879. doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang D., Vallée-Bélisle A., Porchetta A., Plaxco K.W., Ricci F. Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range. Angew. Chem. Int. Ed. 2012;51:6717–6721. doi: 10.1002/anie.201202204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porchetta A., Vallée-Bélisle A., Plaxco K.W., Ricci F. Using distal-site mutations and allosteric inhibition to tune, extend, and narrow the useful dynamic range of aptamer-based sensors. J. Am. Chem. Soc. 2012;134:20601–20604. doi: 10.1021/ja310585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 14.Navani N.K., Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Famulok M., Hartig J.r.S., Mayer G.n. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 18.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamaguchi N., Ellington A., Stanton M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 20.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 22.Padmanabhan K., Padmanabhan K.P., Ferrara J.D., Sadler J.E., Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 24.Sagi J. G-quadruplexes incorporating modified constituents: a review. J. Biomol. Struct. Dyn. 2013:1–35. doi: 10.1080/07391102.2013.775074. [DOI] [PubMed] [Google Scholar]
- 25.Keniry M.A., Owen E.A. Insight into the molecular recognition of spermine by DNA quadruplexes from an NMR study of the association of spermine with the thrombin-binding aptamer. J. Mol. Recognit. 2013;26:308–317. doi: 10.1002/jmr.2274. [DOI] [PubMed] [Google Scholar]
- 26.Radi A.-E., O’Sullivan C.K. Aptamer conformational switch as sensitive electrochemical biosensor for potassium ion recognition. Chem. Commun. 2006:3432–3434. doi: 10.1039/b606804a. [DOI] [PubMed] [Google Scholar]
- 27.Yang X., Li T., Li B., Wang E. Potassium-sensitive G-quadruplex DNA for sensitive visible potassium detection. Analyst. 2010;135:71–75. doi: 10.1039/b913036e. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsuka K., Sato S., Sato Y., Sota K., Ohzawa S., Matsuda T., Takemoto K., Takamune N., Juskowiak B., Nagai T. Fluorescence imaging of potassium ions in living cells using a fluorescent probe based on a thrombin binding aptamer-peptide conjugate. Chem. Commun. 2012;48:4740–4742. doi: 10.1039/c2cc30536d. [DOI] [PubMed] [Google Scholar]
- 29.Kong D.-M., Guo J.-H., Yang W., Ma Y.-E., Shen H.-X. Crystal violet-G-quadruplex complexes as fluorescent sensors for homogeneous detection of potassium ion. Biosens. Bioelectron. 2009;25:88–93. doi: 10.1016/j.bios.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Hu K., Huang Y., Zhao S., Tian J., Wu Q., Zhang G., Jiang J. Ultrasensitive detection of potassium ions based on target induced DNA conformational switch enhanced fluorescence polarization. Analyst. 2012;137:2770–2773. doi: 10.1039/c2an35416k. [DOI] [PubMed] [Google Scholar]
- 31.Kim B., Jung I.H., Kang M., Shim H.-K., Woo H.Y. Cationic conjugated polyelectrolytes-triggered conformational change of molecular beacon aptamer for highly sensitive and selective potassium ion detection. J. Am. Chem. Soc. 2012;134:3133–3138. doi: 10.1021/ja210360v. [DOI] [PubMed] [Google Scholar]
- 32.Li T., Wang E., Dong S. G-Quadruplex-based DNAzyme as a sensing platform for ultrasensitive colorimetric potassium detection. Chem. Commun. 2009:580–582. doi: 10.1039/b815814b. [DOI] [PubMed] [Google Scholar]
- 33.Takenaka S., Juskowiak B. Fluorescence detection of potassium ion using the G-Quadruplex structure. Anal. Sci. 2011;27:1167. doi: 10.2116/analsci.27.1167. [DOI] [PubMed] [Google Scholar]
- 34.Wilcox J.M., Rempel D.L., Gross M.L. Method of measuring oligonucleotide-metal affinities: interactions of the thrombin binding aptamer with K+ and Sr2+ Anal. Chem. 2008;80:2365–2371. doi: 10.1021/ac701903w. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. mFold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markham N.R., Zuker M. In: Bioinformatics, volume II. Keith J.M., editor. vol. 453. Humana Press; Totowa, NJ: 2008. pp. 3–31. (Structure, Function and Applications). [Google Scholar]
- 37.Mullen M.A., Assmann S.M., Bevilacqua P.C. Toward a digital gene response: RNA G-Quadruplexes with fewer quartets fold with higher cooperativity. J. Am. Chem. Soc. 2012;134:812–815. doi: 10.1021/ja2096255. [DOI] [PubMed] [Google Scholar]
- 38.Fang X., Pan T., Sosnick T.R. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 39.Aldaye F.A., Palmer A.L., Sleiman H.F. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 40.Ratmeyer L., Vinayak R., Zhong Y.Y., Zon G., Wilson W.D. Sequence specific thermodynamic and structural properties for DNA·RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 41.De Rache A., Kejnovská I., Vorlíčková M., Buess-Herman C. Elongated thrombin binding aptamer: AG-Quadruplex cation-sensitive conformational switch. Chem. A Eur. J. 2012;18:4392–4400. doi: 10.1002/chem.201103381. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Chen L., Ma H., Zhou T., Li X. Aptamer biosensor for label-free impedance spectroscopy detection of potassium ion based on DNA G-quadruplex conformation. Biosens. Bioelectron. 2013;48:108–112. doi: 10.1016/j.bios.2013.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains Supplementary Fig. 1.
This document contains Supplementary Fig. 2.
This document contains Supplementary Fig. 3.
This document contains Supplementary Fig. 4.


