Abstract
Background
Breast cancer is the most common malignancy in Iranian women. Mutations in BRCA1 gene is one of the important genetic predisposing factors in breast cancer. This gene is a tumor suppressor that plays an important role in regulating the functions of RAD51 protein for strand invasion in homologous recombination repair.
Methods
The BRCA1 gene has amplified in the DNA isolated from breast cancer patients' leukocytes, using Polymerase Chain Reaction technique. The PCR products have sequenced using an automated DNA sequencer and subsequently obtained data have aligned with the human BRCA1 DNA sequences available online.
Results
In this study, we have considered nine different mutations on 60 examined chromosomes from 30 patients, living in Kerman province. A deletion of one adenine (c.1017delA) and insertion of one cytosine (c.969InsC) have found as the most frequent (20%) mutation in this survey. A substitution of thymine for adenine (c.999T>A) has detected as the second common BRCA1 gene defect (6.7%). The other mutations have identified as single nucleotide replacement including: c.792A>C, c.825G>C, c.822T>A, c.1068A>G, c.969A>T and c.966T>C.
Conclusion
The identified BRCA1 mutations were in accordance with the previous reports. To our knowledge, four mutations: (c.969InsC, c.792A>C, c.825G>C, c.822T>A) which have identified in this study, have not been previously reported in the literature. A larger cohort study would help identifying all relevant BRCA1 mutations in this population.
Keywords: Breast cancer, BRCA1 gene, DNA mutational analysis
Introduction
Breast cancer is the most frequent malignancy among Iranian women [1] even worldwide [2]. The major factors that increase breast cancer risks include therapeutic radiation to chest in patients less than30 years old, BRCA1 or BRCA2 mutations, and familial history of breast or ovarian cancer in three or more first-degree relatives, respectively [3]. The Minor important risk factors include late parity or null-parity (age<30 yrs), early menarche (age<12 yrs) or late menopause (age>55 yrs), combined hormone replacement therapy, post-menopausal obesity, alcohol consumption (2 drinks per day), white race, breast density, and sedentary lifestyle [3].
It has been estimated that 3-5% of breast cancers caused by BRCA1 or BRCA2 have inherited dominantly [4]. Variants in CHEK2, PTEN, TP53, ATM, STK11/LKB1, CDH1, NBS1, RAD50, BRIP1 and PALB2 genes have shown to be associated with high or moderate risks of breast cancer. However, a small part of familial hereditary breast cancers could be explained by these variants [5].
Easton and colleagues [6] have identified five novel breast cancer susceptibility genes that elucidate 3.6% of the increased risk seen in multi-case families. The most significant association with increased risk of breast cancer has found in FGFR2 (Fibroblast Growth Factor Receptor 2) gene with a single nucleotide polymorphism.
The most inherited cases of breast cancer have associated with mutations in BRCA1 and BRCA2 genes. The lifetime risk for breast cancer has often considered being between 40-85% in germ-line mutation (s) in each of two genes [7]. BRCA1 is a tumor suppressor gene, located on the long arm of chromosome 17 and contains 23 exons coding a 1863 amino acid polypeptide [8]. It has observed that BRCA1 plays an important role in regulating the actions of RAD51, an essential protein for strand invasion in homologous recombination repair [9]. A specific regulation of homologous recombination by BRCA1 might maintain genomic integrity and suppress tumor development in proliferating cells [10].
So far, more than 3,400 distinct mutations, alterations and polymorphisms have been reported for both genes [11]. An updated list of BRCA1 gene mutations could be accessed online at Breast Cancer Information Core (BIC) database [12]. Knowledge of BRCA1 mutations have provided better identification, management, and genetic counselling of patients who have affected or predisposed to breast cancer.
It has shown that the ratio of breast to ovarian cancer incidence significantly depends on the location of the mutation [13]. More than half of the BRCA1 mutations have occurred in the largest coding region (exon 11) of the gene.
Herein, we have investigated DNA mutations on exon 2 and part of exon 11 of BRCA1 gene in a cohort of Kerman patients suffering from breast cancer. This is the first study about breast cancer genes in Kerman, which is a vast province, approximately 11% of the whole country, with more than 2.7 million population located in south center of Iran (Figure 1).
Figure 1.
Provincial map of Iran
Materials and Methods
A group of 30 patients (22 women and 8 men) with breast cancer have recruited from referrals of cancer clinic, using: family history, clinical examination and paraclinical tests. Most of the patients (83.3%) were affected with ductile carcinoma. Lobular carcinoma has reported in two patients (13.3%) and a woman had modular carcinoma (3.4%). The subjects have selected according to the National Comprehensive Cancer Network [14] guidelines (BRISK-1) for familial breast cancer (Table 1). The isolated cases have referred to the genetic laboratory for BRCA1 mutation detection between May 2006 and February 2011. A genetic counsellor has explained the aims and objectives of this study to the patients. After obtaining a consent sheet, 8 ml whole blood from their brachial vein have drawn into tubes containing 200 µl EDTA (Ethylene Diamine Tetra-acetic Acid). Genomic DNA has isolated from leukocytes of the whole blood using salt-saturation method as previously prescribed [15]. Using isolated genomic DNA from the patients, Polymerase Chain Reaction (PCR) has performed to amplify DNA fragments containing exon 2 and partial regions covering exon 11 of BRCA1 gene. Primers used for amplification of exon 2 yielding a 443 bp product have listed in Table 2. Two DNA fragments of exon 11, 309 bp and 300 bp, have obtained using the previously published primer sequences [16].
Table 1.
National Comprehensive Cancer Network criteria for familial breast cancer
| Family History |
| • Early-age-onset breast cancer |
| • Two breast primaries or breast and ovarian/fallopian tube/primary peritoneal cancer in a single individual or two or more breast primaries or breast and ovarian/fallopian tube/primary peritoneal cancers in close relative(s) from the same side of family (maternal or paternal) |
| • A combination of breast cancer with one or more of the following: Thyroid cancer, sarcoma, adrenocortical carcinoma, endometrial cancer, pancreatic cancer, brain tumors, diffuse gastric cancer, dermatologic manifestations or leukemia/lymphoma on the same side of family |
| • Member of a family with a known mutation in a breast cancer susceptibility gene |
| • Populations at risk |
| • Male breast cancer |
| • Ovarian/fallopian tube/primary peritoneal cancer |
| Known BRCA1/2, p53, PTEN, or other gene mutation associated with breast cancer risk |
Table 2.
DNA primer sequences of exon 2 and part of exon 11 of BRCA1 gene
| Exon | Primer sequence | PCR product size |
| 2 | Forward: 5'-AAACCTTCCAAATCTTCAAA-3' | 443 bp |
| Reverse: 5'-GTCTTTTCTTCCCTAGTATGT-3' | ||
| 11A | Forward: 5'-AACACCACTGAGAAGCGTGCAG-3' | 309bp |
| Reverse: 5'-CTCACACAGGGGATCAGCATTC-3' | ||
| 11B | Forward: 5'-CAACATAACAGATGGGCTGGAAG-3' | 300bp |
| Reverse: 5'-ACGTCCAATACATCAGCTACTTTGG-3' |
PCR has performed using the following conditions: 35 cycles of denaturation at 94˚C for 45s, annealing at the optimal temperature of each primer pair for 30s and extension at 72˚C for 30s. PCR products have visualized on a 1% agarose gel and run on an automated DNA sequencer. The sequenced data have aligned with human BRCA1 DNA sequences available online (Gene Bank: U14680.1) and the results have compared to those recorded in the BIC database.
Results
In this study, isolated DNA from 30 patients (25 women and 5 men) with breast cancer has analysed for detection of any mutations in exon 2 and a part of exon 11 in the BRCA1 gene. Between the 30 considered patients, we have perceived; 2 patients with bilateral breast cancer, 3 with ovarian cancer previous records, and 4 who have born from consanguineous marriages. The average patient age was 51 yrs with a median age of 50 yrs. Breast cancer family history has reported in 36.7% of patients and none of the studied subjects have mentioned a history of other disease.
All of the identified mutations have located within the amplified parts of exon 11 of BRCA1 gene (Table 3). No mutation has found in 16 cases (53.5%). Among the 28 examined chromosomes, nine different types of mutation have detected in 14 patients. Three out of the nine gene defects were synonymous: c.999T>A, c.966T>C and c.969A>T. A deletion of one adenine (c.1017delA) and insertion of one cytosine (c.969InsC) have found to be the most frequent (20%) mutations in this survey (Table 3). Substitution of thymine for adenine, c.999T>A, has detected as the second common BRCA1 gene defect (6.7%). The other mutations that have identified as single nucleotide replacement included: c.792A>C, c.825G>C, c.822T>A and c.1068A>G. The sequenced graph of the c.1068A>G mutation has shown in figure 2.
Table 3.
BRCA1 Gene Mutations identified in the breast cancer patients from Kerman Province, Iran
| Mutation | Nt | Coding | Amino acid | % | N | Reference |
| TGT>AGT | 939 | c.822 | p.Cys274 Ser | 3.3 | 1 | – |
| ACT>ACA | 1118 | c.999 | p.Thr333Thr | 6.7 | 2 | – |
| CAG>CGG | 1186 | c.1068 | p.Gln356Arg | 3.3 | 1 | BIC:17667 |
| AAG>A-G | 1135 | c.1017 | p.Lys340Stop | 10 | 3 | BIC: 1160 |
| GCT>GCC | 1085 | c. 966 | p.Ala322Ala | 3.3 | 1 | – |
| GGA>GGT | 1088 | c.969 | p.Gly323Gly | 3.3 | 1 | – |
| GGG>CGG | 942 | c. 825 | p.Gly275Arg | 3.3 | 1 | – |
| INS C | 1088 | c. 969 | p.Gly323Stop | 10 | 3 | – |
| AGT>CGT | 909 | c.792 | p.Ser264Arg | 3.3 | 1 | – |
| Unknown | – | – | – | 53.5 | 16 | – |
| Total | – | – | – | 100 | 30 | – |
N; Number, %; Percent, Nt; Nucleotide
Figure 2.
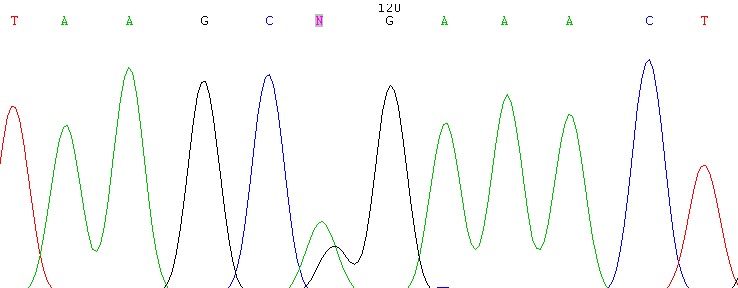
Substitution of Adenine to Guanine at Nucleotide 1186 in exon 11 of BRCA1 Gene
Discussion
This is the first report of describing mutations in the BRCA1 gene in Kerman population. Since hereditary forms of breast cancer have often seen in patients with bilateral involvement, early onset, and positive family history, these criteria have taken into consideration in selecting the study subjects. Breast cancer has multi-factorial inheritance and affect men less than women. Therefore, the recurrence risk in relatives of an affected man is higher than the recurrence risk in relatives of an affected woman. Since the BRCA1 gene consists of 23 exons which span more than 80kb of genomic DNA [17], then its mutations would be numerous and widespread. Therefore, mutational analysis by direct sequencing has limited to parts of coding regions of BRCA1 gene.
Breast cancer is a multi-factorial malignancy that results from complex interactions between a number of predisposing factors including the genotype in one or more loci and a variety of environmental exposures. Therefore, the disease might be resulted from other predisposing factors rather than mutations in BRCA1. Since these factors could be responsible for the malignancy, no mutation has found in more than 50% of the studied subjects in our limited work. Exon11contains 60% of BRCA1 coding sequence and we have identified all of the nine different variants, including 4 novel mutations (c.969InsC, c.792A>C, c. 825G>C, c.822T>A) in this region (Table 3).
In our study, two distinct frame-shift mutations have detected: an insertion of a single cytosine at nucleotide 1088 (c.969InsC) and a deletion of an adenine at nucleotide 1135 (c.1017delA). Both of these BRCA1 gene mutations introduce a premature stop codon. These mutations change amino acid chains by altering the reading frame of the gene. All of the affected subjects with these mutations were at high risk for familial breast cancer. Bilateral breast cancer involvements have observed in one out of the three patients with the insertion (c.969InsC) gene defect (Table 3). Another patient from this group has diagnosed with ovarian cancer at the age of 28 yrs. Among the patients carrying an adenine deletion, a man and a woman have mentioned family history of breast cancer. It was not surprising to find the highest prevalence (20%) of these frame-shift gene defects in our study (Table 3), since most of the BRCA1gene defects include premature termination codon have caused by out of frame deletions and insertions in the coding sequence [18]. The deletion of adenine (c.1017delA) has been previously reported by Pohlreichin on BIC database [12]. This mutation has reported from Czech Republic in 2004 for the first time [19]. Our finding is in accordance with the previous report.
We found three variations c.999T>A (6.7%), c. 966T>C (3.3%) and c.969A>T (3.3%) that code for threonine, alanine, and glycine, respectively. These genetic differences do not influence the related protein chain and consider as neutral mutants (Table 3).
Among the seven cases of this group, one patient was a man and two of them have mentioned family history of breast cancer. The c.999T>A variation was the second most frequent mutation in our study. These variations have classified as clinically benign synonymous sequence variants because allelic frequency data from population studies have not currently been available [20].
In one patient whose breast cancer has diagnosed at the age of 41, we have identified a missense mutation (c.1068A>G) that results in a p.Gln356Arg amino acid change. The c.1068A>G mutation has been previously reported in several patients by DiCioccio and colleagues at BIC database [12]. This is a common gene defect in European and American populations. A missense mutation (c. 825G>C) that causes a p.Gly275Arg amino acid change has identified in a patient who was affected with breast cancer. Her sister has diagnosed with breast cancer in her late 40 s. The affected men with the breast cancer were carriers of two different novel missense mutations in the BRCA1 gene (c.792A>C and c.822T>A) that result in p.Ser264Arg and p.Cys274 Ser amino acid changes, respectively. Breast cancer in males is very rare and has reported in less than 1% of all cancers in men [21] then they have diagnosed at the later-stage of the disease in comparison to women.
In conclusion, although we have found a few novel mutations in BRCA1 gene in Kerman population, none of the detected gene defects have reported in other provinces of Iran. However, some of detected mutations have been previously reported in patients from other parts of the world. This study is not sufficient to show all probable mutations in the genes which involved in breast cancer or even in the BRCA1 gene in Kerman; thus, a large cohort study would facilitate in identifying all relevant BRCA1 or BRCA2 mutations in this population.
Acknowledgments
We thank Dr Anuradha Kumari (Oregon Health and Service University, Portland, OR, USA) for critical reading of this article, and we have also indebted to Afzalipour Hospital Kerman, Iran for financial support.
Footnotes
Conflicts of Interest
There is no conflict of interest in this article. Kerman University of Medical Sciences supported this work financially.
Authors' Contribution
Nasrollah Saleh-Gohari designed the study, analysed the data and wrote the paper. Marzye Mohammadi-Anaie performed PCR tests and contributed to literature review. Behjat Kalantari-Khandani selected and referred patients from cancer clinic to genetic laboratory. All authors read and approved the final manuscript.
REFERENCES
- 1.Mousavi SM, Mohagheghi MA, Mousavi-Jerrahi A, Nahvijou A, Seddighiet Z. Outcome of breast cancer in Iran: a study of Tehran Cancer Registry data. Asian Pac J Cancer Prev. 2008;9(2):275–8. [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics. CA Cancer J Clin. 2010;60 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Warner E. Breast-cancer screening. N Engl J Med. 2011;365:1025–32. doi: 10.1056/NEJMcp1101540. [DOI] [PubMed] [Google Scholar]
- 4.Mackaya J, Szecseib CM. Genetic counseling for hereditary predisposition to ovarian and breast cancer. Ann Oncol. 2010;17(21):334–8. doi: 10.1093/annonc/mdq365. [DOI] [PubMed] [Google Scholar]
- 5.Van der Groep P, van der Wall E, van Diest PJ. Pathology of hereditary breast cancer. Cell Oncol (Dordr). 2011;34(2):71–88. doi: 10.1007/s13402-011-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald MG, Macdonald DJ, Krainer M, Hoover I, O'Neil E, Unsal H, et al. Germ-line BRCA1 mutations in Jewish and non-Jewish women with early-onset breast cancer. N Engl J Med. 1996;334:143–9. doi: 10.1056/NEJM199601183340302. [DOI] [PubMed] [Google Scholar]
- 9.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 10.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7(2):263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Zarkadoulia EA, Ioannidou EN, Peppas G, Christodoulou Ch, Rafailidis PI. The effect of psychosocial factors on breast cancer outcome: a systematic review. Breast Cancer Res. 2007;9(4):R44. doi: 10.1186/bcr1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.http://research.nhgri.nih.gov/bic
- 13.Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, et al. Germ-line mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat Genet. 1995;11:428–33. doi: 10.1038/ng1295-428. [DOI] [PubMed] [Google Scholar]
- 14.http://www.nccn.org/index.asp
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215–6. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehdipour P, Atiri M, Pour-Farzad F. Detection of BRCA1 gene mutation in the breast cancer probands by non-radioactive PCR-SSCP method. Arc Irn Med. 1999;2(2):67–70. [Google Scholar]
- 17.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 18.Anczukow O, Buisson M, Salles MJ, Triboulet S, Longy M, Lidereau R, et al. Unclassified variants identified in BRCA1 exon 1: consequences on splicing. Genes, Chromosomes & Cancers. 2008;47:418–26. doi: 10.1002/gcc.20546. [DOI] [PubMed] [Google Scholar]
- 19.Pohlreich P, Stribrna J, Kleibl Z, Zikan M, Konopasek B, Novotny J, et al. Mutations and splicing variants of the BRCA1 gene in Czech breast and ovarian cancer families. Proc Am Soc Clin Oncol. 2002;21 [Google Scholar]
- 20.Keshavarzi F, Javadi GR, Zeinali S. BRCA1 and BRCA2germline mutations in 85 Iranian breast cancer patients. Springer Science+Business Media B.V. 2011 [Google Scholar]
- 21.Tahmasebi S, Akrami M, Omidvari S, Salehi A, Talei A. Male Breast Cancer; Analysis of 58 Cases in Shiraz, South of Iran. Breast Dis. 2010;31(1):29–32. doi: 10.3233/BD-2009-0293. [DOI] [PubMed] [Google Scholar]


