Abstract
Background and Purpose
Accurate grading of cerebral glioma using conventional structural imaging techniques remains challenging due to the relatively poor sensitivity and specificity of these methods. The purpose of this study was to evaluate the relative sensitivity and specificity of structural MRI and MR measurements of perfusion, diffusion, and spectroscopic parameters for glioma grading. A secondary objective was to evaluate a whole-brain MR spectroscopic imaging method for evaluation of brain tumors.
Materials and Methods
Fifty six patients with radiologically suspected untreated glioma were studied with T1- and T2-weighted MR imaging, DCE-MR imaging, DTI, and volumetric whole-brain MR spectroscopic imaging. ROC analysis was performed using the relative CBV, ADC, FA, and multiple spectroscopic parameters to determine optimum thresholds for tumor grading and to obtain the sensitivity, specificity, PPV, and NPV for identifying high-grade gliomas. Logistic regression was performed to analyze all the parameters together.
Results
The relative CBV individually classified glioma as low and high grade with a sensitivity and specificity of 100% and 88% respectively based on a threshold value of 3.34. On combining all parameters under consideration, the classification was achieved with 2% error and sensitivity and specificity of 100% and 96% respectively.
Conclusion
Individually, CBV measurement provides the greatest diagnostic performance for predicting glioma grade; however, the most accurate classification can be achieved by combining all of the imaging parameters. The whole-brain MR spectroscopic imaging method provided data from of a large fraction of the tumor volumes.
INTRODUCTION
Gliomas are the most common primary cerebral neoplasm and major cause of morbidity and mortality in adults, with malignancy that ranges from low grade to anaplastic and glioblastoma multiforme (GBM). Histopathology is the gold standard for grading of glioma, although suffers from inherent sampling error associated with stereotactic biopsy and is unable to evaluate residual tumor tissue after cytoreductive surgery. MRI has played a significant role in noninvasive detection and classification of gliomas; however, the conventional MRI methods mostly fail to discriminate GBMs from solitary metastases1, CNS lymphomas, or other glioma grades, and for this reason MR measures reflecting tissue function, such as MRS, DTI, and PWI, have been investigated for glioma classification.
1H-MRS enables evaluation of brain metabolites such as N-acetyl aspartate (NAA), total choline (Cho), and total creatine (Cre). Additional signals include lactate, which becomes visible in the presence of anaerobic metabolism, and lipids, which may be observed in regions of cellular breakdown caused by necrosis. Typical characteristics of elevated cho, decreased NAA, and the presence of lipids and lactate have been shown to be useful in tumor grading, although reports indicate a range of specificity and sensitivity levels.2,3 While many of these previous studies were performed using single-voxel spectroscopy or multi-voxel single slice spectroscopic imaging (MRSI) with PRESS volume selection, there are no previous reports that have used a more recently-developed method of whole-brain volumetric MRSI and there have been only a limited number of studies carried out at the increasingly-available field strength of 3Tesla. A secondary aim of this study is to evaluate the potential advantage of obtaining metabolite maps that cover a larger volume of the cerebrum.
DTI is sensitive to the rate and direction of diffusion of water within the tissue. Studies have shown that DTI reveals larger peritumoral abnormalities in gliomas, which are not apparent on conventional MRI, and that FA and ADC have been shown to distinguish high grade from low grade tumors.4,5 DCE-MRI measures a composite of tumor perfusion, vessel permeability, and extravascular-extracellular space volume, volume transfer coefficient (ktrans), rate transfer constant between the extracellular extravascular space (EES) and the plasma (kep) leakage (ve), and plasma volume (vp). These pharmacokinetic indices of DCE-MRI provide quantitative measurement of the integrity of the blood-brain barrier and of tissue perfusion. Several studies have shown that DCE can characterize different grades of glioma based on these hemodynamic and pharmacokinetic indices.6–8
Information gained from these multi-parametric imaging techniques is largely complimentary and combinations of these methods have been investigated for glioma classification,2,3,11 and their efficacy characterized in terms of the sensitivity, specificity, PPV, and NPV. Studies have shown that rCBV measurements and metabolite ratios both individually and in combination can increase the sensitivity and PPV when compared with conventional MRI alone in determining glioma grade. In this study the sensitivity, specificity, PPV, and NPV of perfusion MRI, whole brain MRSI, and DTI have been evaluated for glioma grading, and to determine which technique or combination of techniques is of value for classification of low grade from high grade glioma.
MATERIALS AND METHODS
Eighty-seven treatment-naïve patients (61men, 26women, aged 14–69 years) who had received a preliminary diagnosis of intracranial space-occupying lesion on CT/MRI were included in this study. Thirteen were subsequently excluded as they were confirmed as non-gliomas on histopathology. Five studies were dropped due to motion in the data sets, seven because of low quality of spectral data in the region of the tumor, and another six were excluded due to incomplete study data where either the perfusion imaging, T2-weighted MRI, or the DTI was not available. Hence a total of 56 patients were considered for analysis. The institutional ethics committee and the research committee approved the study protocol and informed consent was obtained from the patients or their care providers.
Sample Size Estimation
The sensitivity and specificity of the imaging results were calculated using ROC analysis. To estimate the sample size required for this analysis it was assumed that the area under the ROC curve under the null hypothesis will be 0.8; under the alternate hypothesis it will be 0.99; and that the ratio between the standard deviation of responses in the positive and negative groups was 1. For 90% power and 0.05 level of significance, the minimum sample size required for positive and negative responses was 16 each. The data were assumed as discrete responses having binary outcome (as high or low grade).
MR Imaging
Subjects underwent an hour-long MR protocol that included conventional MRI, whole brain MRSI, DTI and DCE-MRI on a 3-T Signa HDxt MRI scanner (General Electric, Milwaukee, WI) using a 12-channel head coil. Conventional imaging included T2-weighted FSE with TE/TR of 101.8/3500 msec, a signal average (NEX) of 1, FOV of 260×260mm2 and 3-mm slice thickness without any interslice gap; and a T1-weighted image (FSPGR BRAVO) with TE/TR/NEX= 3.01msec/8.02msec/1, FOV of 256×256mm2 and 1-mm slice thickness.
MRSI data were obtained using a volumetric spin-echo MRSI sequence with echo-planar readout and TR/TE=1710/70msec, FOV:280×280×180mm3, 100[read]×50[phase]×18[slice] spatial samples over a 135-mm slab, and acquisition time of 26min. This included frequency-selective water suppression and inversion recovery nulling of the lipid signal with TI=198msec. This sequence included both a spin-echo excitation for the metabolite signal and a low flip-angle gradient-echo excitation for a water reference MRSI signal, which was acquired in an interleaved fashion. The FSPGR BRAVO and MRSI acquisitions were performed at the same angulation, with the slice or slab orientations of all acquisitions parallel to the anterior commissure-posterior commissure (AC-PC) plane, or angulated at +15° from the AC-PC, which was found to facilitate improved magnetic field homogeneity over a larger volume of the cerebrum.
MRSI data were processed using the MIDAS package10. Metabolite image reconstruction included spatial smoothing and interpolation to 64×64×32voxels, with a resultant voxel volume of approximately 1mL. The automated spectral fitting of relative metabolite concentration resulted in maps for NAA, Cre, Cho, and lactate, although due to the spectral overlap with lipid signals that cannot be fully resolved by the spectral analysis procedure this latter result is termed Lactate+Lipid(LL). Additional maps were obtained for the metabolite ratios and measures of spectral quality, including spectral linewidth and Cramer-Rao bounds. Individual metabolite values are reported in institutional units and metabolite ratio values indicate the relative concentrations and not the widely-reported area under the spectral peak. The processing steps included manual creation of a mask that delineated the tumor and surrounding edema; calculation of the MRSI voxel tissue content based on a four-compartment tissue segmentation of the high-resolution T1-weighted MRI into grey- and white-matter, CSF, and an ‘other’ tissue category; and normalization of the fitted metabolite signals to institutional units using the water reference MRSI. The signal normalization assumed a fixed water content and T1 for each tissue type to derive the 100% water equivalent signal; however, because the water content and T1 in the tumor region is unknown these were estimated by deriving a bias-field correction map that minimized image intensity variations over the tumor region and the resultant image used for intensity scaling was smoothed to minimize the effect of local signal variations.
DTI data was acquired using a dual spin-echo single-short echo-planar sequence with ramp sampling, TR/TE/FOV/NEX/slice thickness/interslice gap=10s/100ms/240mm/1/3mm/0, 46slices, image matrix = 256×256, and diffusion weighting b-factor of 1000s/mm2 applied in 12 directions in addition to the reference measurement with b=0s/mm2. DTI data was processed using in-house developed software to obtain eigen-values(λ1, λ2 and λ3) and three orthonormal eigenvectors (e1, e2, e3).11 The tensor field data were then used to compute the DTI metrics such as ADC and FA for each voxel.11
DCE-MRI was performed using a 3-dimensional spoiled gradient recalled echo (SPGR) sequence (TR/TE/flip angle/NEX/slice thickness/FOV/matrix size=5.0msec/2.1msec/10°/0.7/6 mm/240×240mm/128×128mm, number of phases32). At the start of the fourth acquisition, Gd-DTPA-BMA(Omniscan; GE Healthcare, Piscataway, NJ) was administered IV through a power injector (Mallinckrodt Optistar LE) at 5mL/s and a dose of 0.2mmol/kg body weight, followed by a 30-mL saline flush. A series of 384 images over 32 time points for 12 slices were acquired (temporal resolution 5.65seconds). Before 3-dimensional SPGR, 2 inversion recovery FSE images (TR/TE/NEX/slice thickness/FOV/matrix size 940msec/8msec/0.75/6mm/240×240 mm2/128×128mm) with inversion times of 800 and 1,600 msec were acquired for the same slice position to quantify the voxel-wise tissue longitudinal relaxation time, T10. Voxel-wise tissue longitudinal relaxation time, T10, was calculated from the 2 inversion recovery sequences indicated above. Quantitative analysis of the concentration-time curve was performed to calculate the CBV and a corrected CBV map generated by removing the contrast agent leakage effect due to the disrupted BBB.11,12 The relative CBV was then obtained by dividing the mean value of CBV in a ROI by the value obtained from a ROI placed on the normal contralateral side of the brain.
A post-contrast T1-weighted image (FSPGR BRAVO) with TE/TR/NEX=2.98/7.79msec/1 was acquired with FOV of 256×256mm2 and 1-mm slice thickness after the acquisition of the perfusion MR imaging data.
The metabolite maps, DTI metrics, and DCE derived CBF map were co-registered. Image values were then obtained for all image types according to the following methods. For the metabolite maps ROIs were placed within the lesion corresponding to the maximal Cho signal and minimal NAA+Cre, and from contralateral normal-appearing white-matter (NAWM). Mean values of NAA, Cre, Cho, and LL in the tumor and Cre and Cho in the NAWM (CreNormal and ChoNormal respectively) were recorded from 12 interpolated voxels at each ROI, corresponding to a tissue volume of approximately 2mL. The values of CreTumor/CreNormal and ChoTumor/ChoNormal ratio were then calculated for each tumor ROI. Initial evaluation indicated that all studies included voxels with zero NAA; therefore results were not obtained for the widely-reported maximum Cho/NAA ratio. If LL was present it was confirmed that this was also present in the tumor ROIs. Values were accepted only if the fitted spectral line width was less than 13Hz and the Cramer-Rao bound for any one of NAA, Cre, and Cho was under 10%. Since the initial analysis indicated that regions of zero metabolite signal were observed for both necrotic tissue regions and cystic tumor regions, which could be readily distinguished based on the T2 image, an additional image metric obtained from the intensity of the T2-weighted MRI relative to the value in contralateral white matter was also included. For the DTI metrics (FA and ADC), mean values were obtained for ROIs corresponding to regions with high FA and low MD values, and for the DCE derived rCBV values the values were obtained for ROI corresponding to maximum corrected CBV.
In addition to obtaining metabolite values normalized using tissue water, these values were also obtained using normalization with contralateral values obtained from normal appearing white matter. Similarly, contralateral white matter CBV was obtained for determination of rCBV.
An experienced radiologist who was blinded to the MR perfusion, DTI and MR spectroscopic results, reviewed the conventional MR images and graded each tumor according to the two-tier imaging grading system: low- versus high-grade gliomas. The grading was based on: tissue contrast enhancement, border definition, mass effect, signal intensity heterogeneity, hemorrhage, necrosis, degree of edema, involvement of the corpus callosum and tumor crossing the midline.13
To evaluate the potential for whole-brain MRSI and the multiplane-DCE sequence to sample lesions throughout the cerebrum, the number of studies for which the MRI-observed tumor volume was considered to be well-, partially-, or inadequately-sampled by the MRSI or DCE study was recorded. For full-sampling, this evaluation required that >75% of the lesion volume, as indicated by the post-contrast T1- and T2-weighted images, was obtained with a sufficiently good spectral line width based on visual analysis.
Statistical Analysis
ROC curve analyses were first used to determine the cutoff values of individual imaging metrics, with the histologically confirmed grades taken as gold standard. Optimal threshold values were obtained by 1) minimizing the observed number of tumors misclassified (C2 error = fraction of misclassified tumors) and 2) maximizing the average of the observed sensitivity and specificity (C1 error = 1−(sensitivity + specificity)/2). Based on these, sensitivity, specificity, PPV, and NPV were calculated for characterization of gliomas into high and low grade. Logistic regression was performed using forward LR method based on threshold values obtained for each imaging measure.
RESULTS
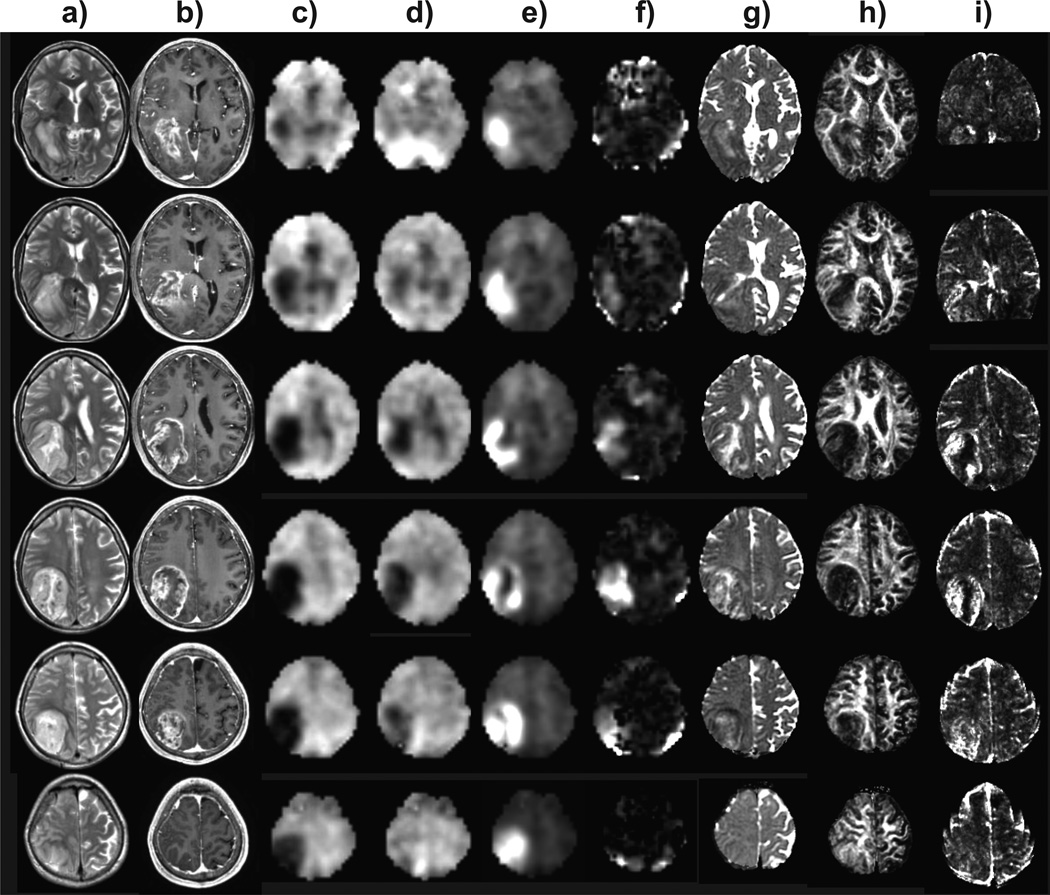
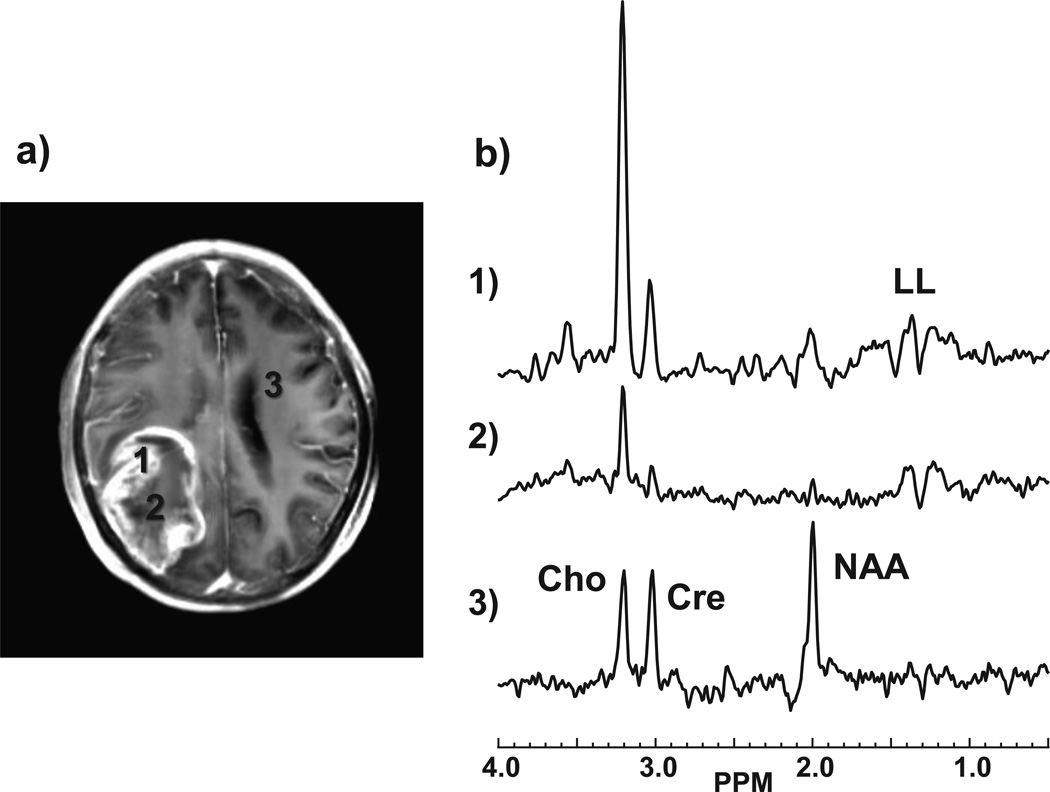
Of 56 subjects included in the study, histopathology indicated 32 with high grade and 24 low grade glioma. Contrast enhancement was seen in 35%, 89%, and 100% of grade II, III, and IV tumors respectively, which is consistent with earlier studies.14–16 In Figs. 1 and 2 are shown example images and spectra for a GBM in the right parietal region of a 48 year old male subject. Findings include the absence of NAA (Fig. 1c) over much of the T2-hyperintense region, which extends outside of the volume indicated by the T1 contrast enhancement, together with reduced Cre and strong increase of Cho, as shown in Fig. 2b. Increased LL, which was visually attributed to be primarily representative of lactate, can be seen in Figs. 1e and 2b within the central region. Results also show decreased FA and locally increased CBV. This result also demonstrates the ability of the whole-brain MRSI method to sample a wide volume of the brain and including the cortical surface at the position of the tumor.
Figure 1.
Example multiparametric results for a grade 4 glioblastoma multiforme. Multi-slice axial images are shown that correspond to every second slice of the volumetric MRSI study, at 11.2 mm spacing. Each column shows results for a) T2-weighted MRI, b) post-contrast T1-weighted MRI, c) NAA, d) Cre, e) Cho, f) LL, g) ADC, h) FA, and i) CBV.
Figure 2.
Example spectra corresponding to the GBM study shown in Fig. 1. The locations of the selected spectra are indicated on the post-contrast T1 MRI (a) which has been integrated over ten 1-mm slices to correspond to the slice thickness of the MRSI study. The spectra (b) correspond to regions of: 1) highest Cho; 2) smallest NAA+Cre; and 3) normal appearing white matter.
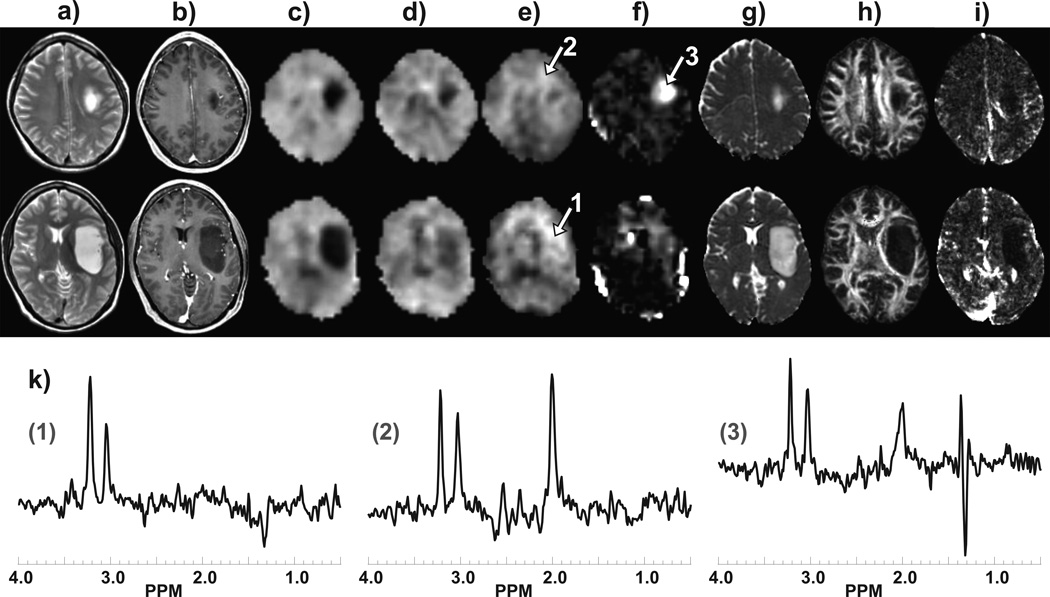
In Fig. 3 are shown images and spectra from two slices for a left insula grade II astrocytoma for a 35 year old male. MRSI results demonstrate an absence of NAA and reduced Cre, similar to the previous example, together with a moderate increase of Cho and a region with high lactate. The findings include increased ADC and decreased FA and CBV that are confined to the region indicated by altered T1 and T2 contrast.
Figure 3.
Example images and spectra at two slices for a subject with grade II astrocytoma. Results are shown for a) T2-weighted MRI, b) post-contrast T1-weighted MRI, c) NAA, d) Cre, e) Cho, f) LL, g) ADC, h) FA, and i) CBV. In k) are shown sample spectra corresponding to the number regions indicated in (e) and (f), with spectral assignments as shown in Fig. 2b.
Based on radiologist's reading, conventional MRI could discriminate low grade glioma from high grade with sensitivity, specificity, PPV, and NPV as 84%, 67%, 77%, and 76% respectively with minimum C1 and C2 error as 24% and 23% respectively. For parameters obtained from T2-weighted FSE, DTI, DCE-MRI and whole brain MRSI, threshold values were obtained separately for minimum C1 and C2 error and are shown in Tables 1, 2 and 3. Best performance for a single imaging measure was obtained for rCBV, for which minimum C1 and C2 error was obtained with a threshold value of 3.34 and provided sensitivity and specificity of 100% and 88% respectively (Table 1).
Table 1.
Threshold values of DCE derived CBV and DTI metrics for differentiating between low- and high-grade gliomas by placing ROIs on areas with maximum CBV, maximum FA and minimum ADC.
| Parameters | Based on minimum C1 error | Errors | Based on minimum C2 error | Errors | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | |
| rCBV | 3.34 | 1.0 | 0.88 | 0.91 | 1 | 0.06 | 0.05 | 3.34 | 1.0 | 0.88 | 0.91 | 1 | 0.06 | 0.05 |
| FA | 0.30 | 0.91 | 0.71 | 0.80 | 0.85 | 0.19 | 0.18 | 0.28 | 0.94 | 0.67 | 0.79 | 0.89 | 0.20 | 0.18 |
| ADC | 0.00098 | 0.94 | 0.75 | 0.83 | 0.90 | 0.16 | 0.14 | 0.00098 | 0.94 | 0.75 | 0.83 | 0.90 | 0.16 | 0.14 |
Table 2.
Threshold values of various metabolites for differentiating between low- and high-grade gliomas by placing ROIs on areas with minimum creatine and minimum NAA
| Based on minimum C1 error | Errors | Based on minimum C2 error | Errors | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | |
| tCho | 2215.32 | 0.78 | 0.42 | 0.64 | 0.59 | 0.40 | 0.38 | 2215.32 | 0.78 | 0.42 | 0.64 | 0.59 | 0.40 | 0.38 |
| tCre | 4847.01 | 0.66 | 0.79 | 0.81 | 0.63 | 0.28 | 0.28 | 5365.19 | 0.72 | 0.71 | 0.77 | 0.65 | 0.29 | 0.28 |
| NAA | 2979.04 | 0.66 | 0.67 | 0.72 | 0.59 | 0.34 | 0.34 | 3302.88 | 0.72 | 0.58 | 0.70 | 0.61 | 0.35 | 0.34 |
| Cho/Cr | 0.49 | 0.75 | 0.62 | 0.73 | 0.65 | 0.31 | 0.30 | 0.39 | 0.94 | 0.42 | 0.68 | 0.83 | 0.32 | 0.28 |
| Lactate | 3666.28 | 0.78 | 0.58 | 0.71 | 0.67 | 0.32 | 0.30 | 3666.28 | 0.78 | 0.58 | 0.71 | 0.67 | 0.32 | 0.30 |
| CreT/CreN | 0.41 | 0.56 | 0.79 | 0.78 | 0.58 | 0.32 | 0.34 | 0.41 | 0.56 | 0.79 | 0.78 | 0.58 | 0.32 | 0.34 |
| ChoT/ChoN | 0.57 | 1 | 0.42 | 0.70 | 1 | 0.29 | 0.25 | 0.57 | 1 | 0.42 | 0.70 | 1 | 0.29 | 0.25 |
| T2T/T2N | 1.73 | 0.94 | 0.62 | 0.77 | 0.88 | 0.22 | 0.20 | 1.73 | 0.94 | 0.62 | 0.77 | 0.88 | 0.22 | 0.20 |
Table 3.
Threshold values of various metabolites for differentiating between low- and high-grade gliomas by placing ROIs on areas with high Choline
| Parameters | Based on minimum C1 error | Errors | Based on minimum C2 error | Errors | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | Threshold | Sensitivity | Specificity | PPV | NPV | C1 | C2 | |
| Cho | 5704.98 | 0.69 | 0.58 | 0.69 | 0.58 | 0.36 | 0.36 | 3662.43 | 1.00 | 0.21 | 0.63 | 1.00 | 0.40 | 0.34 |
| Cre | 8954.16 | 0.47 | 0.79 | 0.75 | 0.53 | 0.37 | 0.39 | 8954.16 | 0.47 | 0.79 | 0.75 | 0.53 | 0.37 | 0.39 |
| NAA | 3858.13 | 0.38 | 0.83 | 0.75 | 0.50 | 0.40 | 0.43 | 11670.49 | 0.94 | 0.08 | 0.58 | 0.50 | 0.49 | 0.43 |
| Cho/Cr | 0.87 | 0.34 | 1.00 | 1.00 | 0.53 | 0.33 | 0.38 | 0.52 | 0.69 | 0.62 | 0.71 | 0.60 | 0.34 | 0.34 |
| Lactate | 4580.80 | 0.62 | 0.58 | 0.67 | 0.54 | 0.40 | 0.39 | 2650.64 | 0.88 | 0.29 | 0.62 | 0.64 | 0.42 | 0.38 |
| CreT/CreN | 1.26 | 0.31 | 0.96 | 0.91 | 0.51 | 0.36 | 0.41 | 1.14 | 0.34 | 0.92 | 0.85 | 0.51 | 0.37 | 0.41 |
| ChoT/ChoN | 2.13 | 0.69 | 0.88 | 0.88 | 0.68 | 0.22 | 0.23 | 1.76 | 0.88 | 0.62 | 0.76 | 0.79 | 0.25 | 0.23 |
| T2T/T2N | 1.59 | 0.94 | 0.58 | 0.75 | 0.88 | 0.24 | 0.21 | 1.59 | 0.94 | 0.58 | 0.75 | 0.88 | 0.24 | 0.21 |
The combination of spectral parameter, rCBV, FA, ADC and T2Tumor/T2Normal where ROIs were placed on region with high choline resulted in sensitivity, specificity, PPV, and NPV of 100%, 88%, 91%, and 100%, respectively for minimum C1 as well as C2 error. When ROIs were placed on region with minimal NAA and creatine, the combination of spectral parameter, rCBV, FA, ADC and T2Tumor/T2Normal provided sensitivity, specificity, PPV, and NPV of 100%, 96%, 97%, and 100%, respectively for minimum C1 as well as C2 error (Table 4).
Table 4.
tCho, tCre, NAA, Cho/Cr, Lactate, CreT/CreN, ChoT/ChoN, T2T/T2N, rCBV, FA and ADC together for differentiating between low- and high-grade gliomas
| ROIs placed on region with |
Threshold based on minimum C1 error |
Errors | Threshold based on minimum C2 error |
Errors | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | C1 | C2 | Sensitivity | Specificity | PPV | NPV | C1 | C2 | |
| HighCholine | 1.0 | 0.88 | 0.91 | 1 | 0.06 | 0.05 | 1.0 | 0.88 | 0.91 | 1 | 0.06 | 0.05 |
| Low NAA &LowCreatine | 1.00 | 0.96 | 0.97 | 1 | 0.02 | 0.02 | 1.00 | 0.96 | 0.97 | 1 | 0.02 | 0.02 |
The MRSI data showed considerable overlap of spectral characteristics across all tumor grades, with 97% of studies having a region with no NAA signal and 68% (58%, 50%, and 88% for grades II, III, and IV respectively) indicating the presence of lactate, as confirmed by visual identification of the lactate doublet in the spectra. Of 61 studies for which MRSI data was suitable for evaluation, the extent to which the whole-brain MRSI acquisition sampled the tumor volume with adequate quality indicated that 72% of the studies provided >75% coverage of the tumor and 20% of studies sampled between 25% and 75% of the tumor volume.
DISCUSSION
The major finding of this study is that that a combination of parameters from DCE-MRI, DTI, and whole brain MRSI enables classification of gliomas into high and low grade with accuracy near to the classification based on histopathology. Results suggest that DCE-MRI derived rCBV is most efficient in classification of high grade glioma from low grade as compare to DTI derived metrices and spectral maps obtained from whole brain MRSI.
Several reports have demonstrated the advantages of multiparametric MR measures for evaluation of gliomas at 1.5T and 3T.1–3,9 Costanzo et al.9 have shown that multiparametric MR assessment of glioma, based on 1H-MRSI, PWI and DWI, can discriminate infiltrating tumor from surrounding vasogenic edema or normal tissues. Fink et al.3 have compared multi-voxel and single-voxel proton MR spectroscopy, DSC MRI, and DWI for distinguishing recurrent glioma from post-radiation injury where they have found that DSC and multi-voxel MRS Cho/Cr peak-area and Cho/NAA peak- height appear to outperform DWI for distinguishing glioma from post-treatment effects. Catalaa et al.2 evaluated perfusion, diffusion, and spectroscopy values in enhancing and non-enhancing lesions for patients with newly diagnosed gliomas of different grades to find increased cell density and increased vascularity within enhancing lesions and increased cellularity in non-enhancing region. However, these studies did not evaluate the ability to classify gliomas into low and high grade using quantitative threshold values based on ROC analysis. Analyses of single imaging measures as well as multiparametric combinations have been used for this purpose. For studies carried out at 3T, MRS had been evaluated for grading of cerebral gliomas at different TE values17 and ROC analysis was conducted for glioma classification using pharmacokinetic indices.7 Minimum peritumoral ADC derived from DTI was used to obtain a cutoff value of 1.302×10−3 mm2/s using ROC curve analysis for distinguishing between GBM and metastasis.4 Combining two techniques such as DTI and DSC-MRI in grading of preoperative nonenhancing gliomas has been reported to discriminate low and high grade with specificity of 92.3% and sensitivity of 86.7%.5 DCE-MR and spectroscopic imaging were combined to obtain specificity and sensitivity as 93.3% and 60.0% respectively to discriminate high and low grade glioma.13
In the present study, conventional MRI was able to classify gliomas into low and high grade with a sensitivity and specificity of 84% and 67% respectively.18–20 Among all the parameters considered, rCBV most efficiently characterized the grades with high sensitivity and specificity and minimal error, which is consistent with previous studies.8 For regions with minimal NAA+Cre, the ratio of signal intensity of T2 with normal contralateral white matter and Creatine were most effective for classification of glioma as compared to the other metabolite maps obtained from MRSI. While ROIs placed on regions with maximal Cho gave the ratio of the T2 MRI signal intensity in the tumor to normal contralateral white matter and ChoTumor/ChoNormal as the most effective classifier. The combination of rCBV, FA, ADC and spectroscopic parameters was able to discriminate high and low grade with only 6% and 5% of C1 and C2 error respectively when ROIs were placed in region with maximum rCBV, FA, ADC and choline. Combinations of the same parameters obtained from ROIs placed in regions with maximum rCBV, FA, ADC and minimal NAA+Cre were able to discriminate high and low grade with 2% of C1 and C2 error and sensitivity and specificity as 100% and 96% respectively. This considerable improvement in discrimination between grades is due to the complimentary information provided from perfusion weighted, diffusion weighted and spectroscopic parameters.
This is the first study evaluating the whole-brain MRSI method for tumor characterization. The availability of the volumetric MRSI measurement with relatively good spatial resolution provided sufficient coverage of the brain with 92% of the studies analyzed considered to have sampled a portion of the tumor volume considered sufficient to make a ROI based measurement. A potential benefit of the improved spatial sampling and relatively good resolution over conventional PRESS-volume selected MRSI methods is improved accuracy for ROI selection. Nevertheless, the diagnostic performance of the MRSI method used in this study remains comparable to previous reports carried out at 1.5T using two-dimensional PRESS volume selected MRSI13 and reporting metabolite ratios. In comparison to the study of Law et al.13 for Cho/Cre, this study shows comparable sensitivity and a small improvement in specificity. An additional observation is that use of signal-normalized individual metabolite values resulted in a diagnostic performance similar to that obtained from the Cho/Cr or using normalization by the values in contralateral while matter.
A new finding of this study is the potential of Cre as a diagnostic marker. Although widely used as a concentration standard, there is regional and individual variability in concentration21 and it has been shown to be decreased in tumors22, however higher Cre values relative to NAWM values were associated with shorter survival23. The results of this study indicate that when ROIs were selected based on minimum creatine and NAA, the classifications based on either the signal normalized Cre or normalized to the value in NAWM achieved the highest specificity of the MRS measures.
This study is the first to apply a ROC analysis to individual metabolite values normalized to tissue water. While this addresses the known limitation of taking metabolite ratios to Cre, a remaining limitation is that the signal normalization did not included measured water density and T1 values, for which alterations from normal values could affect measurements both within the tumor or NAWM. It is, however, notable that performance for Cho alone was poorer than that of Cho/Cre and ChoT/ChoN.
CONCLUSION
The results of this study demonstrate that the application of a comprehensive multiparametric MR protocol enables grading of gliomas with almost 100% accuracy. This approach will provide physicians treating patients with primary glioma with additional diagnostic information.
Abbreviation Key
- FA
Fractional anisotropy
- NPV
Negative predictive value
- PPV
Positive predictive value
- DCE
Dynamic contrast enhanced
- ROC
Receiver operating characteristic
REFERENCES
- 1.Chiang IC, Kuo YT, Lu CY, et al. Distinction between high-grade gliomas and solitary metastases using peritumoral 3-T magnetic resonance spectroscopy, diffusion, and perfusion imagings. Neuroradiology. 2004;46:619–627. doi: 10.1007/s00234-004-1246-7. [DOI] [PubMed] [Google Scholar]
- 2.Catalaa I, Henry R, Dillon WP, et al. Perfusion, diffusion and spectroscopy values in newly diagnosed cerebral gliomas. NMR Biomed. 2006;19:463–475. doi: 10.1002/nbm.1059. [DOI] [PubMed] [Google Scholar]
- 3.Fink JR, Carr RB, Matsusue E, et al. Comparison of 3 Tesla proton MR spectroscopy, MR perfusion and MR diffusion for distinguishing glioma recurrence from posttreatment effects. J Magn Reson Imaging. 2012;35:56–63. doi: 10.1002/jmri.22801. [DOI] [PubMed] [Google Scholar]
- 4.Lee EJ, Brugge K, Mikulis D, et al. Diagnostic value of peritumoral minimum apparent diffusion coefficient for differentiation of glioblastoma multiforme from solitary metastatic lesions. AJR. 2011;196:71–76. doi: 10.2214/AJR.10.4752. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Tian W, Kolar B, et al. MR diffusion tensor and perfusion-weighted imaging in preoperative grading of supratentorial nonenhancing gliomas. Neuro Oncol. 2011;13:447–455. doi: 10.1093/neuonc/noq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law M, Yang S, Babb JS, et al. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25:746–755. [PMC free article] [PubMed] [Google Scholar]
- 7.Jia Z, Geng D, Xie T, et al. Quantitative analysis of neovascular permeability in glioma by dynamic contrast-enhanced MRimaging. Journal of Clinical Neuroscience. 2012;19:820–823. doi: 10.1016/j.jocn.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Awasthi R, Rathore RKS, Soni P, et al. Discriminant analysis to classify glioma grading using dynamic contrast-enhanced MRI and immunohistochemical markers. Neuroradiology. 2011;54:205–213. doi: 10.1007/s00234-011-0874-y. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo AD, Scarabino T, Trojsi F, et al. Multiparametric 3T MR approach to the assessment of cerebral gliomas: tumor extent and malignancy. Neuroradiology. 2006;48:622–631. doi: 10.1007/s00234-006-0102-3. [DOI] [PubMed] [Google Scholar]
- 10.Maudsley AA, Darkazanli A, Alger JR, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awasthi R, Verma SK, Haris M, et al. Comparative Evaluation of Dynamic Contrast-Enhanced Perfusion With Diffusion Tensor Imaging Metrics in Assessment of Corticospinal Tract Infiltration in Malignant Glioma. J Comput Assist Tomogr. 2010;34:82–88. doi: 10.1097/RCT.0b013e3181ae29f0. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Haris M, Rathore DKS, et al. Quantification of physiological and hemodynamic indices using T1 dynamic contrast-enhanced MRI in intracranial mass lesions. J Magn Reson Imaging. 2007;26:871–880. doi: 10.1002/jmri.21080. [DOI] [PubMed] [Google Scholar]
- 13.Law M, Yang S, Wang H, et al. Glioma Grading: Sensitivity, Specificity, and Predictive Values of Perfusion MR Imaging and Proton MR Spectroscopic Imaging Compared with Conventional MR Imaging. AJNR Am J Neuroradiol. 2003;24:1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 14.Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high grade gliomas. AJNR. 2005;26:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 15.Chaichana KL, McGirt MJ, Niranjan A, et al. Prognostic significance of contrast-enhancing low-grade gliomas in adults and a review of the literature. Neurol Res. 2009;31:931–939. doi: 10.1179/174313209X395454. [DOI] [PubMed] [Google Scholar]
- 16.Lote K, Egeland T, Hager B, et al. Prognostic significance of CT contrast enhancement within histological subgroups of intracranial glioma. J Neurooncol. 1998;40:161–170. doi: 10.1023/a:1006106708606. [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, Chang KH, Nab DG, et al. 3T 1H-MR Spectroscopy in Grading of Cerebral Gliomas: Comparison of Short and Intermediate Echo Time Sequences. AJNR. 2006;27:1412–1418. [PMC free article] [PubMed] [Google Scholar]
- 18.Moller-Hartmann W, Herminghaus S, Krings T, et al. Clinical application of proton magnetic resonance spectroscopy in the diagnosis of intracranial mass lesions. Neuroradiology. 2002;44:371–381. doi: 10.1007/s00234-001-0760-0. [DOI] [PubMed] [Google Scholar]
- 19.Dean BL, Drayer BP, Bird CR, et al. Gliomas: classification with MR imaging. Radiology. 1990;174:411–415. doi: 10.1148/radiology.174.2.2153310. [DOI] [PubMed] [Google Scholar]
- 20.Knopp EA, Cha S, Johnson G, et al. Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology. 1999;211:791–798. doi: 10.1148/radiology.211.3.r99jn46791. [DOI] [PubMed] [Google Scholar]
- 21.Maudsley AA, Domenig C, Govindaraju V, et al. Mapping of brain metabolite distributions by volumetric proton MRSI. Magn. Reson. Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean MA, Sun A, Bradstreet TE, et al. Repeatability of edited lactate and other metabolites in astrocytoma at 3T. J Magn Reson Imaging. 2012;36:468–475. doi: 10.1002/jmri.23673. [DOI] [PubMed] [Google Scholar]
- 23.Hattingen E, Delic O, Franz K, et al. Quantitative short echo time 1H-MR spectroscopy of untreated pediatric brain tumors: preoperative diagnosis and characterization. Neurol Res. 2010;32:593–602. [Google Scholar]