Abstract
Quantitation of individual mAbs within a combined antibody drug product is required for preclinical and clinical drug development including pharmacokinetics (PK), toxicology, stability and biochemical characterization studies of such drugs. We have developed an antitoxin (XOMA 3AB) consisting of three recombinant monoclonal antibodies (mAbs) that potently neutralizes the known subtypes of type A botulinum neurotoxin (BoNT/A). The three mAbs bind non-overlapping BoNT/A epitopes with high affinity. XOMA3AB is being developed as a treatment for botulism resulting from BoNT/A. To develop antibody-specific assays, we cloned, expressed, and purified BoNT/A domains from E. coli. Each mAb bound only to its specific domain with affinity comparable to the binding to holotoxin. MAb specific domains were used to develop an ELISA for characterization of the integrity and binding activity of the three mAbs in the drug product. An electrochemiluminescence bridging assay was also developed that is robust to interference from components in serum and we demonstrate that it can be used for PK assays. This type of antigen engineering to generate mAb-specific domains is a general method allowing quantitation and characterization of individual mAbs in a mAb cocktail that bind the same protein and is superior to anti-idiotype approaches.
Keywords: botulinum neurotoxin, botulism, oligoclonal antibodies, protein domain, protein characterization, protein purification
Introduction
Monoclonal antibody (mAb)-based drugs have proven remarkably successful in the clinic, but currently all of these approved drugs consist of single mAbs. [1] In order to enhance efficacy or broaden specificity, combinations of mAbs are now in preclinical and clinical development for a wide range of diseases [2]. The most advanced of these is CL184, a combination of two mAbs designed for the post-exposure prophylaxis against rabies [3] now in Phase II clinical trials. Other mAb combinations under investigation include mAbs for influenza [4], for anthrax [5, 6], and for cancer [7].
Regulatory agency requirements for characterization of combinations of mAbs (oligoclonal antibody) and polyclonal antibodies are based on several existing guidelines[8–10]. Because oligoclonal and polyclonal therapies typically have a unique mechanism of action requiring the presence of all of the mAb components, they are clinically evaluated as the mixture of antibodies. Despite this, analytical methods to characterize and measure the individual components of the mAb mixtures are required for use in product characterization, for production of clinical material, for pharmacokinetic (PK) studies, and for monitoring of drug product stability.
We are developing XOMA 3AB [11], an equimolar mixture of three human/humanized mAbs binding non-overlapping epitopes on botulinum neurotoxin type A (BoNT/A) with high affinity, for the treatment of botulism due to BoNT/A. Botulism is characterized by flaccid paralysis, which if not rapidly fatal requires prolonged hospitalization in an intensive care unit and mechanical ventilation. BoNTs are classified by the Centers for Disease Control and Prevention as one of the 6 highest-risk threat agents for bioterrorism, due to the high potency and lethality of the toxin [12]. It is estimated that major civilian exposure to BoNT would have catastrophic effects. One study calculated that aerosol exposure of 100,000 individuals to toxin, as could occur with an aerosol release over a metropolitan area, would result in 50,000 cases with 30,000 fatalities [13]. Such exposure would result in 4.2 million hospital days and an estimated cost of $8.6 billion.
The mainstay of treatment for adults with botulism is antitoxin produced by hyperimmunizing horses, equine antitoxin [14, 15]. In the St. John study, the most important factors reducing mortality and cost were early availability of antitoxin and mechanical ventilation [13]. Equine antitoxin, however, has a significant incidence of side effects, including hypersensitivity reactions including serum sickness and anaphylaxis [14, 15]. An alternative to equine antitoxin is serotype specific monoclonal antibody (mAb) based antitoxin, however, individual mAbs do not have the requisite potency for development [16]. Combining the three mAbs in the XOMA 3AB cocktail increases the potency of BoNT neutralization by at least three orders of magnitude compared to the potency of the individual antibodies ([16] and unpublished data). Extremely high affinity antibodies are required to treat botulism due to the extreme lethality of the toxin [17].
Oligoclonal and polyclonal antibodies that target the same protein are especially challenging for analytical method development since ELISA assays using the target protein for capture will not distinguish between individual mAbs. Therefore, epitope-based capture is desirable for these cocktails. An alternative, and common, strategy is to generate and use anti-idiotype antibodies specific for each of the three mAb components of XOMA 3AB [18–20]. However, this approach would require accomplishing the challenging task of generating sets of mAbs or polyclonal antibody binding unique epitopes in the binding site of each of the three mAbs, with high affinity and with specificity only for that mAb and not the other two. Furthermore, the anti-idiotype antibodies would be required to have affinities of less than 5 nM, the anticipated effective serum concentration of the drug in humans. Frequently, anti-idiotype antibodies do not have this level of sensitivity [18]. In contrast, the domains developed here largely retain the high affinity of the BoNT/A for each of the component mAbs, allowing construction of highly sensitive PK assays. Furthermore, this approach does not require generation of additional sets of antibodies.
Here, we report expression and purification of BoNT/A domains, which are each bound by one of the three mAbs and describe their use for measuring the three mAbs in the drug mixture. ELISA methods were developed for characterization of the integrity and purity of drug product. An electrochemiluminescence (ECL) assay that is robust to interference from components in serum was also developed and was used to determine the PK of each mAb in the three mAb combination in rats. Besides allowing individual mAb quantification in the mixture, the use of mAb specific non-toxic domains also eliminates exposure of laboratory workers to the highly toxic BoNT and obviates the burdensome requirements for handling select agents such as BoNT.
Materials and Methods
Cloning of BoNT/A1 domains
The three corresponding BoNT/A1 toxin domains (HCC, HCN, LC) (Figure 1) were cloned in E. coli BL21 DE3 using the pET expression system. Each domain construct had a C-terminal SV5-tag [21], hexahistidine tag and N-terminal Met, Ala amino acids introduced by the cloning sites. Domains were determined based on the published crystal structure of BoNT/A [22] and have been previously described [23]. The HCC domain consisted of 204 amino acids, residues 1092 to 1296 with a calculated molecular weight (MW) of 26.9kDa and a calculated pI of 9.25. The HCN domain consisted of 216 amino acids, residues 876 to 1092, with a calculated MW of 29.2kDa and a pI of 8.69. The LC-HN domain consisted of 860 amino acids, residues 1 to 860, with a calculated MW of 102kDa and a calculated pI of 5.42. The BoNT/A1 N-terminal subdomain (HCN) of the receptor binding domain (HC), C-terminal subdomain (HCC) of the HC, and the light chain (LC) fused to the translocation domain (HN) were cloned for expression in E. coli BL21 DE3 using the pET expression system. The HCC, HCN and LC--HN DNA fragments were prepared by digesting pYD2 based plasmids containing BoNT/A HCC, HCN, or LC-HN [23] with NcoI and PmeI (blunt end cutter) followed by gel purification of the insert DNA. The pET21d vector was first digested by EcoRI, followed by Klenow enzyme treatment (New England Biolabs) to create a blunt end. The vector was digested by NcoI and vector and insert were ligated through the NcoI site on one side and the blunt end on the other. The resulting HCN, HCC, and LC-HN constructs had an additional Met and Ala amino acids at their N-termini from the cloning strategy used and C-terminal SV5 [21] and hexahistidine tags from the pET vector. Clones (pET/HCC, pET/HCN, and pET/LC-HN) containing the correct construct were identified by DNA sequencing.
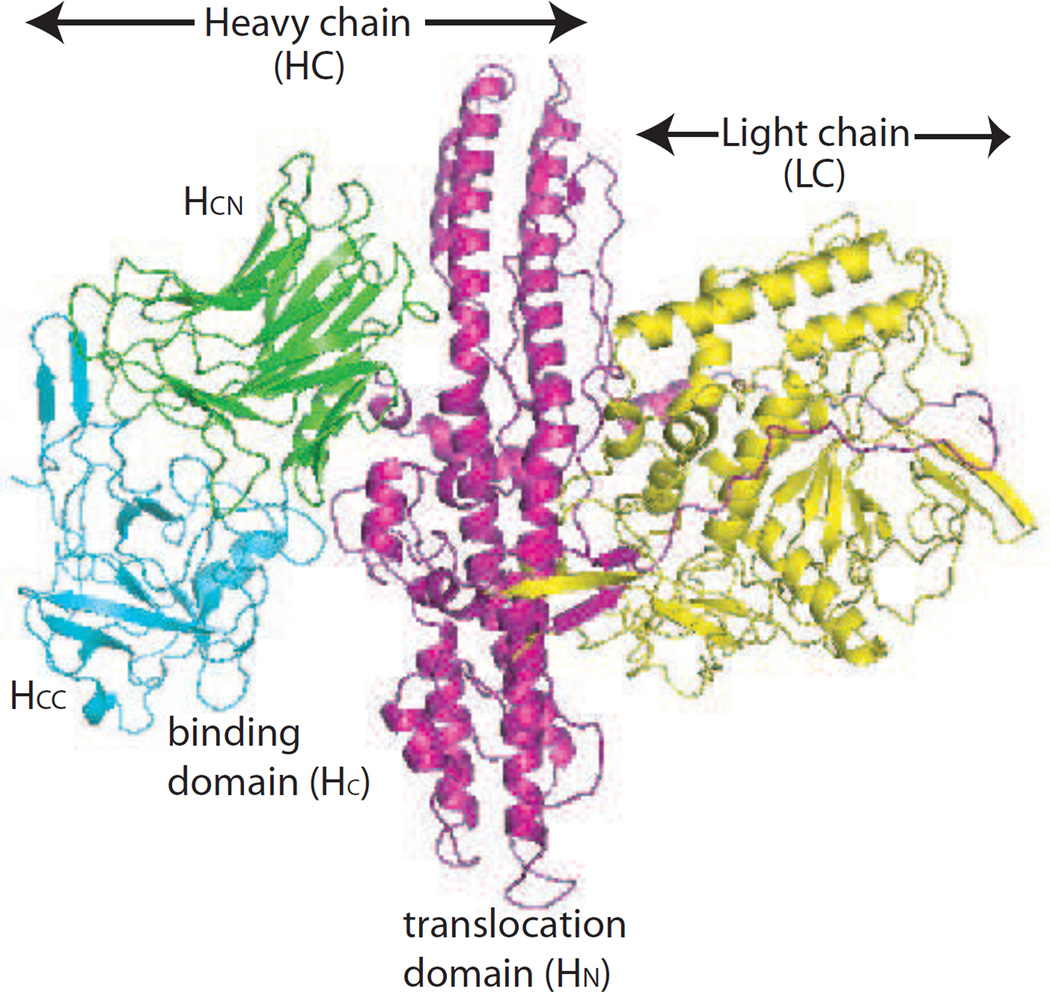
Figure 1. Structure of BoNT/A and BoNT/A domains.
BoNT/A consists of a heavy chain (HC, magenta) and a light chain (LC, yellow). The HC consists of the receptor binding domain (HC) and the translocation domain (HN). The HC consists of a C-terminal domain (HCC) and an N-terminal domain (HCN).
Expression and purification of BoNT/A domains
E. coli expressing each domain were grown at 5 to 50 mL scale, expression was induced with Isopropyl-β-D-thio-galactoside (IPTG), and bacteria lysed and analyzed by SDS-PAGE to determine if proteins were located in the cytoplasm or in inclusion bodies. The induction temperature, duration of induction, and IPTG concentration were optimized. Cultures were then scaled to 10 L in a fermenter (New Brunswick, BioFlo 4500). Small scale purifications were performed to determine the optimal type and order of orthogonal column chromatography for purification. A scalable purification scheme was subsequently developed for each domain (Figure 2).
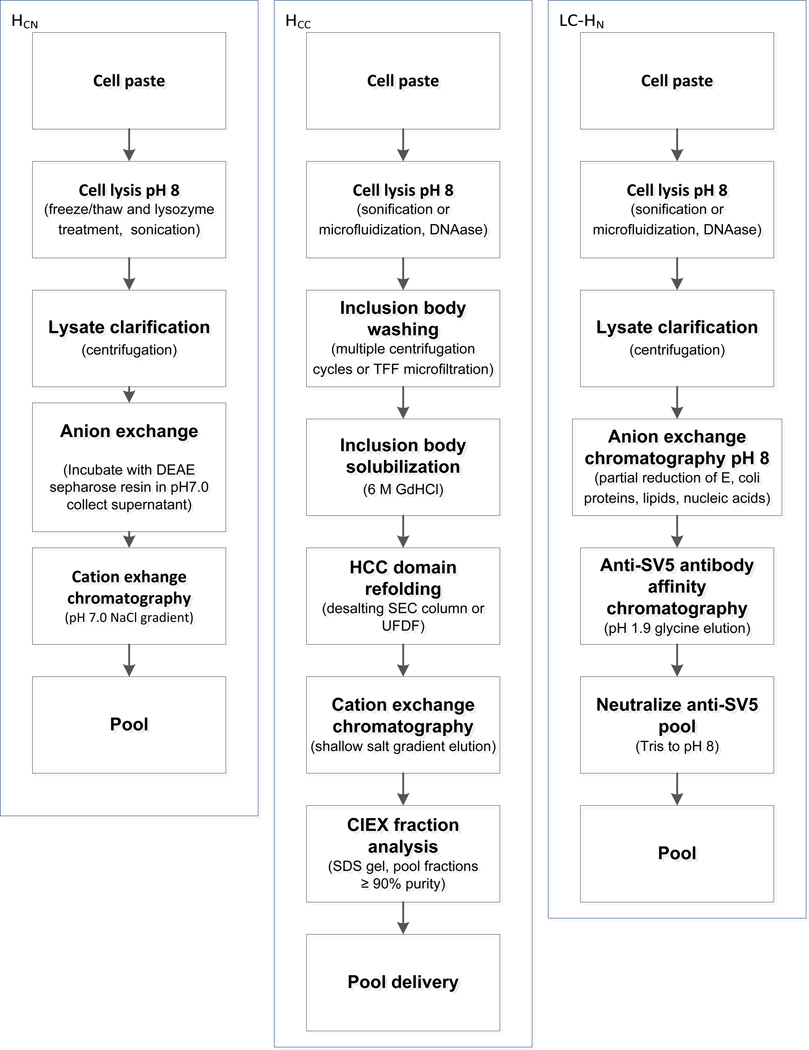
Figure 2. Scalable domain purification methods.
Three separate purification strategies were developed to purify the BoNT/A HCC, HCN, and LC-HN domains at the required scale. See text for details.
Expression and purification of BoNT/A HCC
The HCC domain was expressed in the insoluble fraction and was purified from inclusion bodies. pET/HCC in E. coli BL21 DE3 was grown to an optical density of 2.0 in a 10 L bioreactor and expression induced by the addition IPTG to a final concentration of 1mM. Cultures were grown overnight at 30°C after which bacteria were harvested by centrifugation at 5,000 g for 20 min. Bacterial cell paste was stored frozen at −80°C. For purification, bacteria were thawed and resuspended in 5 mL of lysis buffer (50 mM Tris-HCl, 50 mM NaCl, 5% v/v glycerol, pH8.0) per gram of wet cell paste. Proteinase inhibitor cocktail (Sigma-Aldrich) was added to the lysate at 0.25 mL per gram of wet cell paste along with DNAseI at 5 µg/mL. Bacteria were lysed mechanically by sonication. Lysate was centrifuged at 15,000g for 15 min. HCC was recovered as inclusion bodies in the pellet. The inclusion bodies were washed three times by resuspension in 1:20 diluted lysis buffer and centrifugation at 15,000g for 15 min. The inclusion bodies were solubilized in 6 M Guanidine HCl solution (100 mM NaH2PO4, 10 mM Tris-HCl, 6 M Guanidine-HCl, pH 8.0, 5 mL per gram cell paste weight) by vortex and incubation at room temperature for 1 hour or at 4°C overnight. Refolding was achieved by a desalting step by size exclusion chromatography (SEC) with a Hiprep 26/10 desalting column (GE Healthcare Biosciences) for smaller scale preparation or by ultrafiltration-diafiltration using an AKTAcrossflow™ (GE Healthcare Biosciences) for large scale preparation. In either scale, PBS was used as the buffer to refold HCC. After refolding, the domain was purified by Ni-NTA immobilized metal affinity chromatography (IMAC). A Kvick cassette filter with a 10 kDa select pore size and 0.11 m2 surface area (GE Lifescience UFELA0010010SE) was used. Crossflow was controlled by using a trans membrane pressure of 1 bar and with a crossflow rate of 750 mL/min. The diafiltration process was terminated when the permeate reached a conductivity reading of less than 40 mS/cm. The retentate was recovered and then filtered by a 0.22 µm Steritop filter (Millipore). The filtered retentate was further concentrated and diafiltrated using the AKTAcrossflow™ and the same Kvick cassette filter and control parameters. The diafiltration buffer was 20 mM phosphate buffer, pH7.0. Folded domain was purified using a shallow salt gradient on a cation exchanger (MonoS 4.6/100, GE Lifesciences). The running buffer was 20 mM phosphate buffer pH7.0 and the high salt buffer was 20 mM phosphate buffer pH 7.0, 1 M NaCl. Fractions containing purified HCC were pooled to achieve greater than 95% purity as determined by both SEC and RP-HPLC integrating the area under the A280 curve. Purified HCN domain was buffer exchanged into 5 mM sodium phosphate, 120 mM sucrose, pH 7, and stored at −70°C.
Expression and purification of BoNT/A HCN
The BoNT/A HCN was expressed in the soluble fraction. The purification scheme took advantage of the high pI of the HCN (9.25), using cation exchange chromatography at pH 7.0 to remove most of the E. coli proteins followed by IMAC to purify the HCN.
pET/HCN in E. coli BL21 DE3 was grown to an optical density of 2 in a 10 L bioreactor and expression induced by the addition of 1 mM IPTG. Cultures were grown overnight at 18°C after which bacteria were harvested by centrifugation at 5,000g for 20 min. Approximately 60 grams of bacterial cell paste was stored frozen at −80°C. For purification, bacteria were thawed at room temperature. 300 mL of lysis buffer (50 mM Tris, 50 mM sodium chloride, 5 % v/v glycerol, pH 8; 5 mL per gram cell paste) at ambient temperature was prepared for use by the addition of 0.6 mL EDTA (from a 500 mM stock), 0.238 mL benzonase recombinant nuclease to degrade nucleic acids and reduce viscosity (EMD Chemicals, 1000 Units per gram cell paste), 5.5 mL protease inhibitor cocktail (Sigma-Aldrich P8849, 0.093 mL per gram cell paste), and 0.056 mL Ready-Lyse recombinant lysozyme (Epicentre R1802M, 28,000 Units per gram cell paste). Bacterial cells were resuspended in the prepared lysis buffer and stirred on a magnetic stir plate for one hour. Raw lysate was then chilled in an ice water bath and clarified by centrifugation at 15,000 x g for 20 min in a refrigerated centrifuge at 5°C. Clarified domain lysates were purified using a binding mode cation exchange chromatography step. Lysate was diluted 1:1 by volume with ice cold 20 mM sodium phosphate, pH 7, with mixing, and adjusted to pH 7 with 1 N hydrochloric acid. The pool was 0.2 micron filtered, and kept on ice while it was loaded onto a 2.2 cm × 15 cm (57 mL bed volume) cation exchange column (GE Healthcare SPXL) equilibrated in 20 mM sodium phosphate, pH 7. After washing the column to baseline with equilibration buffer the domain was eluted with a 10 column volume linear gradient to 200 mM sodium chloride at constant pH. To remove C-terminal clipped species corresponding to HCN residues 2–243 and 2–239, partially purified HCN preparations (SPXL pools) were loaded onto a 2.2 cm × 16 cm Ni-NTA Superflow resin column (Qiagen, 61 mL bed volume), and eluted with an imidazole gradient ranging from 20 to 250 mM imidazole at pH 7.5, with 25 mM sodium phosphate and 1 M NaCl in all buffers to inhibit non-specific ionic binding. Purity of the eluted fractions was assessed by SDS-PAGE, SE-HPLC, and reverse phase HPLC, and the fractions were pooled to achieve 95% purity by SE-HPLC. For long term storage, purified HCN domain was buffer exchanged into 5 mM sodium phosphate, 120mM sucrose, pH 7, and stored at −70°C.
Expression and purification of BoNT/A LC-HN
The LC-HN domain was expressed in the soluble fraction and was purified from the bacterial cytosol. pET/LC-HN in E. coli BL21 DE3 was grown to an optical density of 2 in a 10 L bioreactor and expression induced by the addition of 1 mM IPTG. Cultures were grown overnight at 18°C after which bacteria were harvested by centrifugation at 4000g for 20 min. Bacterial cell paste was stored frozen at −80°C. For purification, bacteria were thawed and approximately 7 grams cell paste was resuspended in 35 mL of lysis buffer (50 mM Tris, 50 mM sodium chloride, 5 % v/v glycerol, pH 8). Bacteria were lysed by the addition of 1.4 mL of Lysozyme (Sigma, 10 mg/mL stock concentration), and 175 µL of DNaseI (Sigma, 1 mg/mL stock concentration), and 1.75 mL of protease inhibitor cocktail (Sigma P8465). Lysis was promoted by sonication on ice for 6 minutes, 10 seconds on/10 seconds off. Crude lysate was clarified by centrifugation at 15,000g for 15 minutes. Clarified lysate was partially purified on an anion exchange column (GE Healthcare HiTrap QXL), equilibrated with 20 mM Tris pH 8. Clarified lysate was diluted 5-fold with equilibration buffer to reduce conductivity, and applied to the QXL column. The column was washed with 5 column volumes of equilibration buffer followed by a 5 column volume wash with 20 mM Tris, 80 mM NaCl pH 8. A 5 column volume linear gradient from 80 mM to 400 mM sodium chloride, in 20 mM Tris pH 8, was used to elute the domain. The entire UV absorbing eluate peak was collected as a single pool and further purified by affinity chromatography using anti-SV5 mouse mAb immobilized on chromatography resin (Sigma A7345). The affinity column was equilibrated in PBS, and the QXL pool was loaded directly. The column was washed with 5 column volumes of PBS and eluted with 0.1 M glycine-HCl pH 1.9. The eluted domain pool was immediately neutralized by addition of 1 M Tris-HCl, pH 8. Other BoNT domains were purified using IMAC due to its lower cost and the observation that some domains were unstable at the low pH required for elution from the SV5 column. For long term storage, purified LC-HN domain was buffer exchanged into 5 mM sodium phosphate, 120 mM sucrose, pH 7, and stored at −70°C
Expression and purification of BoNT/A mAbs A, B, and C
BoNT/A mAbs A (binds HCC), B (binds HCN), and C (binds LC-HN) were expressed from stable Chinese Hamster Ovary (CHO) cell lines and purified by Protein A chromatography followed by flow-through anion exchange and hydrophobic interaction chromatographies.
Size exclusion chromatography and reverse phase chromatography for BoNT/A domain
SE-HPLC
All HPLC analysis was performed with the Agilent 1200 Series LC System. The sample loop was 100µL with an injection volume of 10–50µL, depending on the sample total protein concentration (0.5–2mg/mL). All HPLC data were analyzed with the Agilent ChemStation Version B.03.02. SE-HPLC was performed with a TosoH TSK gel Super SW2000 column (4.6 × 300 mm) packed with 4µm particles containing 250 Å pores. The injected samples were eluted isocratically in 20 minutes with elution buffer: 20 mM sodium phosphate, 0.2 M ammonium sulfate, pH 6.8 at a flow rate of 0.35 mL/min. Peaks were detected with by UV at 280 nm and 214 nm. Alternatively, a TosoH TSK gel G3000 column (7.8 × 300 mm) was used with a flow rate of 0.5 mL/minute and 30 minute analysis time.
Antibody binding by SE-HPLC and concentration measurement
BoNT/A HCC, HCN, or LC-HN and their corresponding antibody were mixed in an apparent molar ratio of 2:1 in an HPLC sample vial based on the concentrations determined by UV A280 nm using the theoretical molar extinction coefficient and molecular weight in Supplemental Table 1. The domain and the corresponding mAb concentrations during binding assay were in the range of 10–20µM and 5–10µM respectively. The binding reaction buffer was essentially the same as that of the individual domain, which was 5mM sodium phosphate, 120mM sucrose, pH 7. The BoNT/A domain/antibody mixtures were left at ambient temperature for approximately 10 minutes and then kept at 2–8°C auto-injector until HPLC testing (within 24 hours). The samples were analyzed using the SE-HPLC method described above. The G3000 column was found to give superior resolution for the binding assay mixture samples.
RP-HPLC
An Agilent 300SB CN column was used with 0.1% (v/v) TFA in water as buffer A and 0.1% (v/v) TFA in acetonitrile as buffer B. The column compartment was heated to 60°C and kept at 60°C during the analysis. For HCC and HCN, the peaks were eluted with a gradient from initial 22% B to 60% B in 25 minutes. For LC-HN, the peaks were eluted with a gradient from initial 38% B to 68% B in 30 minutes. The peaks were detected with the Agilent multi-wavelength UV detector at 280 nm and 214 nm.
BoNT/A domain formulation
Pre-formulation screening studies were conducted in solution at 5°C and 25°C with each of the three domains prior to proceeding to lyophilization for HCC and HCN ( LC-HN is stored frozen at −70°C). The effects of pH (weakly acidic, neutral, and weakly basic), ionic strength (Na Cl concentration from 0 to 1.0 M), and excipients (mannitol, sorbitol, and sucrose) on stability were assessed. Lyophilization was conducted with the VirTis Genesis 25XL lyophilizer (SP Scientifc, Stone Ridge, NY) and controlled with the Encore software.
Measurement of affinity and binding specificity of mAbs for BoNT/A domains
Binding affinities (KD) and rate constants (kon, koff) were measured using a KinExA 3000 flow fluorimeter (Sapidyne Instruments Inc., Boise Idaho) as previously described [24–26]. While time consuming due to equilibration times of up to 3–4 days, KinExA provides accurate binding constants for high affinity binding interactions. Studies of BoNT/A domain reaction mixtures were performed in PBS (pH 7.4) at room temperature with 1 mg/mL bovine serum albumin (BSA), and 0.02% (w/v) sodium azide added as a preservative. Antibody solution was serially diluted into a constant concentration of either BoNT/A or the corresponding BoNT/A domain sufficient to produce a reasonable signal, where the antibody concentration was varied at least tenfold above and below the value of the apparent KD and antigen concentrations were no more than fourfold above the KD to ensure a KD controlled experiment. Samples were equilibrated, then 13 dilutions were passed over a flow cell containing a 4 mm column of NHS-activated Sepharose 4 Fast Flow beads (GE Healthcare) covalently coated with the corresponding antibody to capture the free BoNT/A or BoNT/A domain. An Alexa-647 labeled antibody was passed over the beads to detect the free antigen by binding and producing a signal relative to the amount of free BoNT/A or BoNT/A domain bound to the beads. In the case of the BoNT/A domains a detection antibody that bound the SV5 tag was used, and in the case of full-length BoNT/A a detection antibody binding a non-overlapping epitope was used. Each dilution was tested in duplicate. Sample volumes varied from 3 to 25 mL depending on antibody affinity. The equilibrium titration data were fit to a 1:1 reversible binding model using KinExA Pro Software (version 3.0.6; Sapidyne Instruments) to determine the KD. Binding curves are provided as supplementary figures.
Binding specificity was also measured using Attana A100 Quartz Crystal Microbalance (QCM) (Attana AB, Stockholm, SWEDEN). Label-free toxin domains were tested for binding by using mAb A, mAb B, or mAb C captured on the chip. This method was used as a simple and fast way to detect binding activities of the domains (data not shown).
ELISA binding assay
Plate ELISAs specific for mAb A, mAb B, and mAb C were developed using the purified domains. 96 well microtitre plates (Thermo Scientific) were coated overnight at 2–8°C with 100 µL/well of purified HCC (5 µg/mL) HCN (10 µg/mL) or LC-HN (5 µg/mL) domain. Plates were blocked with 1% (w/v) BSA, 0.05% (v/v) Tween 20 in PBS for 2 hours. Plates were washed one time with 0.05% (v/v) Tween 20 in PBS (PBS-T) by an automated plate washer (Molecular Devices SKAN400). 100 µL of sample or standard mAb was added to each well. The plate was then incubated at room temperature for 2 hrs, washed three times with PBS-T before adding 100 µL/well of 1:10000 dilution of 0.4 mg/mL horseradish peroxidase conjugated goat anti-human IgG (Fc)(Pierce) for 2 hours. Plates were washed again as described above and the reaction developed by the addition of 2,2-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) at 100µL/well [27] [28, 29]. After 8–18 mins., the reaction was stopped and plates read in a Molecular Devices M2e instrument, and plotted using a 4-parameter fit algorithm against the mAb concentration to yield a sigmoidal curve. Every sample or standard was measured in duplicate on the same plate.
Electrochemiluminescent (ECL) assays
Highly sensitive ECL assays [30] were developed to measure the concentrations of the individual component mAbs (mAb A, B, or C) in serum matrix for PK studies. To detect mAb A in rat serum, 96 well standard MA2400 plates (Meso Scale Discovery (MSD), Gaithersburg, MD, Catalog No. L15XA-3) were coated overnight at 2–8°C with 100 µL/well of 0.5 µg/mL HCC domain in phosphate buffered saline (PBS). After blocking with 1% (w/v) BSA in PBS with 0.05% (v/v) Tween-20 for 1 to 4 hours, 50 µL of either serum samples or mAb A or IgG standards were added to each well. Every sample or standard was measured in duplicate using two wells on the same plate. The plate was then shaken at room temperature for 1 hr, washed four times with PBS-T before adding 50 µL/well of 1 µg/mL of Ruthenium (II) tris-bipyridine-(4-methylsulfone) NHS ester (SULFO-TAG™, MSD) labeled goat anti-human IgG (Jackson ImmunoResearch, West Grove, PA). Plates were washed again as described above, the MSD Read Buffer T was then added, and plates were read in a MSD SECTOR Imager 2400 instrument. To detect mAb B in rat serum, the HCN domain was conjugated to biotin (Pierce Biotechnology, Catalog No. 21336). 96-well streptavidin plates (MSD Catalog No. L15SA-1) were blocked at room temperature for 2 hours without shaking in 200 µl/well of 3% BSA in PBS-T. The blocking solution was removed by aspiration. Fifty µL/well of 0.25 µg/mL biotinylated HCN domain in high salt buffer containing 1M NaCl, 1% (w/v) BSA in PBS-T was added with the plates incubated for 1 hour on a shaker. Plates were then washed 4 times in PBS-T. Sample addition, washing, addition of secondary reagents and detection of mAb B binding was performed as described above for mAb A except Read Buffer S was used. To detect mAb C in rat serum, a bridging assay format was employed. 96-well streptavidin plates were blocked at room temperature for 2–4 hours without shaking in 300 µL/well of 3% BSA in PBS-T. Fifty µl of the mixture of biotinylated LC-HN and SULFO-TAG conjugated LC-HN was incubated with 50 µl of mAb C standard or diluted sample in each well of a 96-well polypropylene plate at RT for 2 hours on a shaker. At the end of the incubation period, the blocked plates were washed 4 times in PBS-T. Samples that were incubated with the labeled domains were then transferred to the blocked plates at 50 µL/well and incubated for 1 hour at RT on a shaker. At the end of the incubation, the plates were washed 4 times with PBS-T and 150 µL of Read Buffer T was added to each well. The plates were then read in a MSD SECTOR Imager 2400 instrument. A standard curve was plotted as ECL signal versus concentrations of antibody standards. A regression analysis was performed using a 4-parameter fit and concentrations were determined for samples and controls by interpolation from the standard curve.
For the detection of mAb A, B, and C in Cynomolgus monkey and human serum, ECL assays in bridging format similar to the mAb C for rat serum were employed. Each of the 3 domains was labeled separately with either biotin or SULFO-TAG. Assay procedures including sample preparation, plate blocking, incubation procedures and detection of binding were similar to those descried for mAb C in rat serum above.
To demonstrate the specificity of this assay, one set of spiked human serum (Bioreclamation) samples was prepared consisting of a 1:1:1 mixture of mAbs A, B, and C at 40 ng/mL, 4 µg/mL and 400 µg/mL as determined by ELISA and A280 nm (molar extinction coefficient = 1.57 for mAb A, 1.37 for mAb B and 1.27 for mAb C). The ECL assay described above was used to measure the concentration of each of the individual mAbs. The percent recovery was calculated by dividing the concentration measured by the ECL assay by the nominal concentration.
Rat pharmacokinetic study
To characterize the pharmacokinetic profile of XOMA 3AB, male and female Sprague-Dawley rats (15/sex/group) were given a single intravenous dose of XOMA 3AB at 0.1, 1 or 10 mg/kg administered over 45–60 sec. Serum samples were collected from cohorts of 3 rats/sex/group at predose and postdose at 5 minutes, 6 and 24 hours on Day 1, and once on Days 2, 5, 8, 11, 15, 22, 29, 36, 43, 57 and 72. Serum levels of mAb A, mAb B, and mAb C were determined using a ECL assay described above. Serum samples for the immunogenicity analysis were collected from 3 rats/sex/group to assess the presence of anti-drug antibodies on Days 0, 29, 36, 43, 57, and 72 (termination).
Treatment of the animals was in accordance with regulations outlined in the USDA Animal Welfare Act (9 CFR, Parts 1, 2 and 3) and the conditions specified in the Guide for the Care and Use of Laboratory Animals (ILAR publication, 1996, National Academy Press), and was approved by the laboratory’s Intuitional Animal Care and Use Committee.
Results
XOMA 3AB consists of a humanized mAb (mAb B) and two fully human mAbs (mAb A and mAb C). Each mAb binds BoNT/A with high affinity (Table 1). mAb B binds the C-terminal subdomain of the BoNT/A receptor binding domain (HCC), mAb A binds the N-terminal subdomain of the BoNT/A receptor binding domain (HCN), and mAb C binds a complex epitope requiring both the translocation domain (HN) and light chain (LC) of BoNT/A (LC-HN) (Figure 1).
Table 1. Binding affinities and kinetics of mAbs for BoNT/A and for purified BoNT/A domains.
The equilibrium binding constant (KD) and the association rate constant (kon) were measured by flow fluorimetry in a KinExA. The dissociation rate constant koff was calculated from KD and kon
| BoNT/A1 Holotoxin | BoNT/A1 Domain | ||||||
|---|---|---|---|---|---|---|---|
| mAb | KD (pM) |
kon(M−1s−1) | koff(s−1) | Domain | KD (pM) |
kon(M−1s−1) | koff(s−1) |
| mAb A | 1.56 | 1.09 7 | 1.70 −5 | HCC | 1.33 | 1.00 7 | 1.33 −5 |
| mAb B | 6.16 | 5.04 6 | 3.10 −5 | HCN | 258.84 | 6.57 6 | 1.70 −3 |
| mAb C | 11.41 | 5.03 5 | 5.74 −6 | LCHN | 11.1 | 6.99 5 | 7.76 −6 |
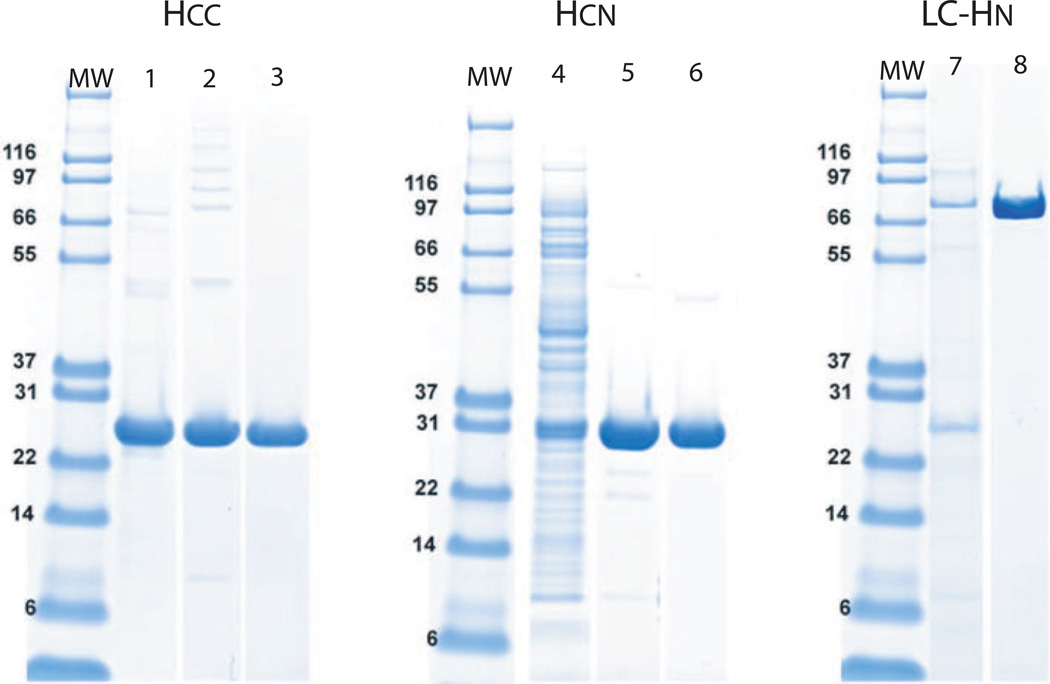
Characterization of purity, specificity, and affinity of BoNT/A domains
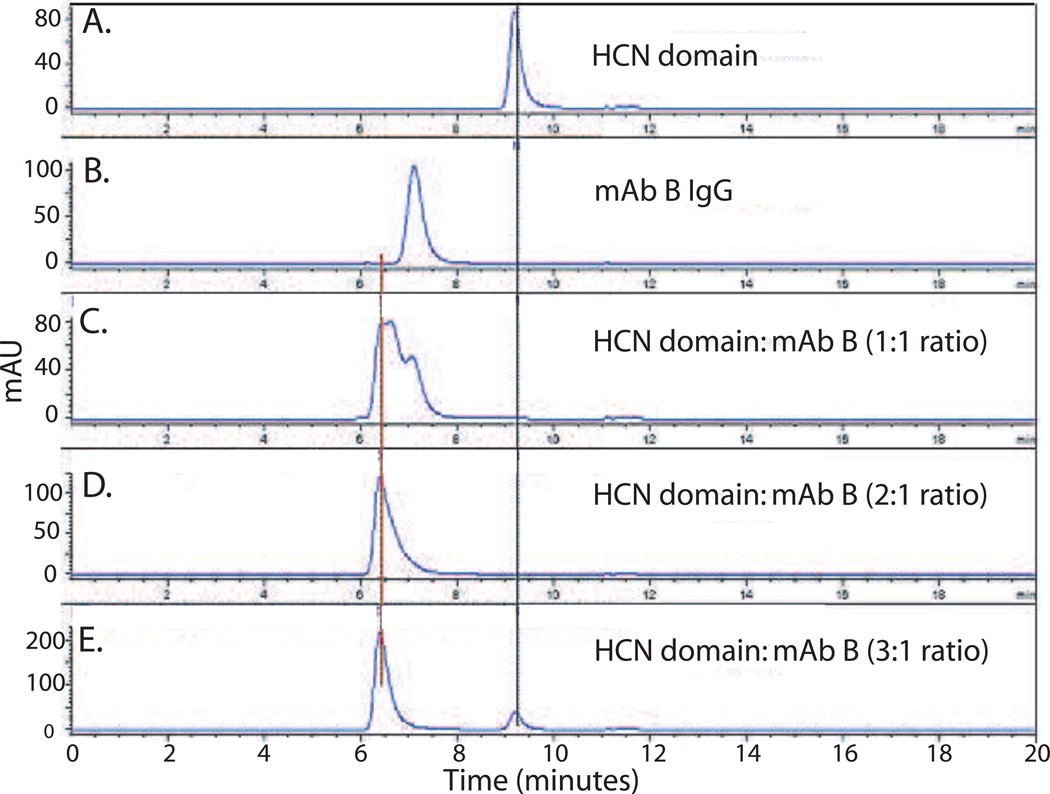
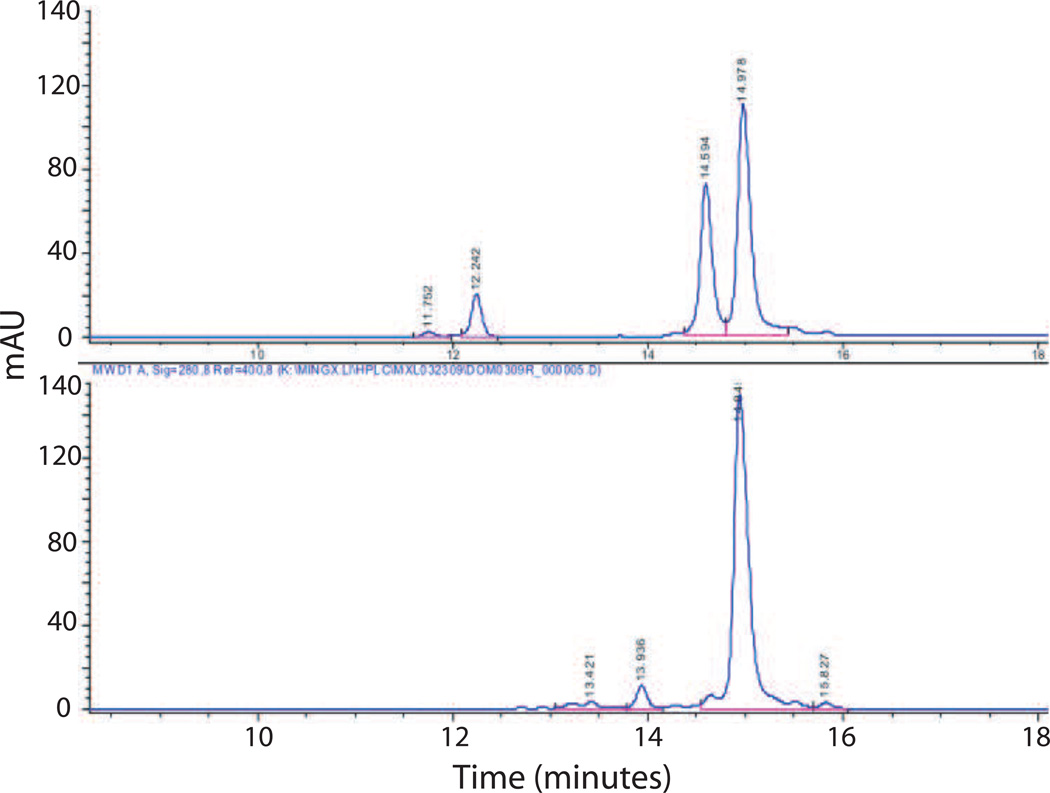
A suite of analytical methods was developed to characterize the purity, specificity, and binding activity and affinity of the domain proteins, and these characteristics were used to define a set of specifications and action limits to ensure consistency across multiple batches. Using these schemes, a purity of greater than 95% by SE-HPLC was achieved for each of the domains (see gel comparison of lanes 3, 6 and 8 in Figure 3) with yields 40–100 mg, 40–60 mg, and 1–10 mg/gm of E. coli paste for the HCC, HCN, and LC-HN domains respectively. Rapid SE-HPLC assays were developed for each domain that simultaneously determined domain product monomer content and impurities as well as antibody binding activity and specificity. For example, the purity of the HCN domain can be determined by the peak area under the SE-HPLC elution profile curves (Figure 4). A single peak is seen for the HCN domain which can be distinguished from the peak for mAb B (Figure 4A vs. B). Incubation of mAb B with HCN at a 1:1 ratio results in the appearance of a peak which elutes earlier than the mAb B peak which as determined by light scattering is consistent with the size of a complex of mAb B:HCN (Figure 4C) In addition, the HCN peak disappears, indicating that all of the HCN is immunoreactive and can be bound by mAb B. Since there are two antibody binding sites per mAb B IgG, excess mAb B is seen in this elution trace. As the ratio of HCN:mAb B is increased, the mAb B peak disappears, as only the immune complex is present (Figure 4 D–E). Similar assays were developed for HCC and LC-HN with results comparable to the results seen for HCN with respect to purity and immunoreactivity (data not shown). SE-HPLC was also used to confirm the specificity of each of the BoNT/A domains for their respective IgG by showing that each IgG only bound their respective domains and did not bind the other two domains (see Supplementary Figure 1 for example).
Figure 3. Analysis of the purity of BoNT/A HCN, Hcc and LC-HN domains by SDS-PAGE.
Coomassie-stained SDS-PAGE gels were run to characterize the purity of domain fractions. Molecular weight markers (MW) are in the left lane of each gel. Lanes 1–3 HCC (Lane 1, HCC after refolding from denatured inclusion bodies; Lane 2, excluded elution shoulder fractions showing prominent “ladder” containing improperly oxidized HCC species identified by RP-HPLC (see Figure 5); Lane 3, HCC main peak). Lanes 4–6 HCN (Lane 4, clarified HCN lysate; Lane 5, partially purified HCN ion exchange pool; Lane 6, Ni-NTA IMAC pool). Lanes 7–8 LC-HN (Lane 7, partially purified ion exchange pool; Lane 8, purified LC-HN pool).
Figure 4. Analysis of purity and immunoreactivity of BoNT/A HCN domain by SE-HPLC.
A. The purity and presence of aggregation can be identified by the elution profile of BoNT/A HCN. B. mAb B elutes before HCN, due to its larger size. C–E. When HCN and mAb B are mixed together, the HCN peak disappears, indicating that it is near 100% immunoreactive. At higher molar ratios of HCN, the mAb B peak disappears.
RP-HPLC was also developed for each domain to assess polarity-based purity and breakdown products. This assay indicated that in the case of early batches of HCC, two major peaks were present (Figure 5). However, when we conducted mass spectrometric analysis of these early batches of HCC samples that contained various ratios of the two RP-HPLC peaks by direct infusion into a ThermoFinnigan LCQ Duo Ion Trap mass spectrometer, we found that the mass values for all the different samples were the same within 2 Da. The presence of both disulfide-oxidized and reduced forms of HCC, which contains two cysteines, was confirmed by repeating the RP-HPLC after reducing the HCC sample (Figure 5). Modification of the HCC purification was greater than 95% in the reduced form. RP-HPLC was also used to assess the purity of the HCN and LC-HN domains (data not shown).
Figure 5. Analysis of the purity of BoNT/A HCC by reverse phase HPLC (RP-HPLC).
Top panel: Two major HCC peaks are recognizable. Bottom panel: Addition of reducing agent to HCC results in disappearance of one of the peaks, indicating that the two peaks shown in the top panel represent disulfide-oxidized and reduced HCC (see text for details).
Flow fluorimetry in a KinExA was used to measure the affinity and binding kinetics of mAb A, mAb B, and mAb C for their respective domains which was compared to the affinity of the mAbs for BoNT/A1 holotoxin (Table 1). The affinity of both mAb A and mAb C for their respective domains was comparable to their binding affinity for BoNT/A1 (mAb A 1.33 pM vs 1.56 pM; mAb C 11.1 pM vs 11.41 pM). mAb B bound with high affinity (258.84 pM) to its respective domain, but the binding affinity was approximately 40 fold lower than the affinity for holotoxin (258.84 pM vs 6.16 pM). The parental antibody from which mAb B was derived binds closely to the interface between HCC and HCN [31]. It is possible that the HCC is required to achieve wild-type binding affinity of mAb B. As shown in Levy et al. [23] the mAb B epitope is exclusively on HCN, but close to the boundary with HCC, thus HCC could play a role in stabilizing or modifying the conformation of HCN confirmation.
ELISA binding assays
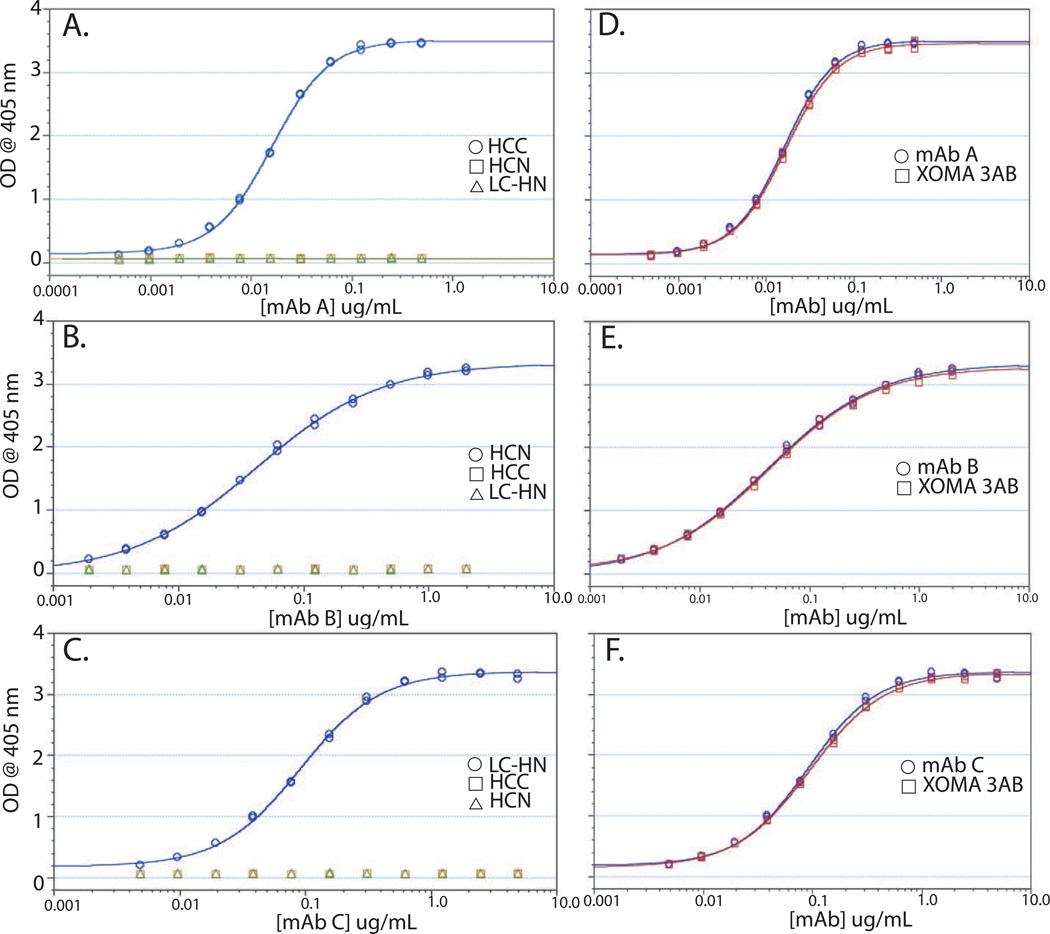
To measure the active concentration of each individual mAb in the three mAb combination, three separate ELISAs were developed. Purified BoNT/A HCN, HCC, or LC-HN were coated onto 96 well ELISA plates and the binding of each mAb determined. mAbs A, B, and C bound specifically to their respective domains with a linear range between 10 ng/mL − 100 ng.mL, 10 – 100 ng/mL, and 5 – 50 ng/mL respectively (Figure 6). No detectable binding of mAb A, mAb B, or mAb C was seen for other domains at concentrations up to 1 – 10 µg/mL (Figure 6). Due to this specificity, the ELISA could be used to measure the amount of each mAb present in XOMA 3AB. ELISA binding curves of XOMA 3AB were superimposable on the binding curves of mAbs A, B, and C assayed on their domain-specific domain (Figure 6).
Figure 6. Domain-specific ELISAs.
Binding of mAb A (A), mAb B (B), and mAb C (C) to plates coated with either HCC, HCN, or LC-HN. Binding of each mAb was only seen to plates coated with their respective domain. Binding of mAb A (D) mAb B (E) or mAb C (F) to plates coated with their respective domain compared to the binding of a combination of all three mAbs (XOMA 3AB).
Formulation of domains for long term storage
SE-HPLC, RP-HPLC, UV spectrometry, and ELISA were used to evaluate the stability of each of domains under both standard (2–8°C) and accelerated/stressed (25°C for liquid and 40°C for lyophilized) storage conditions. Data from the liquid state pre-formulation screening studies indicated that these domains were less stable than their monoclonal antibody counterparts. The stability data further indicated that the three domains exhibited significantly different stability profiles, with the LC-HN domain being relatively the most stable and the HCC domain the least stable among the three. For instance, the HCC domain showed evidence of degradation after only one month in liquid storage at 2–8°C as determined by RP-HPLC (Supplemental Figure 2). Reformulation of the HCC by lyophilization indicated no evidence of degradation after 4 weeks at either 2–8°C or 40°C. In order to enhance the stability and standardize the handling procedure for the domain proteins as critical reagents, we chose the lyophilized formulation for long-term storage of all three domains.
Electrochemiluminescent (ECL) pharmacokinetics assay
Electrochemiluminescent (ECL) binding assays using sandwich and homogeneous bridging formats were developed to allow the quantification of each mAb of XOMA 3AB in rat, cynomolgus monkey and human serum. The mAb A assay showed a lower limit of quantification (LLOQ) of 1.22 ng/mL and a linear range of quantification of 1.22 ng/mL − 5000 ng.mL in rat serum. The mAb B assay had a LLOQ of 2.44 ng/mL with a linear range of quantification between 2.44 and 5000 ng/mL while the mAb C assay has a LLOQ of 0.61 ng/mL and a linear range from 0.61 to 2000 ng/mL in rat serum. No detectable binding of mAb A, mAb B, or mAb C was seen for other domains at concentrations up to 10 µg/mL (data not shown).
Next, the ability of each assay to measure known amounts of mAbs A, B, and C spiked into human serum was determined. Three different amounts of each mAb (400 µg/mL, 4 µg/mL, or 40 ng/mL for mAb A assay and 200 µg/mL, 2 µg/mL, or 120 ng/mL for mAb B and mAb C assays) were spiked into sera and the amount present quantified. In the mAb A specific assay using the HCC domain, measured amounts of mAb A were within 2 to 7% of the spiked amount with a coefficient of variation (CV) of 2.26% to 4.38% (Table 2). No detectable binding was measured for mAb B or mAb C, even at the 400 µg/mL amount. Similar results were seen for mAb B (measured amounts within 0 to 16% of the spiked amounts and CV of 0.570% to 7.26%) and for mAb C (measured amounts within 2 to 13% of the spiked amounts and CV of 0.333 to 3.5%).
Table 2. Measurement of the concentration of mAb A, mAb B, and mAb C spiked into human serum in each of the bridging ECL assays.
The indicated amount of either mAb A, mAb B, or mAb C was spiked into human serum and the amount of each measured in triplicate in a bridging ECL assay using either the HCC domain specific for mAb A, HCN for mAb B or LC-HN for mAb C. Only the target antibody gave a signal in these assays. % recovery = measured concentration of mAb/nominal concentration of mAb. CV = coefficient of variation.
| Assay | mAb A Specific Assay using Labeled HCC Domain |
mAb B Specific Assay using Labeled HCN Domain |
mAb C Specific Assay using Labeled LC-HN Domain |
||||||
|---|---|---|---|---|---|---|---|---|---|
| mAb A | mAb B | mAb C | |||||||
|
Nominal Concentration (ng/mL) |
400000 | 4000 | 40 | 200000 | 2000 | 120 | 200000 | 2000 | 20 |
|
Mean Observed Concentration (ng/mL) |
408000 | 4280 | 38.6 | 199000 | 1930 | 101 | 211000 | 2040 | 17.3 |
|
Mean % Recovery1 |
102 (2.31) |
107 (4.69) |
96.4 (4.02) |
99.7 (7.23) |
96.7 (2.93) |
84.4 (0.481) |
105 (2.47) |
102 (4.16) |
86.7 (0.289) |
| %CV | 2.26 | 4.38 | 4.17 | 7.26 | 3.03 | 0.570 | 2.34 | 4.09 | 0.333 |
Standard deviations shown in parentheses
We next determined the ability of the assay to accurately measure each mAb in an equimolar mixture of each of the three mAbs (XOMA 3AB) spiked into human serum. In the mAb A specific assay using the HCC domain, measured amounts of mAb A were within 0 to 15% of the spiked amount with a coefficient of variation (CV) of 3.93% to 7.13% (Table 3). Similar results were seen for mAb B (measured amounts within 6 to 17% of the spiked amounts and CV of 1.87% to 5.51%) and for mAb C (measured amounts within 9 to 17% of the spiked amounts and CV of 0.781% to 3.35%) (data not shown).
Table 3. Detection of mAb A in a three antibody mixture (mAb A, mAb B, and mAb C) spiked into human serum.
The amount of mAb A present in a known concentration of an equimolar mixture of mAb A, mAb B, and mAb C was determined using the HCC domain ECL assay (5 replicates).
| Nominal Concentration (ng/mL) | 400000 | 4000 | 40 |
|---|---|---|---|
|
Mean Observed Concentration (ng/mL) |
403000 | 4020 | 34.0 |
| Mean % Recovery1 | 101(5.11) | 100(6.35) | 84.9 (3.34) |
| %CV | 4.36 | 7.13 | 3.93 |
Standard deviations shown in parentheses
Pharmacokinetics of mAbs A, B, and C following administration of XOMA 3AB to rats
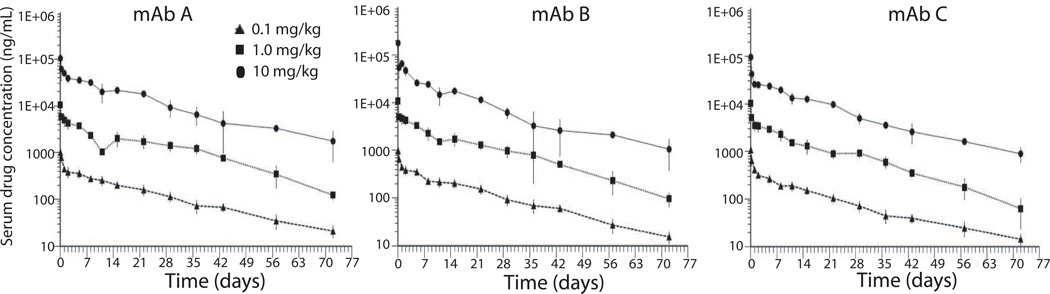
The concentration-time profiles of mAb A, mAb B, and mAb C were determined in rats following a single intravenous administration of XOMA 3AB (Figure 7). All animals survived and there were no adverse clinical signs attributed to treatment. PK assays based on the ECL format were conducted to measure serum levels of each individual mAb. Results using rats that did not develop antibodies to mAb A, B, or C, are presented; the presence of anti-drug antibodies led to extremely rapid elimination of test antibodies. PK evaluation of XOMA 3AB showed that the individual mAbs are slowly eliminated from the serum with mean half-lives ranging from 8.45 day to 17.43 days (Figure 7). Mean residence time (MRT) values also indicate extended exposure after a single dose of XOMA 3AB, and show a trend to be shortened in the high dose group. There were no consistent differences observed in these parameters, half-life and MRT, between male and female animals or among the three mAbs. Indicators of exposure, observed Cmax (maximum concentration) and AUC (area under the concentration-time curve) values, increased approximately linearly with dose.
Figure 7. Pharmacokinetics of mAbs A, B, and C in rats.
The concentration-time curves of mAb A, mAb B and mAb C following a single intravenous administration of XOMA 3AB at 0.1, 1, 10 mg/kg. Each point represents the mean ± SD of serum drug levels from six (3 males and 3 females) or more animals. The data from male and female animals were combined.
In summary, the PK behavior of the three component monoclonal antibodies, mAb A, mAb B, and mAb C, was similar, exhibiting dose dependent exposure and long elimination half-life after intravenous administration of 0.1, 1 and 10 mg/kg in Sprague-Dawley rats.
Discussion
An ever increasing number of mAb combinations are under development for a range of diseases [2–7]. Therapeutic mAb combinations pose additional developmental challenges compared to individual mAbs, including the need to be able to measure the concentration and activity of each mAb independently when combined, both in vitro and in vivo. Here we show that individual domains or subdomains of BoNT/A, a multidomain protein, can be independently expressed and purified at scale and used to develop binding assays that can specifically measure the concentration of the three BoNT/A mAbs in XOMA 3AB both in vitro and in vivo. Companion analytic tools, including SDS-PAGE, KinExA, SE-HPLC, and RP-HPLC, allow monitoring of domain quality over time based on affinity for their respective mAbs, purity, amount of aggregation and immunoreactivity. These tools also allowed identification of long term storage formulations for each of the domains.
Each mAb bound a different epitope contained within either a unique folded domain of BoNT/A (the LC-HN) or within subdomains of the HC (HCC and HCN), increasing the likelihood that the epitopes could be successfully expressed and purified in a stable form [22]. The BoNT/A LC-HN and HCC have been previously expressed and purified from E. coli [32, 33]. In cases where conformational mAb epitopes are located on the same domain, it should prove possible to generate mutants within the mAb epitope that knock out binding of one mAb but not the other. For example we have used yeast display to map the epitopes of three BoNT/A mAbs binding the same structural domain and shown that mutants can be constructed that specifically knock out binding of only one mAb [23].
In conclusion, we developed specific BoNT/A domains and assays for individually measuring in vitro and in vivo binding for each of the three mAb components of XOMA 3AB. These assays can be used for both characterizing the monoclonal and oligoclonal drug products during manufacturing and for monitoring their stability over time to ensure quality of the drug and its component mAbs. Use of the non-toxic domains obviates the need to work with the highly toxic BoNT/A and eliminates the need for select agent registration. The type of antigen engineering described here should be broadly applicable for the generation of sensitive assays for quantitation of individual mAbs in an oligoclonal antibody cocktail that bind the same protein.
Supplementary Material
Acknowledgements
This work was partially supported by NIAID contract number HHSN266200600008C and HHSN272200800028C and Cooperative Agreement U01AI056493. Contributions to the measurement and data for PK are gratefully received from H. Ng, C. Green, T. Harrison, A. D’Andrea and J. Mirsalis from SRI International, 333 Ravenswood Ave., Menlo Park Ca, 94025. We would also like to acknowledge Anwar Zaman for his help with purifying the domains, and Jacob Kung and Dennis Lee for their help with ELISAs.
Abbreviations used
- AUC
area under the concentration-time curve
- BoNT
Botulinum neurotoxin
- BoNT/A1
BoNT sub-serotype A1
- BSA
bovine serum albumin
- Cmax
maximum concentration
- CV
coefficient of variation
- Fab’2
fragment antigen binding
- Fc
fragment crystallizable
- IgG
immunoglobulin G
- IMAC
Ni-NTA immobilized metal affinity chromatography
- LD50
lethal dose 50%
- mAb
monoclonal antibody
- MRT
Mean residence time
- scFv
single chain variable fragment
- BoNT/A HC
C-terminal domain of the BoNT/A heavy chain
- BoNT/A HN
N-terminal domain of the BoNT/A heavy chain
- BoNT/A LC
BoNT/A light chain
- pM
picomolar
- fM
femtomolar
- ECL
electrochemiluminescence
- ELISA
Enzyme-linked immunosorbent assay
- FACS
fluorescent activated cell sorting
- HPLC
high pressure liquid chromatography
- IU
International Unit
- IPTG
Isopropyl-β-D-thio-galactoside
- KD
equilibrium dissociation constant
- kon
association rate constant
- koff
dissociation rate constant
- MFI
mean fluorescent intensity
- MW
molecular weight
- PBS
phosphate buffered saline
- PBS-T
PBS with Tween
- PK
pharmacokinetics
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate PAGE
- SEC
Size-exclusion chromatography
- SE-HPLC
Size-exclusion high pressure liquid chromatography
- XOMA 3AB
an equimolar mixture of three human/humanized mAbs binding non-overlapping epitopes on botulinum neurotoxin type A (BoNT/A)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nature Reviews Drug Discovery. 2010;9(10):767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 2.Logtenberg T. Antibody cocktails: next-generation biopharmaceuticals with improved potency. Trends Biotechnol. 2007;25(9):390–394. doi: 10.1016/j.tibtech.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Bakker AB, et al. First administration to humans of a monoclonal antibody cocktail against rabies virus: safety, tolerability, and neutralizing activity. Vaccine. 2008;26(47):5922–5927. doi: 10.1016/j.vaccine.2008.08.050. [DOI] [PubMed] [Google Scholar]
- 4.Prabakaran M, et al. Combination therapy using chimeric monoclonal antibodies protects mice from lethal H5N1 infection and prevents formation of escape mutants. PLoS One. 2009;4(5):e5672. doi: 10.1371/journal.pone.0005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Novel chimpanzee/human monoclonal antibodies that neutralize anthrax lethal factor, and evidence for possible synergy with anti-protective antigen antibody. Infect Immun. 2009;77(9):3902–3908. doi: 10.1128/IAI.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazor O, et al. Combination of Anthrax-Toxin Neutralizing Antibodies: Analysis of Synergism/Antagonism Effect. The Challenge of Highly Pathogenic Microorganisms. 2010:275–285. [Google Scholar]
- 7.Pedersen M, et al. Sym004: a novel synergistic anti-epidermal growth factor receptor antibody mixture with superior anticancer efficacy. Cancer Research. 2010;70(2):588. doi: 10.1158/0008-5472.CAN-09-1417. [DOI] [PubMed] [Google Scholar]
- 8.Production and quality control of monoclonal antibodies. Directive 75/318/EEC3AB4a. 1995 [Google Scholar]
- 9.Points to Consider in the Manufacture and Testing of Monoclonal Antibody Products for Human Use. doi: 10.1097/00002371-199705000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Schenerman M, et al. CMC Strategy Forum Report: Analysis and structure characterization of monoclonal antibodies. BioProcess Int. 2004;2(2):42–49. [Google Scholar]
- 11.Teshima G, et al. Separation of oxidized variants of a monoclonal antibody by anion-exchange. J Chromatogr A. 2011;1218(15):2091–2097. doi: 10.1016/j.chroma.2010.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiff Y, et al. Measurements of bone turnover markers in premature infants. J Pediatr Endocrinol Metab. 2001;14(4):389–395. doi: 10.1515/jpem.2001.14.4.389. [DOI] [PubMed] [Google Scholar]
- 13.St. John R, Finlay B, Blair C. Bioterrorism in Canada: An economic assessment of prevention and postattack exposure. Can. J. Infect. Dis. 2001;12:275–284. doi: 10.1155/2001/904148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black RE, Gunn RA. Hypersensitivity reactions associated with botulinal antitoxin. Am. J. Med. 1980;69:567–570. doi: 10.1016/0002-9343(80)90469-6. [DOI] [PubMed] [Google Scholar]
- 15.Hibbs RG, et al. Experience with the use of an investigational F(ab')2 heptavalent botulism immune globulin of equine origin during an outbreak of type E origin in Egypt. Clin. Infect. Dis. 1996;23:337–340. doi: 10.1093/clinids/23.2.337. [DOI] [PubMed] [Google Scholar]
- 16.Nowakowski A, et al. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99(17):11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamanna C. The most poisonous poison. Science. 1959;130:763–772. doi: 10.1126/science.130.3378.763. [DOI] [PubMed] [Google Scholar]
- 18.Blasco H, et al. Evaluation of a peptide ELISA for the detection of rituximab in serum. J Immunol Methods. 2007;325(1–2):127–139. doi: 10.1016/j.jim.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Damen CW, et al. Development and validation of an enzyme-linked immunosorbent assay for the quantification of trastuzumab in human serum and plasma. Anal Biochem. 2009;391(2):114–120. doi: 10.1016/j.ab.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Tornetta M, et al. Isolation of human anti-idiotypic antibodies by phage display for clinical immune response assays. J Immunol Methods. 2007;328(1–2):34–44. doi: 10.1016/j.jim.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Hanke T, Szawlowski P, Randall RE. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J Gen Virol. 1992;73(Pt 3):653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 22.Lacy DB, et al. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nature Struct. Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 23.Levy R, et al. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365(1):196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blake RC, 2nd, Pavlov AR, Blake DA. Automated kinetic exclusion assays to quantify protein binding interactions in homogeneous solution. Anal Biochem. 1999;272(2):123–134. doi: 10.1006/abio.1999.4176. [DOI] [PubMed] [Google Scholar]
- 25.Lou J, et al. Affinity maturation of human botulinum neurotoxin antibodies by light chain shuffling via yeast mating. Protein Eng Des Sel. 2010;23(4):311–319. doi: 10.1093/protein/gzq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razai A, et al. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J. Mol. Biol. 2005 doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Szutowicz A, Kobes RD, Orsulak PJ. Colorimetric assay for monoamine oxidase in tissues using peroxidase and 2,2'-azinodi(3-ethylbenzthiazoline-6-sulfonic acid) as chromogen. Anal Biochem. 1984;138(1):86–94. doi: 10.1016/0003-2697(84)90773-5. [DOI] [PubMed] [Google Scholar]
- 28.Uotila M, Engvall E, Ruoslahti E. Monoclonal antiodies to human alphafetoprotein. Mol Immunol. 1980;17(6):791–794. doi: 10.1016/0161-5890(80)90150-9. [DOI] [PubMed] [Google Scholar]
- 29.Al-Kaissi E, Mostratos A. Assessment of substrates for horseradish peroxidase in enzyme immunoassay. J Immunol Methods. 1983;58(1–2):127–132. doi: 10.1016/0022-1759(83)90269-7. [DOI] [PubMed] [Google Scholar]
- 30.Bard AJ, et al. Chemiluminescence, electrogenerated. Encyclopedia of Analytical Chemistry. 2000 [Google Scholar]
- 31.Garcia-Rodriguez C, et al. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25(1):107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 32.Sharma S, Zhou Y, Singh BR. Cloning expression, and purification of C-terminal quarter of the heavy chain of botulinum neurotoxin type A. Protein Expr Purif. 2006;45(2):288–295. doi: 10.1016/j.pep.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Shone C, et al. Bivalent recombinant vaccine for botulinum neurotoxin types A and B based on a polypeptide comprising their effector and translocation domains that is protective against the predominant A and B subtypes. Infect Immun. 2009;77(7):2795–2801. doi: 10.1128/IAI.01252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.