Abstract
MicroRNAs (miRNAs) are small non-coding nucleotide sequences between 17 and 25 nucleotides in length that primarily function in the regulation of gene expression. A since miRNA has thousand of predict targets in a complex, regulatory cell signaling network. Therefore, it is of interest to study multiple target genes simultaneously. Hence, we describe a web tool (developed using Java programming language and MySQL database server) to analyse multiple targets of pre-selected miRNAs. We cross validated the tool in eight most highly expressed miRNAs in the antrum region of stomach. This helped to identify 43 potential genes that are target of at least six of the referred miRNAs. The developed tool aims to reduce the randomness and increase the chance of selecting strong candidate target genes and miRNAs responsible for playing important roles in the studied tissue.
Availability
Keywords: miRNA, Gastric Cancer, Bioinformatics, Web tool, Target Genes
Background
MicroRNAs (miRNAs) are small non-coding nucleotide sequences between 17 and 25 nucleotides in length that primarily function in the regulation of gene expression [1, 2]. Studies have demonstrated that a single miRNA may regulate several mRNAs of multiple functions, and that a single mRNA may be target of several microRNAs [3]. Thus, miRNAs form a complex, regulatory cell-signaling network [2, 4] that results in differentiated gene expression. It is estimated that two-thirds of the human genome is regulated by these small nucleotide sequences. The mechanisms underlying the negative regulation of gene expression by miRNAs are similar in animals and plants, which implies that they are involved in fundamental cellular processes including cell proliferation, development, differentiation and apoptosis [5]. Several studies have described the expression profile of miRNAs in both healthy tissues as for diseases [6– 9].
Altered miRNA expression levels may contribute to disease development in humans. Several reports have linked miRNAs to cancer; the first miRNAs to be characterized were involved in cellular proliferation and death. Human tumors and tumor cell lines exhibit large differences in miRNA expression levels compared with normal tissues [8, 9, 10]. The evidence suggests that differentiated miRNA expression may regulate tumor suppressor genes and oncogenes [11]. To predict putative target gene of a single miRNA in silico, tools like microRNA.org [3] and targetScan [4] are used. The difficulty in this analysis is the amount of predicted targets for a single miRNA, considering that, as mentioned, a single miRNA may have thousands of possible targets. Using online databases available, the TargetCompare web tool proposes a way to analyze all possible common targets of any number of preselected miRNAs, indicating thereby targets with greater chances of being silenced by at least two of the miRNAs pre selected. The tool also associates known diseases linked to these potential target genes.
Implementation:
The Java (http://www.oracle.com/us/technologies/java/) programming language was used for TargetCompare development and it was implemented as shown in Figure 1. The data used to compare putative targets was downloaded from three majors miRNA target prediction datasets: microrna.org [3], targetscan.org [4] and PiTa [12]. To associate the target genes with diseases, the public Genetic Association Database [13] was used. To manage the data, MySQL Database Server (http://www. mysql. com/) was used. The web tool targetCompare is freely available at http://www.lghm.ufpa.br/targetcompare.
Figure 1.
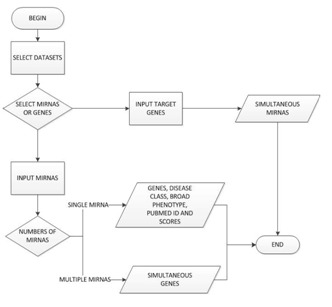
TargetCompare flowchart.
Results & Discussion
Using the developed tool in a single miRNA, it is possible to compare target genes in all three datasets simultaneously (Figure 2). Another way to use the web tool is to query a set of genes to find out which miRNAs target this set of genes (Figure 3). To evaluate the tool, it eas used in a set of the eight miRNAs most highly expressed in antrum region of the stomach [7], we were able to identify 4,748 different genes may be regulated by up to two of the eight miRNAs selected. Using the simultaneous presence of at least six miRNAs as a selection criterion, 43 potential target genes were grouped together Table 1 (see supplementary material). The results obtained with the developed tool suggest that these putative target genes of the eight most highly expressed miRNA in antrum are strong candidates for silencing in the gastric region.
Figure 2.
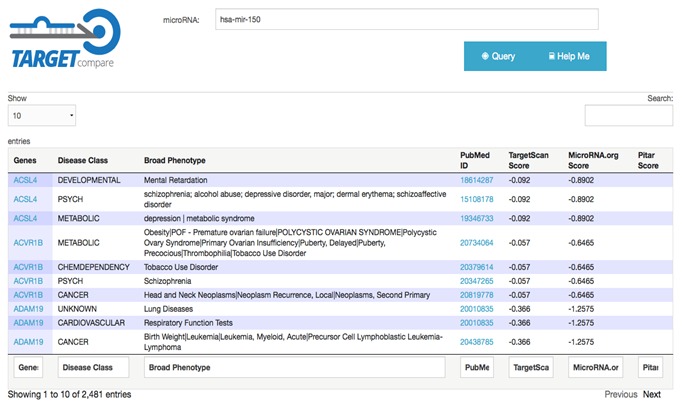
TargetCompare results for a single miRNA. The columns show the disease class associated with the potential target gene, broad phenotype, PubMed ID and the association score of each dataset used
Figure 3.
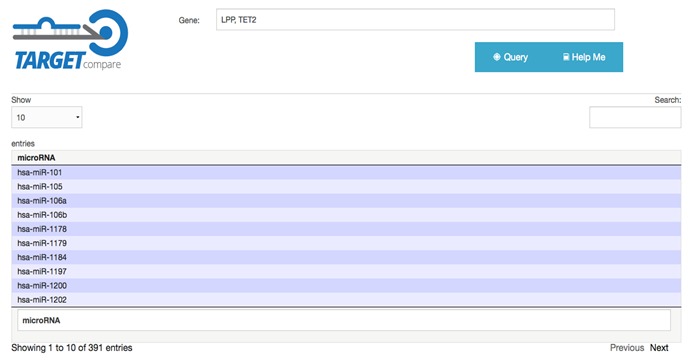
Results for a target gene consult. The column shows all miRNAs that has as target the selected gene in all three target databases
Conclusions
This tool simultaneously evaluates different microRNAs associating them with different classes of diseases. However, the putative target genes need validation. Thus, the described tool is useful to reduce the arbitrariness in the analysis. This increases the chances of selecting target genes having an important role in the studied tissue.
Funding sources
This work was part of the REDE DE PESQUISA EM GENÔMICA POPULACIONAL HUMANA (Bio Computacional/CAPES). Financial support: PROPESP/UFPAFADESP, CNPq, CAPES. FABIANO CORDEIRO MOREIRA supported by CESUPA; IGOR G. HAMOY is supported by Pós-Doc Junior (PDJ) fellowship from CNPq/Brazil (162605/2011- 0); ÂNDREA RIBEIRO-DOS-SANTOS supported by CNPq/Produtividade.
Competing Interests
The authors have declared that no competing interests exist.
Author Contributions
Conceived and designed the experiments: FCM, BD. Analysed the data: FCM, BD. Wrote the first draft of the manuscript: FCM. Contributed to the writing of the manuscript: ARS, IGH. Agree with manuscript results and conclusions: FCM, ARS, IGH. Jointly developed the structure and arguments for the paper: FCM, AMRS. Made critical revisions and approved final version: FCM, ARS, IGH. All authors reviewed and approved of the final manuscript.
Supplementary material
Acknowledgments
The authors would like to thank the staff of the Núcleo de Pesquisas em Oncologias - UFPA who contributed to the realization of this work.
Footnotes
Citation:Moreira et al, Bioinformation 10(9): 602-605 (2014)
References
- 1.Lee RC, et al. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ricarte Fiho JC, et al. Arq Bras Endocrinol Metabol. 2006;50:1102. doi: 10.1590/s0004-27302006000600018. [DOI] [PubMed] [Google Scholar]
- 3.Betel D, et al. Nucleic Acids Res. 2008;36:D149. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, et al. Cell. 2005;120:15. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Meltzer PS. Nature. 2005;435:745. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro-dos-Santos Â, et al. PLoS ONE. 2010;5:e13205. doi: 10.1371/journal.pone.0013205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreira FC, et al. PLoS ONE. 2014;9:e92300. doi: 10.1371/journal.pone.0092300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. Nature Rev Cancer. 2006;6:857. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, et al. Proc Natl Acad Sci USA. 2004;101:2999. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaur A, et al. Cancer Res. 2007;67:2456. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 11.Guo J, et al. J Gastroenterol Hepatol. 2009;24:652. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 12.Kertesz M, et al. Nat Genet. 2007;39:1278. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 13.Becker KG, et al. Nat Genet. 2004;36:431. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


