Abstract
Background
Tumor necrosis factor-α (TNF-α) has been suggested to play a very important role in the development and progression of hepatocellular carcinoma (HCC). Many studies have identified the associations of TNF-α-308 and -238 polymorphisms with HCC risk, but the results remain controversial.
Aim
We conducted this meta-analysis to evaluate the associations between TNF-α-308 and -238 polymorphisms and HCC susceptibility.
Methods
PubMed, Embase, and China National Knowledge Infrastructure electronic databases were searched for all articles on associations between TNF-α-308 and -238 polymorphisms and HCC risk in Asians through September 30, 2013. Odds ratios (ORs) and their 95% CIs were calculated to assess the strength of this association.
Results
A total of 17 case–control studies were identified in our meta-analysis. For the TNF-α-308 G/A polymorphism, 14 studies containing 3154 cases and 3767 controls were included. Overall, the frequency of the A allele was higher in patients with HCC than in the healthy controls (10.2% vs 7.5%), and the A allele and allele carrier were significantly associated with increased risk of HCC in a random effects model (A vs G: OR = 1.57; 95% CI, 1.22–2.01; P = 0.0004; AA + AG vs GG: OR = 1.62; 95% CI, 1.18–2.22; P = 0.003). For the TNF-α-238 polymorphism, 10 research articles were identified. No association was found between the TNF-α-238 G/A polymorphism and risk of HCC in any genetic models (P > 0.05). The sensitivity analysis further strengthened the overall correlations.
Conclusions
Our meta-analysis proved that the TNF-α-308 G/A polymorphism is associated with increased susceptibility to HCC. However, the TNF-α-238 G/A polymorphism is not significantly associated with risk of HCC in Asian populations. Further studies with large sample sizes are needed to confirm these associations among other populations.
Key words: hepatocellular carcinoma, meta-analysis, polymorphism, tumor necrosis factor-α
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third cause of cancer-related mortality worldwide.1 More than 600,000 people die from HCC each year.2 Although several major risk factors of HCC have been shown to contribute to hepatocarcinogenesis, the etiology is still unclear. Chronic inflammation developing through the action of various inflammatory mediators has been recently identified as a cofactor in carcinogenesis.3 Among inflammatory mediators, tumor necrosis factor-α (TNF-α) plays an essential role and has been implicated in inflammation-associated tumors.4
TNF-α, produced by diverse kinds of cells, is 1 of the most important proinflammatory cytokines.5 Researchers have demonstrated that variation between individual TNF-α concentrations are attributed to single nucleotide polymorphisms (SNPs) in the TNF promoter region.6,7 These SNPs have been reported to be associated with breast cancer,8 gastrointestinal cancer,9 prostate cancer,10 lung cancer,11 Graves disease,12 and Alzheimer disease.13
During past years a number of case–control studies have been conducted to investigate the associations between TNF-α polymorphisms and HCC risk in humans, among which TNF-α-308 and -238 G/A polymorphisms are most highlighted and have been considered as risk factors for HCC.14,15 But the findings from previous studies have been controversial. For example, Qin et al16 suggested that the TNF-α-308 G/A polymorphism is associated with a modest decrease in the risk of developing HCC, whereas Teixeira et al17 showed that the TNF-α-308 G/A polymorphism is significantly associated with higher risk of developing HCC. Tian et al18 provided convincing evidence that the TNF-α-238 G/A polymorphism is significantly associated with increased risk of developing HCC, whereas Heneghan et al19 failed to highlight specific associations between the 238 G/A polymorphisms tested and the risk of developing HCC. These discrepancies might be due to genetic trait differences and different linkage disequilibrium. Therefore, to derive a more comprehensive estimation of the associations between TNF-α polymorphisms and HCC risk, we conducted a meta-analysis to assess the association between TNF-α-308 and -238 G/A polymorphisms and HCC susceptibility in Asians.
Materials and Methods
Publication search
A computerized search of PubMed, EMBase, and China National Knowledge Infrastructure electronic databases was conducted to identify relevant articles published between 2003 and 2013 related to HCC risk and TNF-α-308 and -238 G/A polymorphisms. The key words were hepatocellular carcinoma, liver cancer, polymorphism, variant, and tumor necrosis factor or TNF-α as well as their combinations. The corresponding Chinese terms were used in the Chinese database. References of retrieved articles were checked for other eligible studies. The languages of selected articles were restricted to English and Chinese. The search was focused on studies that had been conducted in human beings. Only full-text articles and the most recent studies were included in our meta-analysis.
Selection criteria
The included studies met the following criteria: a case–control or cohort association study; confined to Asian populations; include at least 1 of the 2 SNPs, TNF-α-308, and TNF-α-238; results expressed as odds ratio (OR) and corresponding 95% CI; and genotype distribution of control for a certain polymorphism must be in Hardy-Weinberg equilibrium.
Data extraction
Two investigators independently extracted data and reached a consensus on all of the items. Any disagreement was resolved by discussing with the third expert. The following information was extracted from each article: first author, year of publication, total numbers of case patients and control subjects, and genotyping information. Furthermore, we examined if matching had been used and if the genotyping assay had been validated.
Statistical analyses
Statistical analyses were carried out using Review Manager 5.2 (The Cochrane Collaboration, Oxford, United Kingdom). The crude ORs with 95% CIs were used to measure the associations between TNF-α polymorphisms and risk of HCC, which were calculated according to the method of Woolf. The significance of the pooled ORs was determined by the z test, and a P value < 0.05 was considered statistically significant. The allelic model (A vs G) and genotype genetic models (codominant effects: AA vs GG; dominant effect: AA + AG vs GG; and recessive effect: AA vs AG + GG) were examined to evaluate the G/A allele with the risk of HCC. Heterogeneity of included studies was estimated using Cochran’s Q test and I2 statistics. P values < 0.1 and I2 <50% were considered to be statistically significant.20 The fixed effects model was used when the effects were assumed to be homogenous, whereas the random effects model was used when they were heterogeneous. To assess if our results were substantially influenced by the presence of any individual study, we conducted a sensitivity analysis by systematically removing each study and recalculating the significance of the result. Funnel plots were used to evaluate publication bias. All P values were 2-tailed.
Results
Study characteristics
Two hundred thirteen articles were found with our search criteria. After discarding the duplicate research articles, 154 potential records concerned the topic. Following the title and abstract review, 40 full-text articles were preliminarily identified for further detailed evaluation. Overall, the initial search with the key words and the subject terms identified 17 articles (7 in Chinese and 10 in English) that met the inclusion criteria and were eligible for review. The study selection process is shown in Figure 1.
Fig. 1.
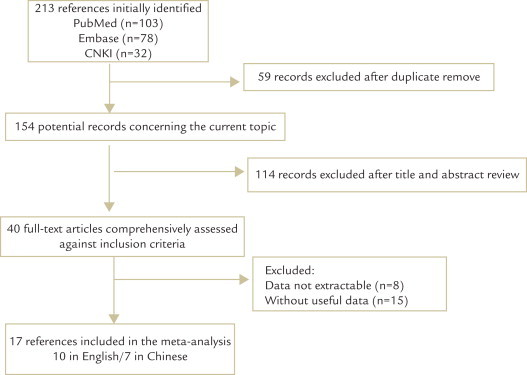
Flow chart of study selection process in our meta-analysis. CNKI = China National Knowledge Infrastructure electronic database.
All 17 studies were identified to evaluate the relationship between the TNF-α-308 and -238 polymorphisms and risk for HCC. For the TNF-α-308 polymorphism, 14 relevant studies (4 in Chinese21–24 and 10 in English19,25–33) with a total of 3154 cases and 3767 controls were included; for the TNF-α-238 polymorphism, 10 research articles (4 in Chinese22,34–36 and 6 in English19,22,25,29,30,32) were identified, including 1388 cases and 2117 controls. Characteristics of the included studies are shown in Table I. All the studies were conducted in persons of Asian descent. Furthermore, the genotype distributions among the control subjects of all studies were consistent with Hardy-Weinberg equilibrium.
Table I.
Main characteristic and genotype distributions of studies included in the meta-analysis.
| First author | Year | Country | Total |
Genotype frequencies (GG/AG/AA) |
||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||
| TNF-α-308 | ||||||
| Heneghan | 2003 | China | 98 | 172 | 88/10/0 | 158/13/1 |
| Wang | 2003 | Japan | 125 | 204 | 109/15/1 | 179/23/2 |
| Ho | 2004 | China | 74 | 289 | 37/34/3 | 225/62/2 |
| Chen | 2005 | China | 572 | 381 | 468/95/9 | 311/67/3 |
| Migita | 2005 | Japan | 48 | 188 | 47/1/0 | 183/5/0 |
| Kummee | 2007 | Thailand | 50 | 150 | 42/6/2 | 123/15/12 |
| Jeng | 2007 | China | 108 | 108 | 80/28/0 | 100/8/0 |
| Sakamoto | 2008 | Japan | 209 | 275 | 205/4/0 | 270/5/0 |
| Song | 2009 | China | 81 | 78 | 53/28/0 | 66/12/0 |
| Jeng | 2009 | China | 200 | 200 | 149/51/0 | 188/12/0 |
| Yang | 2012 | China | 772 | 852 | 619/145/8 | 683/160/9 |
| Wang CK | 2012 | China | 620 | 625 | 496/118/6 | 510/108/7 |
| Shi | 2012 | China | 73 | 116 | 51/20/2 | 84/30/2 |
| Wang | 2012 | China | 124 | 129 | 96/28/0 | 114/15/0 |
| TNF-α-238 | ||||||
| Heneghan | 2003 | China | 98 | 172 | 96/2/0 | 168/4/0 |
| Wang | 2003 | Japan | 125 | 204 | 111/13/1 | 178/24/2 |
| Huang | 2007 | China | 100 | 150 | 88/12/0 | 143/7/0 |
| Kummee | 2007 | Thailand | 50 | 150 | 44/5/1 | 96/4/50 |
| Jeng | 2007 | China | 108 | 108 | 102/6/0 | 106/2/0 |
| Jeng | 2009 | China | 200 | 200 | 194/6/0 | 198/2/0 |
| Jung | 2009 | Korea | 227 | 365 | 193/34/0 | 336/28/1 |
| Wang | 2010 | China | 230 | 513 | 209/20/1 | 455/57/1 |
| Chen | 2011 | China | 126 | 126 | 120/6/0 | 115/11/0 |
| Wang | 2012 | China | 124 | 129 | 117/7/0 | 125/4/0 |
TNF-α= tumor necrosis factor-α.
Association between the TNF-α-308 G/A polymorphism and HCC
The main results of meta-analysis of the association between the TNF-α-308 G/A polymorphism and HCC risk are shown in Table II. Overall, the frequency of the A allele is higher in patients with HCC than in healthy controls (10.2% vs 7.5%). As shown in Figures 2 and 3, our results showed that there was a significant association between the TNF-α-308 G/A polymorphism and increased risk of HCC under allelic and dominant effects (A vs G: OR = 1.57; 95% CI, 1.22–2.01; P = 0.0004; AA + AG vs GG: OR = 1.62; 95% CI, 1.18–2.22; P = 0.003) in a random-effects model, indicating that being an A carrier may be a risk factor for developing HCC among Asians. However, under codominant effects and recessive effects, no significant associations were found (AA vs GG: OR = 1.15; 95% CI, 0.70–1.89; P = 0.57; AA vs AG + GG: OR = 1.12; 95% CI, 0.68–1.84; P = 0.65) in a fixed model.
Table II.
Results of pooled odds ratios (ORs) in this meta-analysis.
| Genotype | Tumor necrosis factor-α-308 |
Tumor necrosis factor-α-238 |
||||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | Pheterogeneity/I2 | OR (95% CI) | P | Pheterogeneity/I2 | |
| A vs G | 1.57 (1.22– 2.01) | 0.0004 | 0.0002/67% | 1.06 (0.59– 1.92) | 0.85 | <0.00001/78% |
| AA vs GG | 1.15 (0.70–1.89) | 0.57 | 0.38/7% | 0.40 (0.05–2.92) | 0.36 | 0.06/59% |
| AA + AG vs GG | 1.62 (1.18–2.22) | 0.003 | <0.00001/77% | 1.13 (0.68–1.89) | 0.64 | 0.001/67% |
| AA vs AG + GG | 1.12 (0.68–1.84) | 0.65 | 0.55/0% | 0.39 (0.05–3.02) | 0.37 | 0.05/61% |
Fig. 2.
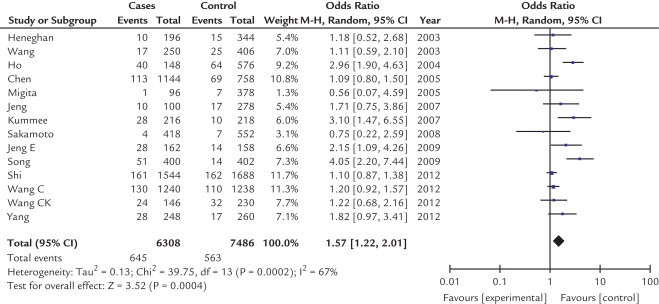
Forest plot of the association between A allele and hepatocellular carcinoma according to tumor necrosis factor-α-308 polymorphism in an Asian population. M-H = xxxxxxxx.
Fig. 3.
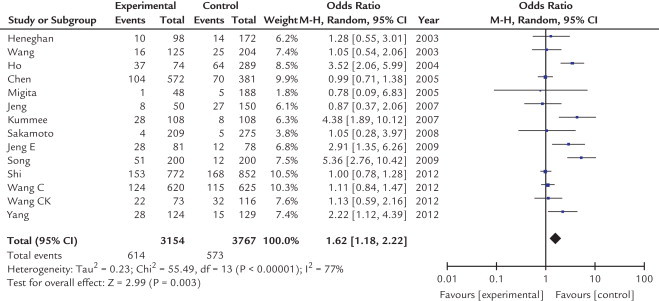
Forest plot of the association between A carriers (AA + AG) and hepatocellular carcinoma according to tumor necrosis factor (TNF)-α-308 polymorphism in 14 studies using random-effect model in an Asian population. AA + GA, AA, and GA genotypes of TNF-α-308 polymorphism; and GG, GG genotype of TNF-α-308 polymorphism. M-H = xxxxxxxx.
Association between the TNF-α-238 G/A polymorphism and HCC
Table II shows the OR for HCC of TNF-α-238 G/A polymorphism with the variant genotypes. The heterogeneity between studies was significant and a random-effects model was employed. As shown in Figures 4 and 5, no significant associations were found in any genotypes (A vs G: OR = 1.06; 95% CI, 0.59–1.92; P = 0.85; AA vs GG: OR = 0.40; 95% CI, 0.05–2.92; P = 0.36; AA + AG vs GG: OR = 1.13; 95% CI, 0.68–1.89; P = 0.64; AA vs AG + GG: OR = 0.39; 95% CI, 0.05–3.02; P = 0.37). This demonstrated that this polymorphism was not associated with HCC risk.
Fig. 4.
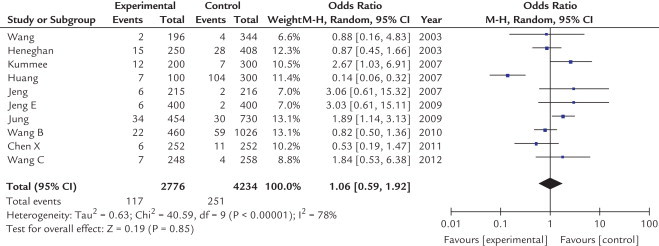
Odds ratios and 95% CIs of hepatocellular carcinoma according to A allele of tumor necrosis factor-α-238 polymorphism. M-H = xxxxxxxx.
Fig. 5.
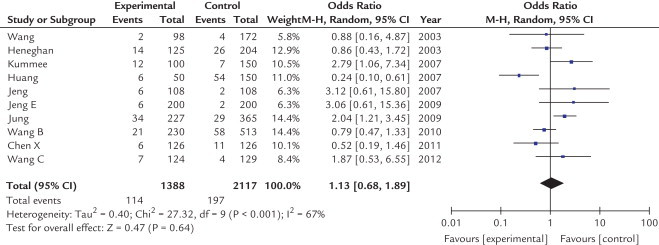
Odds ratios and 95% CIs of hepatocellular carcinoma according to tumor necrosis factor-α-238 polymorphism in 10 studies using a random-effect model. AA + GA, AA, and GA genotypes of tumor necrosis factor-α-238 polymorphism and GG, GG genotype of tumor necrosis factor-α-238 polymorphism. M-H = xxxxxxxx.
Sensitivity analysis
The influence of a single study on the overall meta-analysis estimate was investigated by omitting 1 study at a time, and the omission of any study made no significant difference, indicating that our results were statistically reliable.
Publication bias
The shape of the funnel plots was symmetrical, suggesting that there was no evidence of publication bias among these studies. Figure 6 displayed funnel plots for the associations between the TNF-α-308 polymorphism and HCC. The statistical results still did not show publication bias.
Fig. 6.
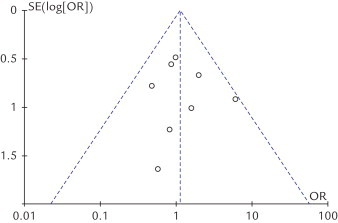
Funnel plots analysis to detect publication bias of this meta-analysis. Each point represents an independent study for the indicated association.
Discussion
Carcinogenesis of HCC is a complex process. The risk factors include chronic infection of hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, carcinogen exposure, excessive alcohol consumption, and a variety of genetic factors.37 During recent decades, inflammation has been considered an important factor involved in carcinogenesis. TNF-α, the most crucial inflammatory cytokine, plays a very important role in inflammation and in the pathogenesis of cancer.38 Studies have demonstrated that TNF-α was not only involved in the pathogenesis of both hepatitis and liver cirrhosis,39,40 but also involved in the development of HCC.41,42 Moreover, TNF-α-308 and -238 polymorphisms have been reported to be related to risk of HCC because the G/A polymorphism can change the transcription of TNF-α and TNF-α production. However the results were inconsistent due to the rather small size and unified ethnicity. To provide further investigation into these controversial points, our meta-analysis was conducted to achieve a more reliable and comprehensive conclusion.
We investigated the association between TNF-α-308 and -238 polymorphisms and risk of HCC. A total of 17 studies were included, 14 for the TNF-α-308 polymorphism and 10 for the TNF-α-238 polymorphism. The results showed that the variant genotypes AA/AG of TNF-α-308 G/A were associated with a significantly increased HCC risk when compared with the GG genotype. Our result is consistent with a previous meta-analysis.43 However this association was limited to Asian populations. For the TNF-α-238 G/A polymorphism, no association was found among Asian people. This result was consistent with Zhou et al43 and Yang et al,44 but not consistent with Wei et al45 and Cheng et al.46 In this study, heterogeneity was found for TNF-α-308 and -238 polymorphisms, so a random-effects model was selected and sensitivity analysis was performed. By removing each study from our meta-analysis each time and rerunning the model to determine the effect on each overall estimate, we found quite little change. This confirmed the stability of our results.
Previous studies found that circulating TNF-α levels were elevated in patients with HCC.47,48 It is reasonable to speculate that the high circulating TNF-α levels found in patients with HCC may be attributed to its SNPs. TNF-α may stimulate the release of other inflammatory cytokines and also induce the production of other fibrogenic factors, such as tumor growth factor-β, interleukin-1, and interleukin-6,49 which can cause or aggravate liver damage.6 Because HCC mostly occurs in Asia where HBV and HCV infections are endemic, the association between TNF-α-308 and -238 G/A polymorphisms and HCC are very important to identify persons in Asia at greater risk of developing HCC. Our meta-analysis was only conducted among an Asian population. Furthermore, the majority of HCCs are related to HBV and HCV infection, but most studies did not provide the number of patients with HBV or HCV infection, number of controls with HBV/HCV infection, or the number of patients with HHC. This suggests that clarifying the independent role of each polymorphism on HBV, HCV, and HCC is quite necessary and might explain the discrepancies with other studies. Moreover, a previous meta-analysis with relatively small sample size may have had insufficient statistical power to detect a slight effect or may have generated a fluctuated risk estimate.
Several limitations exist in our meta-analysis. First, environmental and genetic factors, such as HBV and HCV infection, should be considered because HCC is a multifactorial disease. Second, some other single-nucleotide polymorphisms of cytokines should be identified to determine the interacting functions. Third, there was significant heterogeneity among the studies in overall comparison models. According to the problems we addressed, further studies are needed to detect the independent role of each polymorphism.
Conclusions
Our meta-analysis suggests that the TNF-α-308 polymorphism is associated with risk of HCC, although a significant association was not found between the TNF-α-238 polymorphism and HCC. Larger and more rigorous analytical studies are required to evaluate gene–environment interactions and to clarify the interaction between the TNF-α polymorphism and HBV infection status.
Conflicts of Interest
The authors have indicated that they have no conflicts of interest regarding the content of this article.
Acknowledgments
The authors thank their colleagues at the Department of Gastroenterology, The Second Affiliated Hospital, University of Zhejiang Traditional Chinese Medicine, for providing ongoing support of this project.
Dr Guo-Guang Lou offers kind helps for literature search, study design and writing. Dr Ying-Chao Liu and Le-Qian are also acknowledged for their helps in data collection and data interpretation. This project is completed under the instructions of Professor Bo-Dong Lv.
References
- 1.Waly Raphael S., Yangde Z., Yuxiang C. Hepatocellular carcinoma: focus on different aspects of management. ISRN Oncol. 2012;2012:421673. doi: 10.5402/2012/421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J.D., Roberts L.R. Epidemiology and management of hepatocellular carcinoma. Infect Dis Clin North Am. 2010;24(4):899–919. doi: 10.1016/j.idc.2010.07.004. viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson G.M., Nakada M.T., DeWitte M. Tumor necrosis factor-alpha in the pathogenesis and treatment of cancer. Curr Opin Pharmacol. 2004;4:314–320. doi: 10.1016/j.coph.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Hajeer A.H., Hutchinson I.V. Influence of TNFalpha gene polymorphisms on TNFalpha production and disease. Hum Immunol. 2001;62:1191–1199. doi: 10.1016/s0198-8859(01)00322-6. [DOI] [PubMed] [Google Scholar]
- 7.Hajeer A.H., Hutchinson I.V. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50:216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Pooja S. Role of ethnic variations in TNF-alpha and TNF-beta polymorphisms and risk of breast cancer in India. Breast Cancer Res Treat. 2011;126:739–747. doi: 10.1007/s10549-010-1175-6. [DOI] [PubMed] [Google Scholar]
- 9.Roselli M. TNF-alpha gene promoter polymorphisms and risk of venous thromboembolism in gastrointestinal cancer patients undergoing chemotherapy. Ann Oncol. 2013;24:2571–2575. doi: 10.1093/annonc/mdt251. [DOI] [PubMed] [Google Scholar]
- 10.McCarron S.L. Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res. 2002;62:3369–3372. [PubMed] [Google Scholar]
- 11.Seifart C. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu L.Q. Tumor necrosis factor alpha (TNF-alpha) polymorphisms in Chinese patients with Graves׳ disease. Clin Biochem. 2010;43:223–227. doi: 10.1016/j.clinbiochem.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Di Bona D. Systematic review by meta-analyses on the possible role of TNF-alpha polymorphisms in association with Alzheimer׳s disease. Brain Res Rev. 2009;61:60–68. doi: 10.1016/j.brainresrev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Kato N. Large-scale search of single nucleotide polymorphisms for hepatocellular carcinoma susceptibility genes in patients with hepatitis C. Hepatology. 2005;42:846–853. doi: 10.1002/hep.20860. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.J., Lee H.S. Single nucleotide polymorphisms associated with hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Intervirology. 2005;48:10–15. doi: 10.1159/000082089. [DOI] [PubMed] [Google Scholar]
- 16.Qin H. Tumour necrosis factor-alpha polymorphisms and hepatocellular carcinoma: a meta-analysis. J Int Med Res. 2010;38:760–768. doi: 10.1177/147323001003800304. [DOI] [PubMed] [Google Scholar]
- 17.Teixeira A.C. Alleles and genotypes of polymorphisms of IL-18, TNF-alpha and IFN-gamma are associated with a higher risk and severity of hepatocellular carcinoma (HCC) in Brazil. Hum Immunol. 2013;74:1024–1029. doi: 10.1016/j.humimm.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 18.Tian X. Comprehensive assessment of the association between tumor necrosis factor alpha G238A polymorphism and liver cancer risk. Tumour Biol. 2013 doi: 10.1007/s13277-013-1012-8. [DOI] [PubMed] [Google Scholar]
- 19.Heneghan M.A. Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer. 2003;34:19–26. doi: 10.1385/IJGC:34:1:19. [DOI] [PubMed] [Google Scholar]
- 20.Rabinowitz D. Adjusting for population heterogeneity: a framework for characterizing statistical information and developing efficient test statistics. Genet Epidemiol. 2003;24:284–290. doi: 10.1002/gepi.10231. [DOI] [PubMed] [Google Scholar]
- 21.Song S., Gao Y., Chen S. Interaction of TNF-a-308 and environmental risk factors in primary liver can-cer. Journal of Guangdong Pharmaceutical College. 2009;25:196–201. [Google Scholar]
- 22.Jeng J.E. Independent and additive interactive effects among tumor necrosis factor-alpha polymorphisms, substance use habits, and chronic hepatitis B and hepatitis C virus infection on risk for hepatocellular carcinoma. Medicine (Baltimore) 2009;88:349–357. doi: 10.1097/MD.0b013e3181c10477. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y. Correlation of polymorphism of TNF-α gene promoter with susceptibility to hepatocellular carcinoma in Guangxi. China Oncology. 2012;22:35–41. [Google Scholar]
- 24.Wang C. Correlation between TNF-α polymorphisms and family clustering of primary liver cancer in Guangxi. Chin J of Oncol Prev and Treat. 2012;4:149–152. [Google Scholar]
- 25.Wang Y. Interleukin-1beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology. 2003;37:65–71. doi: 10.1053/jhep.2003.50017. [DOI] [PubMed] [Google Scholar]
- 26.Ho S.Y. Increased risk of developing hepatocellular carcinoma associated with carriage of the TNF2 allele of the -308 tumor necrosis factor-alpha promoter gene. Cancer Causes Control. 2004;15:657–663. doi: 10.1023/B:CACO.0000036173.99930.75. [DOI] [PubMed] [Google Scholar]
- 27.Chen C.C. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol. 2005;34:1310–1318. doi: 10.1093/ije/dyi191. [DOI] [PubMed] [Google Scholar]
- 28.Migita K. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505–510. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Kummee P. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in a Thai population. J Viral Hepat. 2007;14:841–848. doi: 10.1111/j.1365-2893.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 30.Jeng J.E. Tumor necrosis factor-alpha 308.2 polymorphism is associated with advanced hepatic fibrosis and higher risk for hepatocellular carcinoma. Neoplasia. 2007;9:987–992. doi: 10.1593/neo.07781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto T. Interaction between interleukin-1beta -31T/C gene polymorphism and drinking and smoking habits on the risk of hepatocellular carcinoma among Japanese. Cancer Lett. 2008;271:98–104. doi: 10.1016/j.canlet.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Jung K.W. TNFalpha promoter polymorphism is a risk factor for susceptibility in hepatocellular carcinoma in a Korean population. Clin Chim Acta. 2009;407:16–19. doi: 10.1016/j.cca.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Shi H.Z. Association Between EGF, TGF-beta1 and TNF-alpha Gene Polymorphisms and Hepatocellular Carcinoma. Asian Pac J Cancer Prev. 2012;13:6217–6220. doi: 10.7314/apjcp.2012.13.12.6217. [DOI] [PubMed] [Google Scholar]
- 34.Huang H. The Relationship of Polymorphism of Tumor Necrosis Factor-A Gene Promoter-238G/A with Primary Hepatic Carcinoma in Patients. Progress of anatomical sciences. 2007;13:11–13. [Google Scholar]
- 35.Wang B. A study of TNF-alpha-238 and − 308 polymorphisms with different out-comes of persistent hepatitis B virus infection in China. Pathology. 2010;42:674–680. doi: 10.3109/00313025.2010.523696. [DOI] [PubMed] [Google Scholar]
- 36.Chen X. Association of TNF-alpha genetic polymorphisms with hepatocellular carcinoma susceptibility: a case-control study in a Han Chinese population. Int J Biol Markers. 2011;26:181–187. doi: 10.5301/JBM.2011.8580. [DOI] [PubMed] [Google Scholar]
- 37.Kew M.C. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol (Paris) 2010;58:273–277. doi: 10.1016/j.patbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Wang S.S. Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-kB transcription factors and non-Hodgkin lymphoma risk. PLOS ONE. 2009;4:e5360. doi: 10.1371/journal.pone.0005360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 40.Aravalli R.N., Steer C.J., Cressman E.N. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 41.Seitz H.K., Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 42.Wullaert A. Hepatic tumor necrosis factor signaling and nuclear factor-kappaB: effects on liver homeostasis and beyond. Endocr Rev. 2007;28:365–386. doi: 10.1210/er.2006-0031. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y. The TNF-alpha, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol. 2011;137:947–952. doi: 10.1007/s00432-010-0959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P. The TNF-alpha-238 polymorphism and cancer risk: a meta-analysis. PLoS One. 2011;6:e22092. doi: 10.1371/journal.pone.0022092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng K. Tumor Necrosis Factor-alpha 238 G/A Polymorphism and Risk of Hepatocellular Carcinoma: Evidence from a Meta-analysis. Asian Pac J Cancer Prev. 2013;14:3275–3279. doi: 10.7314/apjcp.2013.14.5.3275. [DOI] [PubMed] [Google Scholar]
- 46.Wei Y. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Dig Dis Sci. 2011;56:2227–2236. doi: 10.1007/s10620-011-1617-y. [DOI] [PubMed] [Google Scholar]
- 47.Morsi M.I. Evaluation of tumour necrosis factor-alpha, soluble P-selectin, gamma-glutamyl transferase, glutathione S-transferase-pi and alpha-fetoprotein in patients with hepatocellular carcinoma before and during chemotherapy. Br J Biomed Sci. 2006;63:74–78. doi: 10.1080/09674845.2006.11732724. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y.Y. Increased serum concentrations of tumor necrosis factor-alpha are associated with disease progression and malnutrition in hepatocellular carcinoma. J Chin Med Assoc. 2003;66:593–598. [PubMed] [Google Scholar]
- 49.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]


