Abstract
Alzheimer׳s disease (AD) is one of the most common dementias showing slow progressive cognitive decline. Progression of intracerebral accumulation of beta amyloid (Aβ) peptides by the action of amyloid binding alcohol dehydrogenase (ABAD), a mitochondrial enzyme and β-site amyloid precursor protein cleaving enzyme 1 (BACE1) and the degradation of Acetylcholinesterase (AChE) the main pathological characteristics of AD. Therefore, it is of interest to evaluate the importance of fisetin (a flavonol that belongs to the flavonoid group of polyphenols) binding with AChE, ABAD and BACE1 proteins. Docking experiment of fisetin with these proteins using two different tools namely iGEMDOCK and FlexX show significant binding with acceptable binding values. Thus, the potential inhibitory role of fisetin with AD associated proteins is documented.
Keywords: Alzheimer׳s disease, fisetin, AChE, ABAD, BACE1, docking
Background
Alzheimer׳s disease (AD) is the most common form of dementia. It is a harmful neurological disorder that affects the aged people that is increasing substantially with the symptoms of memory loss, decaylanguage and problems with visual spatial search [1, 2]. About 1%-4% of the population are affected by AD every year among the age group of 65 to 70 years, while this may exceed 6% over 85 years of age [3, 4]. Pathologically AD may be characterized by progressive intra-cerebral accumulation of beta amyloid (Aβ) peptides [5]. It is also found that the tau protein [6] contributes to neuronal, synaptic, and cognitive malfunction [7]. Recent studies showed that Aβ progressively accumulates in synaptic mitochondria and impairs mitochondrial structure and function including membrane potential, membrane permeability transition pore, respiration, energy metabolism, oxidative stress, mitochondrial dynamics, and calcium homeostasis [8– 15]. Amyloid binding alcohol dehydrogenase (ABAD), a mitochondrial enzyme responsible for mitochondrial dysfunction and in the pathogenesis of AD is known [8]. This enzyme has attracted considerable interest because of its ability to interact with Aβ which mediates mitochondrial and synaptic dysfunction [16, 17] . Hence, antagonizing Aβ-ABAD interaction is a strategy to improve the learning memory in AD [17, 18]. Moreover, the β- secretase, widely known as β-site amyloid precursor protein cleaving enzyme 1 (BACE1), initiates the production of the toxic amyloid β (Aβ) that plays a crucial early part in AD pathogenesis. Due to its apparent rate limiting function, BACE1 appears to be a prime target to prevent and lowering the Aβ generation in AD [19–24].
According to the ‘cholinergic hypothesis’ cholinergic function is required for short-term memory, the cholinergic deficit in AD was also believed to be responsible for much of the short-term memory deficit [25, 26]. Cholinergic neurons, such as choline acetyltransferase (ChAT) and acetylcholinesterase (AChE), which are enzymes responsible for synthesis and degradation of Ach, are respectively decreased in the cortex and hippocampus, areas of the brain involved in cognition and memory [27, 28]. So blocking the acetylcholinesterase enzyme is now a comprehensive target for the treatment of AD [29].
The use of medicinal plant derived secondary metabolites or plant based compounds as lead a molecule is clearly established in recent years [30]. The biological effect of these compounds in slowing down the progress of AD or dementia is known [31]. The application and use of molecular docking analysis in predicting ligand-protein target is well established in recent years [32]. Therefore, it is of interest to evaluatethe importance of fisetin (a flavonolthat belongs to the flavonoid group of polyphenols) binding with AD associated AChE, ABAD and BACE1 proteins using molecular docking and analysis.
Methodology
Data and databases:
The data from databases used in this study include PDB (Protein Data Bank) [33] and ZINC [34]. ZINC is a free database of commercially available compounds for virtual screening. It contains over 35 million purchasable compounds in ready-todock; 3D formats which is provided by the Shoichet Laboratory in the Department of Pharmaceutical Chemistry at the University of California, San Francisco (UCSF) [34].
Docking Tools:
The docking tools used in this study include Flexx (LeadIT 2.1.6) and iGEMDOCK. FlexX a fully automated docking program available on LeadIT 2.1.6 package was used to dock compound into the active site of the enzymes. FlexX considers ligand flexibility by changing the conformations of the ligand in the active site while making the protein rigid [35]. iGEMDOCK is a graphical environment for recognizing pharmacological interactions and virtual screening. For postscreening analysis, iGEMDOCK can enrich the hit rate and provide biological insights by deriving the pharmacological interactions from screening compounds [36].
Ligand and protein preparation:
The plant derived flavonoid compound fisetin (ZINC00039111) was downloaded from the ZINC databases. The compound is obtained from the ZINC databases in .MOL2 format. This is prepared by adding hydrogen and applying suitable geometry by using Accelrys Discovery Studio Visualizer 3.5. 3D as shown in Figure 1 (a). The three target enzymes i.e., ABAD (PDB id: 1SO8), BACE1 (PDB id: 2QP8) and AChE (PDB id: 1EVE) were downloaded in pdb format from protein data bank (http://www.rcsb.org/pdb). The enzymes are prepared by using receptor preparing wizard available in LeadIT 2.1.6 package for Flexx Docking and all crystallographic water molecules were removed.
Figure 1.
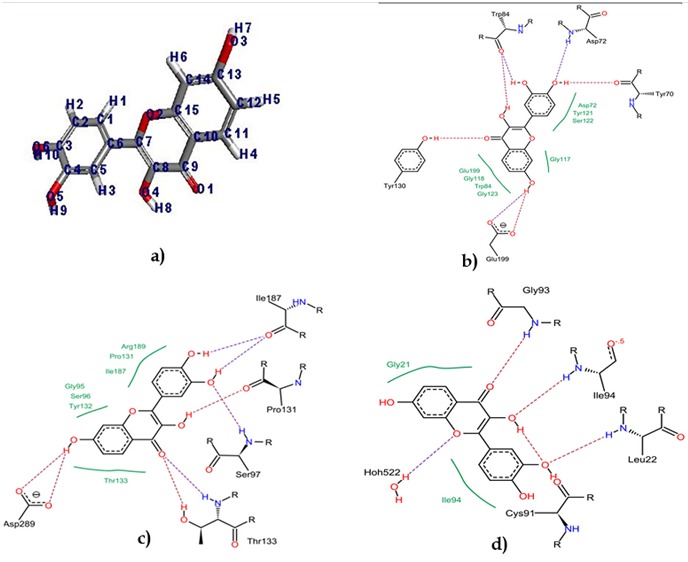
a) 3D structure of fisetin, Interaction of fisetin with b) Acetylcholinesterase c) BACE1 and d) ABAD enzyme.
ADMET tools:
The adverse properties such as absorption, distribution, metabolism, excretion and the toxicity of fisetin were calculated using the ADMET SAR database. They provide the latest and most comprehensive manually curated data for diverse chemicals associated with known ADMET profiles.
Molecular docking with Flexx:
FlexX uses an incremental build up algorithm where ligands are docked starting with a base fragment. Base fragments are generated by severing all noncyclic bonds in a given ligand. All base fragments identified for a given ligand serve as starting points for the docking Table 1 (see supplementary material). After placement of a base fragment (in different positions) the complete ligand is constructed by adding the remaining components back on. Each component is added in accordance with a set of predefined, allowed torsion angles, thus allowing for ligand flexibility. At each step the interactions are evaluated and the best solution is selected according to the docking score [37]. In this study, the docking and subsequent scoring were performed using the default parameters of the FlexX program implanted in LeadIT 2.1.6. The active sites of the enzymes were defined to include residues within 6.5 Å radius around bound inhibitor. Final scores for all FlexX solutions were calculated by a consensus scoring function (CScore) and used for database ranking. Finally the best pose with the highest score was selected for investigating the interactions, HYDE assessment and calculating the free energy of binding (DG)[38, 39].
Molecular docking with iGEMDOCK:
Graphical automatic drug design system for docking, screening and post-analysis program iGEMDOCK was used to gain the docking results of the listed compounds with the target (Table 1). The binding sites of the targets were prepared and the energy minimized compound was imported. During docking, at first the molecules were prepared and bonds, bond orders, explicit hydrogen׳s, charges, flexible torsions were assigned to both the protein and ligands. From the docking, wizard ligands were selected and the scoring function used was iGEMDOCK score. If hydrogen bonding is possible, the hydrogen bond energy contribution to the docking score is assigned a penalty based on the deviations from the ideal bonding angle. This option cansignificantly reduce the number of unlikely hydrogen bonds and also internal electrostatic interaction; internal hydrogen bond sp2-sp2 torsions are calculated from the pose by enabling the ligand evaluation terms. The search algorithm is taken as iGEMDOCK and numbers of runs taken are 70 and max interactions were 2000 with population size 200 and with an energy threshold of 100 also at each step least ‘min’ torsions/translations/rotations are tested and the one giving lowest energy is chosen. If the energy is positive (i.e., because of a clash or an unfavourable electrostatic interaction), then additional ‘max’ positions will be tested. If the pose being docked is closer to one of the ligands in the list than specified by the Root Mean Square Deviation (RMSD) threshold, an extra penalty term (the energy penalty) is added to the scoring function. This ensures a greater diversity of the returned solutions since the docking engine will focus its search on poses different from earlier poses found. The energy penalty was set to 100, RMSD threshold was 2.00 and RMSD calculation by atom ID (fast) were set. Docking was conducted between protein and inhibitor which results in binding affinities in kcal/mol and docking run time. The compound which gives lowest binding energy is chosen as the best inhibitor [40]. iGEMDOCK showed better overall performance in docking simulations when compared with other software. The hydrophobic preference and electrostatic preference were set to 1.00. The binding site of the target was identified at a distance 8Å. The empirical scoring function of iGEMDOCK was estimated as:
Fitness = vdW + Hbond + Elec.
Here, the vdW term is vander Waal energy. H-bond and Elect terms are hydrogen bonding energy and electro-static energy, respectively.
Results & Discussions
Flavonoids are the natural plants compounds with variable phenolic structures, found in fruit, vegetables, stems, flowers, tea, and wine [40]. These natural products were known for their beneficial effects on health long before flavonoids were isolated as the effective compounds. They are usually attached with sugar moiety to increase their water solubility. Most of the flavonoids are known to possess various pharmacological activities, such as antioxidant, antiviral, antibacterial and antimutagenic effects [41, 42]. The stereochemistry of binding fisetin on AD has not been yet characterized. In our present studies, we have used two docking engines to analyse the binding affinity of fisetin on the three target enzymes responsible for the AD i.e., (i) ABAD (1SO8) (ii) AChE (1EVE) (iii) BACE1 (2QP8), which may facilitate further development of more potent Alzheimer agents. Table 1 described result of fisetin against three targets of AD using FlexX and iGEMDOCK.
Amyloid binding alcohol dehydrogenase (ABAD):
In FlexX docking method, the compound fisetin interact with the amyloid binding alcohol dehydrogenase(ABAD) enzyme by least binding energy -15.0147kcal/mol.The binding confirmation of fisetin with the enzyme is demonstrated on the Figure 1 (d), shows that theoxygen of the carbonyl groupbinds with the hydrogen atoms of amino acid residue Gly93. Another hydrogen bond is formed between the oxygen of carboxyl group and the amino group of amino acid residue Ile94. Leu22 and Cys91 are also involved in the hydrogen bond with oxygen and hydrogen of the fisetin. In iGEMDOCK, the binding energy of fisetin with amyloid binding alcohol dehydrogenase (ABAD) enzyme is -103.006kcal/mol. The overall binding information of fisetin and ABAD is described in (Table 1). From this calculated score, the relative conformation and arrangement of fisetin shows a significant affinity towards the enzyme amyloid binding alcohol dehydrogenase.
Acetylcholinesterase enzyme (AChE):
Molecular docking analysis of fisetin with AChE, by using FlexX, the docking score is -28.2652kcal/mol. In Figure 1 (b), the interaction between of fisetin and acetylcholinesterase enzyme is showing hydrogen bonds which is formed between the hydrogen and oxygen of hydroxyl group of fisetin with the oxygen of Tyr70 residue and hydrogen of Asp72. The oxygen of carbonyl group of Trp84 residue formed two hydrogen bonds with the hydrogen of two hydroxyl group׳s. Tyr130 and Glu199 residues are also involved in the interaction with fisetin molecule. The binding energy of fisetin calculated by iGEMDOCKis-112.042 kcal/mol (shown in Table 1), described a high affinity towards Acetylcholinesterase enzyme.
BACE1enzyme:
Figure 1 (c) shows the binding of fisetin which is -31.4957 kcal/mol in FlexX by making contact with amino acid residue Ile 187 by forming two hydrogen bonds between the hydrogen׳s of hydroxyl groups and oxygen of the carbonyl group of amino acids. Some other hydrogen bonds are also seen which contributed by Thr133, Ser97, Asp289 and pro131. In iGEMDOCK, the calculated result of the interaction is -70.9739 kcal/mol between fisetin-BACE1enzyme. The binding energy also shows a significant ligand-receptor complex with this enzyme.
ADMET results for Fisetin:
ADMET profile was evaluated using the admetSAR database for fisetin shows the highest binding energy Table 2 & Table 3 (see supplementary material). admetSAR predicted classification and regression values for fisetin and the results seems to have been calculated for different types of models such as blood brain barrier, human intestina labsorption, CaCO2 permeability all of which showed positive results ensuring that the compound passes all the models and have no side effects on absorption. Similarly in case of metabolism, various cytochrome P450 (CYP) substrate and inhibitor models were calculated and the results show that they are nonsubstrate and non-inhibitor except CYP450 1A2 Inhibitor. In terms of toxicity, it is found to be non-carcinogenic. Although some toxicity models show some negative results the regression profiles indicates that they have very low probability values.
Conclusion
The predicted binding of fisetin with AD associated amyloid binding alcohol dehydrogenase, acetylcholinestease, and BACE1 enzymes d is documented in this study. The ADMET properties have been calculated for fisetin and it has been shown that it is non-carcinogenic and non-toxic in nature.
Competing interests
All authors declare that they have no competing interests.
Supplementary material
Acknowledgments
We thankthe Bangladesh Council of Scientific and Industrial Research (BCSIR).
Footnotes
Citation:Dash et al, Bioinformation 10(9): 562-568 (2014)
References
- 1.Henderson AS, Jorm AF. John Wiley & Sons Ltd. 2002 [Google Scholar]
- 2.Alzheimer's Association. Alzheimers Dement. 2012;8:131. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Evans DA, et al. JAMA. 1989;262:2551. [PubMed] [Google Scholar]
- 4.Geldmacher DS, Whitehouse PJ. JNeurology. 1997;48:S2. doi: 10.1212/wnl.48.5_suppl_6.2s. [DOI] [PubMed] [Google Scholar]
- 5.Hardy J, Selkoe DJ. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6.Alonso A, et al. ProcNatlAcadSci USA. 2001;98:6923. [Google Scholar]
- 7.Du H, et al. ProcNatlAcadSci USA. 2010;107:18670. [Google Scholar]
- 8.Caspersen C, et al. FASEB J. 2005;19:2040. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 9.Lustbader JW, et al. Science. 2004;304:448. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 10.Hansson Petersen CA, et al. ProcNatlAcadSci USA. 2008;105:13145. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devi L, et al. J Neurosci. 2006;26:9057. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert A, et al. NeurodegenerDis. 2008;5:157. [Google Scholar]
- 13.Hauptmann S, et al. NeurobiolAging. 2009;30:1574. [Google Scholar]
- 14.Du H, et al. Neurobiol Aging. 2011;32:398. doi: 10.1016/j.neurobiolaging.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J, et al. ProcNatlAcadSci USA. 2009;106:14670. [Google Scholar]
- 16.Takuma K, et al. ProcNatlAcadSci USA. 2009;106:20021. [Google Scholar]
- 17.Takuma K, et al. FASEB J. 2005;19:597. doi: 10.1096/fj.04-2582fje. [DOI] [PubMed] [Google Scholar]
- 18.Yao J, et al. J Neurosci. 2011;31:2313. doi: 10.1523/JNEUROSCI.4717-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R, et al. Lancet Neurol. 2014;13:319. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sathya M, et al. ClinChimActa. 2012;414:171. [Google Scholar]
- 21.Vassar R, et al. J MolNeurosci. 2001;17:157. [Google Scholar]
- 22.Cole SL, et al. Curr Alzheimer Res. 2008;5:100. doi: 10.2174/156720508783954758. [DOI] [PubMed] [Google Scholar]
- 23.Hilpert H, et al. J Med Chem. 2013;56:3980. doi: 10.1021/jm400225m. [DOI] [PubMed] [Google Scholar]
- 24.Bajda M, et al. Int J Mol Sci. 2014;15:5128. doi: 10.3390/ijms15035128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis PT, et al. J NeurolNeurosurg Psychiatry. 1999;66:137. [Google Scholar]
- 26.Bartus RT, et al. Science. 1982;217:408. doi: 10.1126/science.7046051. [DOI] [PubMed] [Google Scholar]
- 27.McGleenon BM, et al. Br J ClinPharmacol. 1999;48:471. [Google Scholar]
- 28.Wilkinson DG, et al. Drugs Aging. 2004;21:453. doi: 10.2165/00002512-200421070-00004. [DOI] [PubMed] [Google Scholar]
- 29.Mehta M, et al. Int J Alzheimers Dis. 2012;12:728983. doi: 10.1155/2012/728983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler MS, et al. J Nat Prod. 2004;67:2141. doi: 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- 31.Ji HF, et al. EMBO Rep. 2009;10:194. doi: 10.1038/embor.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghose, et al. J Comb Chem. 1999;1:55. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, et al. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irwin JJ, et al. J ChemInf Model. 2005;45:177. [Google Scholar]
- 35.Rarey M, et al. J Mol Biol. 1996;261:470. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 36.Hsu KC, et al. BMC Bioinformatics. 2011;15:S33. doi: 10.1186/1471-2105-12-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroemer RT, et al. Curr Protein Pept Sci. 2007;8:312. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- 38.Reulecke I, et al. Chem Med Chem. 2008;3:885. [Google Scholar]
- 39.Schneider N, et al. J Comput Aided Mol. 2012;24:417. [Google Scholar]
- 40.Middleton EJ, et al. AdvExp Med Biol. 1998;439:175. doi: 10.1007/978-1-4615-5335-9_13. [DOI] [PubMed] [Google Scholar]
- 41.Whittern CC, et al. J Am Chem Soc. 1984;61:1072. [Google Scholar]
- 42.Nagao A, et al. Biosci Biotech & Biochem. 1999;63:1787. doi: 10.1271/bbb.63.1787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


