Abstract
Fc receptors play a central role in maintaining the homeostatic balance in the immune system. Our knowledge of the structure and function of these receptors and their naturally occurring polymorphisms, including single nucleotide polymorphisms and/or copy number variations, continues to expand. Through studies of their impact on human biology and clinical phenotype, the contributions of these variants to the pathogenesis, progression, and/or treatment outcome of many diseases that involve immunoglobulin have become evident. They affect susceptibility to bacterial and viral pathogens, constitute as risk factors for IgG or IgE mediated inflammatory diseases, and impact the development of many autoimmune conditions. In this chapter, we will provide an overview of these genetic variations in classical FcγRs, FcRLs, and other Fc receptors, as well as challenges in achieving an accurate and comprehensive understanding of the FcR polymorphisms and genomic architecture.
Introduction
Highly homologous in their extracellular sequences, members of the Fc receptor family have both structural differences as well as allelic variations which impact biological properties and their respective roles in pathophysiology. Investigation over the last two decades has demonstrated regulatory and/or coding single nucleotide polymorphisms (SNP) that change receptor biology through one of three mechanisms: quantitative receptor expression, ligand affinity, or signaling capacity. Emerging data have also demonstrated copy number variation (CNV) in the classical low affinity Fc receptors for IgG. Many of the SNPs and CNVs are associated with pathogenesis, severity, and/or treatment outcome in a range of immune-mediated diseases. Signaling and biology of Fc receptors are discussed in Chapter X and Y. In this chapter, we discuss the germ line variations in the genes encoding Fc receptors and how these variations impact receptor function and association with disease.
Human FcR Polymorphisms: Location and Functional Implications
Single Nucleotide Polymorphisms
Numerous single-nucleotide polymorphisms have been identified through Fc receptor sequence analysis, particularly within the classical low-affinity FcγR cluster located on the long arm of chromosome 1. The allele frequencies of these genetic variants, many of which have not been characterized for function, may differ across different ancestry groups. The more thoroughly studied SNPs with known functional relevance and disease association are presented in Tables 1 and 2.
Table 1.
Genetic variations of classical FcγRs
| Receptor | Genetic variation | Functional property | Disease/trait |
|---|---|---|---|
| FcγRIIa |
R131H (rs1801274) |
H131: higher affinity, can bind IgG2 |
Infections (Bredius et al. 1993; Endeman et al. 2009; Platonov et al. 1998; Jansen et al. 1999; Yee et al. 2000; Sanders et al. 1994; Bredius et al. 1994; Yuan et al. 2005; Loke et al. 2002; Garcia et al. 2010; Diamantopoulos et al. 2013; Forthal et al. 2007), Autoimmune inflammation (International Consortium for Systemic Lupus Erythematosus et al. 2008; Shrestha et al. 2012; Onouchi et al. 2012; Karassa et al. 2002; Magnusson et al. 2004; Kyogoku et al. 2004; Weersma et al. 2010; Dijstelbloem et al. 1999; Norsworthy et al. 1999; Song et al. 1998; Dijstelbloem et al. 2000; Morgan et al. 2006; van der Pol et al. 2000, 2003; Khor et al. 2011), atopy (Wu et al. 2014) |
| rs10919543 | increasing mRNA expression | TA (Saruhan-Direskeneli et al. 2013) | |
| 27Q > W (rs9427397) (rs9427398) | unknown | KD (Breunis et al. 2013) | |
| rs58055840 | unknown | Immunocyte levels (Orru et al. 2013) | |
| rs10800309 | unknown | UC (McGovern et al. 2010; Asano et al. 2009), SS (Lessard et al. 2013) | |
| rs12746613 | unknown | RA (Raychaudhuri et al. 2009) | |
| rs6658353 | unknown | SS (Lessard et al. 2013) | |
| FcγRIIb | I232T (rs1050501) | T232: altered partition to lipid rafts; altered signaling capability | SLE (Kono et al. 2005; Chu et al. 2004; Chen et al. 2006), atopy (Wu et al. 2014) |
| 2B.1/2B.4a (rs3219018) | 2B.4: higher promoter activity/expression | SLE (Su et al. 2004; Blank et al. 2005; Olferiev et al. 2007), KD (Breunis et al. 2013) | |
| rs2125685 | unknown | Periodontitis (Sugita et al. 2012) | |
| FcγRIIc | STP/Q13 (rs10917661) | STP: pseudogene; Q13:expression | ITP (Breunis et al. 2008), SLE (Li et al. 2013), KD (Breunis et al. 2013) |
| CNV | altered protein expression level | ITP (Breunis et al. 2008), SLE (Li et al. 2013) | |
| FcγRIIIa | V158F rs396991 | V158: higher affinity for IgG1, IgG3 | SLE (Wu et al. 1997; Edberg et al. 2002; Koene et al. 1998), RA (Morgan et al. 2006), GPA (Dijstelbloem et al. 1999), Lupus nephritis (Jonsen et al. 2007) |
| CNV | Altered protein expression level | anti-GBM disease (Zhou et al. 2010) | |
| rs445509 | unknown | Periodontitis (Chai et al. 2010) | |
| FcγRIIIb | NA1/NA2b | NA1: higher affinity | SLE (Hatta et al. 1999), ITP (Foster et al. 2001) |
| SH | unknown | unknown | |
| CNV | Altered protein expression level | SLE (Willcocks et al. 2008), ANCA vasculitis (Tse et al. 2000; Niederer et al. 2010), SS (Nossent et al. 2012) |
aPromoter haplotype. 2B.1: -120G-386T; 2B.4: -120C-386A
bCoding haplotype. NA1: 141G 147C 227A 349G; NA2: 141C 147T 227G 349A
TA: Takayasu’s arteritis; KD: Kawasaki disease; UC: ulcerative colitis; SS: Sjögren’s Syndrome; RA: rheumatoid arthritis; SLE: systemic lupus erythematosus; ITP: idiopathic thrombocytopenia purpura; GPA: Granulomatosis with polyangitis (Wegener’s granulomatosis); anti-GBM disease: Anti–glomerular basement membrane (anti-GBM) antibody disease; ANCA: anti neutrophil cytoplasmic antibodies
Table 2.
Genetic variations of FcεR, FcαR, FCRLs and FcRn
| Receptor | Genetic variation | Functional property | Disease/trait |
|---|---|---|---|
| FcεRI-α | −66T/C (rs2251746) | −66T:higher promoter activity/expression | AD (Hasegawa et al. 2003), asthma (Zhou et al. 2012), high IgE (Weidinger et al. 2008; Granada et al. 2012) |
| −315C/T (rs2427827) | −315T:higher promoter activity/expression | Chronic urticarial (Kim et al. 2006; Bae et al. 2007), asthma (Shikanai et al. 1985; Potaczek et al. 2006; Zhou et al. 2012) | |
| FcεRI-β | E237G | unknown | Atopy, asthma (Zhang et al. 2004; Yang et al. 2014), nasal allergy (Kim et al. 2007; Laprise et al. 2000; Nagata et al. 2001) |
| I181L | unknown | Atopy (Li and Hopkin 1997) | |
| −109C/T | unknown | High IgE (Hizawa et al. 2000), asthma (Kim et al. 2006; Yang et al. 2014) | |
| −426C/T −654T/C | −426C and −654T:higher promoter activity/expression | atopy (Nishiyama et al. 2004) | |
| FcεRII | R62 W (rs2228137) | W62:resistance to proteolytic cleavage | Asthma (Laitinen et al. 2000) |
| rs3760687 | unknown | High IgE (Sharma et al. 2014) | |
| FcαRI | S248G | G248: higher IgA-mediated activation | SLE (Wu et al. 2007) |
| FCRL1 | rs4971154 | unknown | T1D (Plagnol et al. 2011) |
| FCRL3 | rs7528684 | Altered gene expression |
SLE, RA, AITD (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013); (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013); T1D (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013); MS (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013); Endometriosis (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013) |
| rs7522061 | unknown | MS (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013) | |
| rs2282288 | unknown | GD (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013) | |
| rs11264798 | unknown | T1D (Osuga et al. 2011; Teles et al. 2011; Szczepanska et al. 2013) | |
| FCRL4 | rs2777963 | unknown | AS (Zeng et al. 2012) |
| rs14335 | unknown | AS (Zeng et al. 2012) | |
| rs10489674 | unknown | AS (Zeng et al. 2012) | |
| FCRL5 | rs12036228 | unknown | AS (Tang et al. 2009) |
| rs6427384 | unknown | AS (Tang et al. 2009) | |
| FcRn | VNTRa | altered promoter activity/expression | unknown |
aVNTR: variable number of tandem repeats
AD: atopic dermatitis; T1D: type-1 diabetes; AITD: autoimmune thyroid disease; MS: multiple sclerosis; GD: Graves’ disease; AS: ankylosing spondylitis
FcγRIIa (FCGR2A)
A nonsynonymous polymorphism (519G > A, rs1801274) in exon 4 encoding the membrane proximal Ig-like domain of FCGR2A leads to an arginine (R) to histidine (H) change at position 131 and alters receptor affinity for ligand. The R131 and H131 alleles are co-dominantly expressed. The FcγRIIa-H131 allele readily binds human IgG2 while the R131 allele does not effectively bind IgG2 (Salmon et al. 1992; Parren et al. 1992). Studies with IgG3 suggest that the H131 allele may bind IgG3 with moderately greater affinity than the R131 allele (Parren et al. 1992; Bredius et al. 1994). Crystallographic analysis and molecular modeling studies suggest that the H131R position is on the contact interface between receptor-IgG (Maxwell et al. 1999). As the most broadly expressed FcγR across a range of cell types in humans, the variation in ligand affinity has functional relevance in determining cellular interactions with IgG antibodies, including the clearance of IgG2 immune complexes. For example, neutrophils from FcγRIIa-H131 homozygous donors are much more effective than neutrophils from R131 homozygous donors in phagocytosing IgG2-opsonized particles (Bredius et al. 1993).
Several FCGR2A SNPs, including rs1801274 encoding R131H (International Consortium for Systemic Lupus Erythematosus 2008), as well as several variants in non-coding regions, including rs10919543 (Saruhan-Direskeneli et al. 2013), rs12746613 (Raychaudhuri et al. 2009), rs10800309 (McGovern et al. 2010; Asano et al. 2009), rs6658353 (Lessard et al. 2013), and rs6427609 (Kettunen et al. 2012), have been associated with disease phenotypes in various genome-wide association studies (GWAS). These disease association studies, based on high through put genotyping technologies, suggest that variation in FcγRIIa biology may contribute to a number of human disease phenotypes. However, not all variants identified through such studies have an obvious function or relationship to biological processes, and direct inference of pathophysiology requires further study. In some cases, SNP-based associations may be tagging linkage disequilibrium (LD) blocks. Given the segmental duplication in the classical low affinity FCGR cluster and the consequent high degree of genomic sequence homology, this region is not technically amenable to efficient genotyping with array-based strategies. Thus, genotyping coverage in genome-wide association studies is not optimal because of difficulty in accurate probe design and position assignment.
FcγRIIb (FCGR2B)
Some nonsynonymous coding SNPs in the FcR cluster affect the signaling capacity of the expressed receptor. In the FCGR2B gene locus, a nonsynonymous T > C SNP (rs1050501) encodes an isoleucine (I) to threonine (T) substitution at position 187 in the transmembrane domain; this variant is also known as I/T232 when the signal peptide is included in the numbering (Kyogoku et al. 2002; Li et al. 2003). The FcγRIIb-187threonine allele, which is less efficient in trans-locating into lipid rafts in the plane of the cell membrane, may result in decreased quantitative participation of FcγRIIb in the assembly of lipid raft-based signaling complexes with a resultant decreased inhibitory potential (Kono et al. 2005; Floto et al. 2005).
Su et al. identified a promoter haplotype (rs3219018) in FcγRIIb that alters receptor expression (Su et al. 2004). The less common promoter haplotype (–386C-120A) showed increased binding of transcription factors GATA4 and Yin-Yang 1, leading to higher receptor expression than found with the more frequent haplotype (–386G-120T) (Su et al. 2004a, b; Blank et al. 2005). Of note, sequence analysis of these promoter variants has revealed nearly identical sequence in the proximal promoter region of FCGR2C, thus underscoring the important consideration of the potential for expression of both receptors.
FcγRIIc (FCGR2C)
FCGR2C, often considered a pseudogene, has received less attention than other Fc receptors. The nonsynonymous SNP (202T > C, rs10917661) in its first extracellular domain changes the common allele (202T), which encodes a translation termination codon at residue position 13, to 202C, which encodes an open reading frame (ORF) for glutamine. The FcγRIIc-ORF allele produces an ITAM-containing activating receptor that has been detected on NK cells (Metes et al. 1998, 1999; Stewart-Akers et al. 2004) and B cells (Li et al. 2013). Functionally, NK cells bearing the ORF allele are capable of clearing anti-FcγRII coated particles through reverse antibody-mediated cellular cytotoxicity (ADCC) (Ernst et al. 2002; Breunis et al. 2008). On B cells, the FcγRIIc-ORF allele counterbalances the negative feedback of FcγRIIb on BCR signaling, resulting in enhanced B cell responsiveness including upstream signaling events such as tyrosine kinase phosphorylation and calcium transients, and integrated cell programs such as antibody production (Li et al. 2013).
FcγRIIIa (FCGR3A)
Similar to FcγRIIa, FcγRIIIa also has co-dominantly expressed alleles that affect receptor affinity for ligand. In the second extracellular domain of FCGR3A, a point substitution of T to G at nucleotide 559 (rs396991) changes the phenylalanine (F) at amino acid position 158 to valine (V). The FcγRIIIa-158V allele (also known as 176 V when the leader sequence is included) displays higher affinity for IgG1 and IgG3 relative to the 158F (176F) allele. The 158 V form is also capable of binding IgG4, while the 158F allele is not (Wu et al. 1997; Koene et al. 1997). NK cells from FcγRIIIa-158 V (high binder) homozygous donors exhibit increased calcium influx, greater CD25 expression, and faster apoptosis than those cells from FcγRIIIa-158F (low binder) homozygous donors (Wu et al. 1997).
FcγRIIIb (FCGR3B)
The GPI-anchored FcγRIIIb, mainly expressed on neutrophils, has three different allotypic variants, known as NA1, NA2, and SH. The neutrophil antigen (NA) variants NA1 and NA2 are a product of five nonsynonymous SNPs in the first Ig-like domain, with an asparagine to serine switch at amino acid position 65 resulting in altered glycosylation and reduced affinity in the NA2 allele (Ravetch and Perussia 1989; Salmon et al. 1990). FcγRIIIb-NA1 exhibits higher affinity and more efficient phagocytosis of IgG1 and IgG3 opsonized particles compared to the NA2 allele (Salmon et al. 1990). The SH allele results from an alanine to aspartic acid substitution at position 78 and is observed in the context of the NA2 allele (Bux et al. 1997). The exact function of the SH allele is not yet known.
FcαRI (FCAR)
FCAR (CD89) encodes the human IgA receptor FcαRI. A common SNP (844A > G) was identified through direct sequencing of the coding region of FCAR (CD89) (Jasek et al. 2004; Wu et al. 2007). This transition changes amino acid codon 248 in the cytoplasmic domain from serine to glycine, resulting in enhanced cellular functions. For example, when equivalently stimulated with human IgA, neutrophils homozygous for the FcαR-G248 allele produce significantly higher levels of IL-6 compared to neutrophils from homozygous FcαR-S248 individuals. In the absence of FcR γ-chain pairing, FcαR-S248 allele fails to induce pro-inflammatory cytokines. In contrast, FcαR-G248 maintains signaling capacity even without the FcRγ, producing both IL-6 and TNFα. The increased activity of the G248 form may reflect, at least in part, its enhanced association with the Src family kinase, Lyn (Wu et al. 2007).
FcεRI (FCER1A/B/G)
The high affinity Fc receptor for IgE, FcεRI, has SNPs in the promoter region of the receptor α-chain (FCER1A). Through mutational screening of the proximal promoter, −95T > C (also referred to as −66) and −344C > T (also referred as –335) SNPs have been identified in several ethnicities (Shikanai et al. 1985; Hasegawa et al. 2003; Potaczek et al. 2006). Functionally, the −95T allele has greater GATA-1 binding, increased transcription of FCER1A message, and enhanced FcεRI protein expression on mast cells compared to the −95C allele (Hasegawa et al. 2003; Nishiyama 2006). Similarly, the −344C to T transition increases the binding of Myc-associated zinc finger (MAZ) transcription factors, resulting in increased protein expression (Kim et al. 2006; Bae et al. 2007). Furthermore, these two SNPs affect proximal promoter activity in an additive manner, with the highest activity attributed to the −95T-344T haplotype (Kanada et al. 2008).
The other two subunits of the IgE receptor, the FcεRIγ and FcεRIβ, have also been screened for genetic variations. Although the FCER1G gene is highly conserved (Wu et al. 2002), the FCER1B gene (also named MS4A2) contains several SNPs in the promoter region. The −426C-654T haplotype has higher binding of Yin-Yang 1 and higher transcription activity relative to the −426T-654C haplotype (Nishiyama et al. 2004).
The low affinity receptor for IgE, FcεRII (CD23), carries a functional SNP at position 62 in exon 4, resulting in an arginine (R) to tryptophan (W) substitution. The less common W62 allele is resistant to proteolytic shedding while the common R62 allele is known to be cleaved by a wide range of proteases and shed from cell surface (Meng et al. 2007). Soluble FcεRII has mitogenic properties, promoting the survival and differentiation of germinal center B cells (Liu et al. 1991). In vitro experiments have also suggested that the R62 W SNP affects IgE production through affecting Erk phosphorylation, which results in altered B cell responsiveness to IL-4 (Chan et al. 2014).
FcRLs
The FCRL genes encoded at the autoimmunity-linked 1q23 locus are highly polymorphic with SNPs and many mRNA splice isoforms identified for each gene locus. However, proteins corresponding to most of the splice isoforms have not been identified (Davis et al. 2002). Numerous SNPs have been identified within the FCRL coding regions, introns, the upstream promoter and the downstream noncoding regions. With the exception of the FCRL3 −T169C promoter SNP (rs7528684), which alters an NF-kB binding site and results in increased expression of the FCRL3 mRNA and protein in PBMC, CD19 + B cells and CD8 + T cells subsets (Kochi et al. 2005; Gibson et al. 2009; Chu et al. 2011), little is known about functional correlates in FCRL family SNPs. Nevertheless, many studies have identified association between autoimmune disease and genetic variation in FCRL genes suggesting an important role in disease. Several case-control studies of FCRL3 polymorphisms in autoimmunity are summarized in a recent review (Chistiakov and Chistiakov 2007).
FcRn (FCGRT)
Although no common functional SNPs have been identified to date in FCGRT, the gene that encodes the neonatal Fc receptor, FcRn, a variable number of tandem repeats (VNTR) region in the promoter region consists of one to five repeats of a 37-bp motif (VNTR1-VNTR5) (Sachs et al. 2006). VNTR3 is the most common allele in Caucasian and Asian populations, followed by VNTR2. In vitro experiments have shown that VNTR3 has stronger transcriptional activity compared to VNTR2, resulting in more FcRn expression. Under acidic conditions, monocytes homozygous for VNTR3 showed increased IgG binding capacity compared to monocytes derived from VNTR2/VNTR3 heterozygous individuals (Sachs et al. 2006).
Copy Number Variations (CNVs)
Allotyping individual for the NA1 and NA2 alleles of FCGR3B led to the earliest observed copy number variation (CNV) in the classical low affinity FCGR cluster. Lack of both alleles identified FCGR3B deficiency (Clark et al. 1990; Huizinga et al. 1990), and duplication of the gene was inferred when all three alleles of FCGR3B (NA1, NA2 and SH) were simultaneously detected in the same individual (Koene et al. 1998). Copy number variation of FCGR3B correlates with the expression level of FcγRIIIb and with the capacity of neutrophils to phagocytose immune complexes (Willcocks et al. 2008).
CNV has also been reported for FCGR2C and FCGR3A. Because FCGR2C and FCGR3B are adjacent in the genome (Fig. 1), CNV of both genes is highly correlated (de Haas et al. 1995; Reilly et al. 1994). Copy number of the FCGR2C-ORF allele correlates with FcγRIIc expression levels and consequently, activation status of NK cells (Breunis et al. 2008) and B cells (Li et al. 2013). Similarly, CNV of FCGR3A correlates with FcγRIIIa expression on NK cells (Breunis et al. 2009).
Fig. 1.
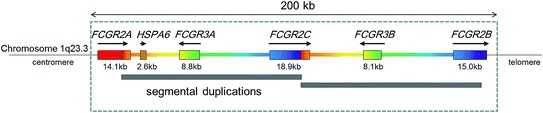
Genomic structure of the classical low-affinity FCGR cluster. Identical colors represent sequence homology. Figure adapted from Li et al. (2009)
Human FcR Polymorphisms: Association with diseases
The central role of Fc receptors in supporting an appropriate humoral immune system has been demonstrated by numerous ex vivo and in vivo studies, in both human and model animals. Often one allele enhances activation and/or net immune system activity while the second allele tends to be less effective in eliciting responses, such as clearance and processing of immune complexes or antibody opsonized particles. Thus, functional FcR polymorphisms may significantly influence effector cell functions, thus providing diversity in host responses pertinent to many infectious, inflammatory and autoimmune diseases. For many SNPs, however, especially when they are in noncoding regions, the direct impact on biological function is not known and the potential influence on pathophysiology is ambiguous. An understanding of these associations and their implications for disease processes awaits further insight into the pertinent genomic architecture of the overall immune response.
Infectious Diseases
Infection with Encapsulated Bacteria
Often working in synergy with the complement system, FcγR-mediated clearance of antibody-coated microbes and FcγR-triggered inflammatory cytokine release are important mechanisms in eliminating infectious agents. Since human IgG2 is relatively inefficient in initiating the complement cascade, the FcγRIIa-131H allele is the primary leukocyte receptor capable of effectively clearing IgG2-coated microbes, which is important in host defense against encapsulated bacteria such as Streptococcus pneumonia, Hemophilus influenza, and Neisseria meningitidis (Bredius et al. 1993; Jefferis and Kumararatne 1990; Endeman et al. 2009; Platonov et al. 1998; Jansen et al. 1999). In the context of Strep pneumonia pneumonia, the FcγRIIa-131R allele, which fails to bind IgG2, may be over-represented in bacteremic patients, and in one study, the most severely infected bacteremic patients, who died within 1 week of hospitalization, were all homozygous for the R131 allele (Yee et al. 2000). Similarly, the FcγRIIa-131R allele is associated with increased infection by Hemophilus influenza and Neisseria meningitidis in multiple bacterial respiratory diseases and sepsis (Endeman et al. 2009; Platonov et al. 1998; Sanders et al. 1994; Bredius et al. 1994; Yuan et al. 2005). Of note, FcγRIIa also binds C-reactive protein with allele sensitivity reciprocal to IgG2 (Stein et al. 2000). High levels of CRP during infection may contribute to the clearance of IgG2-coated microbes by the R131 allele by opsonizing encapsulated bacteria and subsequently activating the complement mediated clearance (Weiser et al. 1998), which may compensate, at least in part, for the lack of FcγR-IgG2 mediated clearance in patients with the R131 allele.
Periodontitis
Periodontitis, an infectious disease caused by pathogenic anaerobic bacteria in the periodontium and the corresponding host response, is influenced by a combination of behavioral, environmental and genetic factors. Several types of FcγR-bearing cells are found in periodontal tissues, including neutrophils, lymphocytes and dendritic cells (Yuan et al. 1999). Functional studies largely focused on neutrophils have demonstrated that neutrophils homozygous for the FcγRIIa-131H allele were more efficient in bacterial phagocytosis, degranulation and elastase release (Nicu et al. 2007). In the same study the homozygous FcγRIIa-H131 patients also showed more bone loss than those with the H/R or R/R allotypes. Kobayashi et al. has also reported that neutrophils carrying the FcγRIIIb-NA2 allele showed lower reactivity to IgG1/IgG3 coated periodontopathic bacteria and induced weaker oxidative burst (Kobayashi et al. 2000).
Association studies calculating the clinical relevance of FcγR polymorphisms in periodontitis have reported mixed results, complicated by the difference in size and ethnicity of the population studied and the inconsistent definitions of disease stage and progression. A recent meta-analysis aggregating 17 studies reported modest association of FcγRIIa-131R with aggressive periodontitis in Asians, relatively strong association of the FcγRIIIb-NA1/NA2 polymorphism with both aggressive and chronic periodontitis, and a statistically insignificant relationship between the FcγRIIIa-F158 V and periodontitis (Song and Lee 2013). In studies of the distribution of the inhibitory FcγRIIb variants, significant enrichment of the FcγRIIb-232T allele in patients with aggressive periodontitis compared to both chronic periodontitis patient and healthy control groups occurs in Japanese periodontitis patients (Yasuda et al. 2003). Furthermore, the composite genotype of FcγRIIb-232T plus FcγRIIIb-NA2 was strongly associated with aggressive periodontitis. The large number of B cells (Yuan et al. 1999) and the elevated antibody level (Horino et al. 1989) in periodontal lesions, as well as our understanding of the biology of the FcγRIIb-232T allele make the link between FcγRIIb-232T and periodontitis biologically plausible.
Besides the well-known polymorphisms, several other SNPs in the FcγR cluster have been identified in association with periodontitis. For example, the FCGR2B-nt645 + 25A/G (rs2125685) SNP in intron 4 was reported in Japanese patients and was related to changes in receptor expression level and severity of periodontitis (Sugita et al. 2012). A little studied SNP in FCGR3A (rs445509) was associated with chronic periodontitis in a Chinese population (Chai et al. 2010). Further study of these variants may elucidate their function and contribution to disease.
Virus Infection
Variants influencing Fc receptor function are also relevant in host defense mechanisms for virus infections. Dengue virus may co-opt Fcγ receptors for cell entry when the antibody-opsonized virus particles are phagocytized by FcγR-bearing myeloid cells, establishing infection in the phagocytes (Moi et al. 2010; Littaua et al. 1990; Garcia et al. 2011). Several studies have suggested the FcγRIIa-R131 allele may have a protective effect in Dengue virus infection (Loke et al. 2002; Garcia et al. 2010). The FcγRIIa-R131H SNP is one important factor in host defense, as it is also reported to be relevant in infections with A/H1N1 influenza (Zuniga et al. 2012), severe acute respiratory syndrome (SARS)- coronavirus (Yuan et al. 2005), and Epstein–Barr virus (Diamantopoulos et al. 2013). In human immunodeficiency virus (HIV) infection, patients with homozygous low affinity R131 allele showed the highest rate of disease progress (Forthal et al. 2007). The FcγRIIIa-V158F genotype also correlates with the development of Kaposi’s sarcoma in HIV-infected patients (Forthal et al. 2007; Lehrnbecher et al. 2000).
Inflammatory and Autoimmune Diseases
Vasculitides
The vasculitides are a group of disorders that involve inflammation of the blood vessels. Although the etiology of vasculitis is often not clear, vascular inflammation can be immunologically mediated, triggered by immune complexes, anti-neutrophil cytoplasmic antibodies, anti-endothelial cell autoantibodies as well as by cell-mediated processes. The classification of the vasculitides is typically based on the size of the affected vessel. Granulomatosis with polyangiitis (GPA), formerly known as Wegener’s granulomatosis, is a type of neutrophil mediated vasculitis affecting small and medium sized vessels. GPA is often characterized by the presence of anti-neutrophil cytoplasmic antibodies (ANCA) (Nolle et al. 1989). Engagement of both ANCA target and Fc receptors on myeloid cells by ANCA elicits production of interleukin-8, a neutrophil chemotactic factor, and a series of effector programs such as oxidative burst, degranulation and release of neutrophil extracellular traps (NETs) (Ralston et al. 1997; Porges et al. 1994; Kessenbrock et al. 2009; Sangaletti et al. 2012). No clear association between GPA susceptibility and the FcγRIIa allotype has been demonstrated although some evidence suggests a relationship to the likelihood of relapsing disease (Edberg et al. 1997; Tse et al. 1999, 2000). FcγRIIIb, the numerically predominant FcγR on neutrophils, is the major receptor interacting with anti-PR3 IgG ANCA (Kocher et al. 1998), and FCGR3B CNV has been associated with GPA (Fanciulli et al. 2007). The FcγRIIIb-NA1 allele, known to induce stronger neutrophil activation than the NA2 allele (Salmon et al. 1990), has similar allele frequencies in GPA and healthy populations, suggesting no role in overall disease risk. However, the presence of the NA1 allele is associated with the development of severe renal damage in GPA patients (Neira et al. 1996; Kelley et al. 2011).
The recent identification of IgA ANCA in GPA, in addition to IgG ANCA, led to the investigation of the involvement of FcαRI in GPA pathogenesis. Indeed, the FcαRI-248G variant, which induces an augmented inflammatory response to IgA, was associated with overall susceptibility to GPA, as well as predisposition to severe renal disease (Kelley et al. 2011).
Kawasaki disease affects medium-sized blood vessels most commonly in children under 5 years of age. Genome wide association studies have identified an association between Kawasaki disease and the FCGR2A locus with the 131H variant conferring elevated disease risk (Shrestha et al. 2012; Onouchi et al. 2012). It is reasonable to speculate the FcγRIIa-131H bearing leukocytes are more pro-inflammatory in the setting of Kawasaki disease, although direct experimental evidence waits to be established. One might also anticipate an association between IgG receptor variants and intravenous immunoglobulin (IVIG), the only proven therapy for Kawasaki disease. Indeed, in Japanese patients, those with the FcγRIIa-131H allele responded more efficiently to IVIG administration. Patients with the 131R allele were more likely to develop coronary lesions even after treatment (Taniuchi et al. 2005). Consistent with the notion that tilting the immune system towards inflammation might be associated with disease expression, the FCGR2C-ORF SNP was recently reported to be enriched in Kawasaki disease patients (Breunis et al. 2013).
Takayasu’s arteritis is a rare form of large vessel vasculitis. A recent GWAS in Turkish and North American Takayasu’s arteritis patients identified a noncoding SNP in the FCGR2A/FCGR3A locus (rs10919543) as a susceptibility marker, which appeared to have a regulatory effect on FCGR2A transcript expression (Saruhan-Direskeneli et al. 2013).
Several other forms of chronic inflammatory diseases have been reported to have associations with the FCGR cluster. The FCGR2A/2C region has been related to susceptibility to ulcerative colitis, one sub-phenotype of inflammatory bowel disease, in two GWA studies (McGovern et al. 2010; Asano et al. 2013). In addition to the well-known FcγR-R131H variant, the rs10800309 variant in this locus awaits further work to determine potential functional relevance.
Systemic Lupus Erythematosus
Systemic Lupus Erythematosus (SLE) is an autoimmune disease characterized by autoantibodies and immune complexes. Although the etiology of SLE is unknown, many genes play a role in the susceptibility to and severity of the disease, and GWAS and candidate genes studies have identified the FCGRs as important contributors to the SLE diathesis (Harley et al. 2009).
A GWAS study of Europeans confirmed the association of FCGR2A (rs1801274; 519G > A encoding R131H) with SLE (International Consortium for Systemic Lupus Erythematosus 2008). This nonsynonymous SNP is a risk factor for lupus nephritis and systemic lupus erythematosus in African Americans (Salmon et al. 1996; Edberg et al. 2002), Caucasians (Manger et al. 2002; Karassa et al. 2002; Magnusson et al. 2004; Kyogoku et al. 2004) and Asians (Siriboonrit et al. 2003; Lee et al. 2002; Chu et al. 2004), as well as for myasthenia gravis in Caucasians (Weersma et al. 2010; van der Pol et al. 2003). Homozygosity for the transmembrane 187T variant of FcγRIIb is also associated with SLE susceptibility in Japanese (Kyogoku et al. 2002), Chinese (Chu et al. 2004) and Thais (Siriboonrit et al. 2003). Interestingly, the 187T allele has a lower frequency in European Americans and is not associated with SLE in either this ancestry group or in African Americans where the frequency of 187T is similar to that of Asians (Li et al. 2003; Magnusson et al. 2004). Whether this difference represents, less statistical power for detection of association in these groups or an epistatic effect is not certain. The FcγRIIb-187T allele may be a risk factor for anti-GBM disease in Chinese (Zhou et al. 2010) while a promoter haplotype, 2B.4 (−386C −120A), which alters FCGR2B gene expression is associated with SLE (Su et al. 2004). In a second patient population, homozygosity of the −386C allele alone (also referred to as −343C) affirmed an association of promoter variants with SLE (Blank et al. 2005). CNV in this receptor cluster, including the FCGR2C-ORF allele, may be associated with SLE in patients of European and African ancestry (Li et al. 2013).
The low IgG binding FcγRIIIa-158F is associated with SLE and with lupus nephritis (Wu et al. 1997; Karassa et al. 2002; Jonsen et al. 2007; Dong et al. 2013) in multiple ancestry groups including Europeans, African Americans (Edberg et al. 2002; Koene et al. 1998), Chinese (Chu et al. 2004), and Japanese (Kyogoku et al. 2002). Interestingly, homozygosity for the high IgG binding –158 V allele is a significant predictor of end-stage renal disease in a multiethnic group of SLE patients (Alarcon et al. 2006). Both FcγRIIIb CNV and NA1/NA2 alleles may be associated with SLE in UK Caucasians (Willcocks et al. 2008), Thais (Siriboonrit et al. 2003), Japanese (Hatta et al. 1999), and Spanish (Gonzalez-Escribano et al. 2002).
The −169C > T SNP (rs7528684) in FCRL3, which alters an NFκB binding site and is associated with FCRL3 mRNA and surface protein expression, is associated with autoimmunity in some ethnic groups. Associated with SLE, RA, and AITD in Japanese (Kochi et al. 2005; Gibson et al. 2009), this variant is not associated with these conditions in other ethnicities suggesting that it is not a general autoimmunity risk factor (Chistiakov and Chistiakov 2007). The −169C > T SNP is not associated with SLE in Chinese (You et al. 2008), Koreans (Choi et al. 2006), or Mexican patients with childhood-onset SLE (Ramirez-Bello et al. 2013), but the association with the presence of autoantibodies in Polish SLE patients suggests a possible role in production of autoantibodies (Piotrowski et al. 2013). Results of meta-analyses differ on whether the −169C > T is associated with SLE in different ethnicities (Breunis et al. 2013; Mao et al. 2010; Song et al. 2013), and the mechanism(s) through which this variant may contribute to SLE remains unclear.
Rheumatoid Arthritis and Juvenile Idiopathic Arthritis
Evidence for the contributions of the classical low-affinity Fcγ receptors to Rheumatoid Arthritis suggests that several polymorphisms may be associated with RA manifestations in different ethnic groups, although associations are not always consistent. While GWAS indicated that FCGR2A is associated with RA (Raychaudhuri et al. 2009), candidate gene studies suggest the FCGR3A is associated with RA (Morgan et al. 2000; Morgan et al. 2003) and a role for FCGR2C is unclear.
The −169C > T promoter SNP in FCRL3 is associated with RA in Caucasians and Chinese (Thabet et al. 2007; Eike et al. 2008; Maehlen et al. 2011; Wu et al. 2010), with JIA in Mexicans (Ramirez-Bello et al. 2013), and with JIA in Norwegian patients (Eike et al. 2008). This SNP has been correlated with increased FCRL3 surface expression on Tregs of patients with erosive RA (Bajpai et al. 2012), and the −169CC genotype may be correlated with radiographic severity in Korean RA patients (Han et al. 2012). A more detailed review of Fc receptor associations and reheumatoid arthritis is discussed in Chapter XX.
Spondyloarthropathies
The rs2777963T > C, rs14335A > G and rs10489674C > T polymorphisms in FCRL4 have been associated with susceptibility and severity of ankylosing spondylitis (AS) in Han Chinese (Zeng et al. 2012). Similarly, in FCRL5 two nonsynonymous SNPs, rs12036228C > T and rs6427384T > C in exon 5 and exon 7, respectively, and their C-T haplotype were found to be associated with ankylosing spondylitis in HLA-B27 positive Han Chinese, suggesting a role in AS (Tang et al. 2009). However, the role, if any, of these SNPs in FCRL4 and 5 expression or function is unclear.
Diabetes Mellitus and Autoimmune Endocrinopathies
Several studies have found association between autoimmune endocrinopathies and SNPs in FCRL family members, although potential underlying mechanisms remain elusive. In a recent study of Type 1 Diabetes (T1D) the C-allele of FCRL1 rs4971154 was strongly associated with the presence of the IA-2A autoantibody in serum suggesting a role in production of autoantibodies (Mao et al. 2010). Although the FCRL3 −169C > T SNP was not associated with T1D in several studies of Caucasians (Eike et al. 2008; Owen et al. 2007; Duchatelet et al. 2008), a recent study of 8,506 T1D patients in the United Kingdom found a strong negative association between the C allele and anti-IA-2A autoantibody- positive T1D (Mao et al. 2010). The mechanism of association remains unclear.
In autoimmune thyroid disease, Owen et al. found modest association of the 3’UTR C > A SNP rs2282288 with Grave’s Disease in Europeans (Owen et al. 2007). The −169TT promoter genotype of rs7528684 was associated with remission in Japanese AITD patients (Inoue et al. 2012), and with protection against Grave’s Disease in Chinese (Gu et al. 2010). A potential role for FCRL3 in production of autoantibodies is supported by the observations that the rs11264798C > G and rs7528684C > T SNPs are associated with thyroid peroxidase autoantibody (TPOA) positivity in GD and anti- IA-2A positivity in T1D (Plagnol et al. 2011), while the rs7522061T > C SNP is associated with anti-876 ZnT8A positivity (autoantibody to the zinc transporter 8 in islet cells) in T1D patients (Howson et al. 2012).
Multiple Sclerosis
The FCRL3 −169C > T SNP (rs7528684) has been associated with multiple sclerosis in a Spanish cohort (Martinez et al. 2007; Matesanz et al. 2008). While the T allele of the nonsynonymous coding SNP (rs7522061), which results in the N28D change, was found to be protective in Spanish, the G allele was a risk factor for MS in patients in the United Kingdom (Matesanz et al. 2008).
Inflammatory Bowel Disease
Despite its association with many autoimmune disorders in different ethnicities, the −169C > T SNP appears not to be associated with risk for ulcerative colitis, Crohn’s disease or primary sclerosing cholangitis (Eike et al. 2008), or with Inflammatory Bowel Disease (Martinez et al. 2007).
Allergic Diseases
Allergic diseases are a type of hypersensitivity characterized by mast cell activation and IgE-mediated inflammation. The high-affinity IgE receptor expressed on mast cells, FcεRI, has long been considered a candidate gene in allergic diseases. Multiple studies have established a consistent genetic association between allergies and the promoter variants of FcεRI α-chain. The −66T > C and/or the −315C > T SNPs are associated with atopic dermatitis, chronic urticaria, asthma, and high serum IgE levels (Hasegawa et al. 2003; Potaczek et al. 2006; Kim et al. 2006; Bae et al. 2007; Zhou et al. 2012; Niwa et al. 2010). The −66T > C SNP was highlighted as the strongest hit in two GWA studies with high IgE levels (Weidinger et al. 2008; Granada et al. 2012). These genetic findings may be explained by functional studies that have demonstrated that both SNPs amplify transcription activity, increasing FcεRI expression on mast cells and basophils (Hasegawa et al. 2003; Kanada et al. 2008), and the well-established observation that surface FcεRI expression correlates positively with circulating IgE levels (MacGlashan 2005). Similarly, several SNPs in the FcεRI β-chain are associated with allergic inflammatory diseases such as atopy, asthma, and nasal allergy (Nishiyama et al. 2004; Zhang et al. 2004; Laprise et al. 2000; Nagata et al. 2001; Li and Hopkin 1997; Hizawa et al. 2000; Kim et al. 2006, 2007; Yang et al. 2014). Functional properties of these SNPs are not known.
The low-affinity IgE receptor on B cells, FcεRII (CD23), is important in regulating IgE production and B cell differentiation. The R62W alteration in the FCER2 gene, that yields increased IgE binding and augmented ERK signaling (Chan et al. 2014), is associated with elevated serum IgE levels and an increased risk of severe asthma exacerbation in children (Laitinen et al. 2000; Koster et al. 2011; Tantisira et al. 2007). A promoter SNP in the FCER2 gene, rs3760687, associated with increased total serum IgE (Sharma et al. 2014), may alter the activity of the transcription factors Sp1 and Sp3, leading to modulation of FcεRII expression (Potaczek et al. 2009).
Even though IgE and IgE receptors have been known to be the major players in allergic inflammation, allergen-specific IgG and FcγRs also play a role (Kaneko et al. 1995; Jonsson et al. 2012; Williams et al. 2012; Lau et al. 2005; Bruhns et al. 2005). In a candidate gene study, both the FcγRIIa-R131H and the FcγRIIb-I187T SNPs have been associated with atopy (Wu et al 2014). In this context, it is conceivable that FcγRIIa-H131 allele may clear allergen-IgG2 immune complexes more efficiently, preventing inflammation and tissue damage. Whether allergen-specific IgG2 levels vary in accordance with FcγRIIa polymorphisms is unknown. Furthermore, the FcγRIIb-187T allele may not be as effective in negatively regulating BCR function, resulting in increased B cell IgE production. Crosstalk between FcγRIIb and FcεRI on mast cells is also a possibility.
Association with Response to Antibody Therapy
The efficacy of therapeutic monoclonal antibodies used in autoimmune diseases to induce ADCC and deplete autoreactive B lymphocytes from circulation depends, at least in part, on the strength of the interaction of activating FcγRs with the therapeutic antibody on the opsonized target cells. The FcγRIIIa -158F/V polymorphism influences the efficacy of rituximab treatment, which targets the CD20 surface protein on B cells, with patients homozygous for the high binding -158 V allele showing the best response (Robledo et al. 2012; Cooper et al. 2012). The precedent that alleles which alter binding and function of FcγRIIa and FcγRIIIa may affect the efficacy of antibody therapy is an important principle in antibody-based therapeutics. A more extensive discussion of the role of Fc receptors in the use of therapeutic antibodies is presented in Chapter XX, “FcR and therapeutic antibodies”.
Conclusions
Genetic variations in human Fc receptors, through their impact on antibody-mediated mechanisms, contribute to individual and population-based host defense and susceptibility to a range of human diseases. Fc receptor polymorphisms modulate the effectiveness of immune system in defense against invading pathogens by regulating immune cell activities. They also impact the handling of immune reactants and the threshold of immune tolerance. Complex clinical phenotypes, such as autoimmunity or allergy, involve multiple genetic and environmental factors, and the subtle regulatory effects of various naturally occurring polymorphisms are compounded in their impact over time. Accurate assessment of the contributions of Fc receptor polymorphisms to immune system function and clinical phenotype requires a careful understanding of the genomic structure, sequence homology, and known physiological responses of Fc receptors in addition to well phenotyped study populations for adequately powered association studies. Such studies have provided important insights into pathogenetic mechanisms and potential novel therapeutic approaches.
Contributor Information
Marc Daeron, Phone: +33 1 45688642, Email: daeron@pasteur.fr.
Falk Nimmerjahn, Phone: +4909131 8525050, Email: falk.nimmerjahn@fau.de.
Robert P. Kimberly, Email: rpk@uab.edu
References
- Salmon JE, et al. Allelic polymorphisms of human Fc gamma receptor IIA and Fc gamma receptor IIIB. Independent mechanisms for differences in human phagocyte function. J Clin Invest. 1992;89(4):1274–1281. doi: 10.1172/JCI115712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, et al. On the interaction of IgG subclasses with the low affinity Fc gamma RIIa (CD32) on human monocytes, neutrophils, and platelets. Analysis of a functional polymorphism to human IgG2. J Clin Invest. 1992;90(4):1537–1546. doi: 10.1172/JCI116022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredius RG, et al. Role of neutrophil Fc gamma RIIa (CD32) and Fc gamma RIIIb (CD16) polymorphic forms in phagocytosis of human IgG1- and IgG3-opsonized bacteria and erythrocytes. Immunology. 1994;83(4):624–630. [PMC free article] [PubMed] [Google Scholar]
- Maxwell KF, et al. Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa. Nat Struct Biol. 1999;6(5):437–442. doi: 10.1038/8241. [DOI] [PubMed] [Google Scholar]
- Bredius RG, et al. Phagocytosis of Staphylococcus aureus and Haemophilus influenzae type B opsonized with polyclonal human IgG1 and IgG2 antibodies. Functional hFc gamma RIIa polymorphism to IgG2. J Immunol. 1993;151(3):1463–1472. [PubMed] [Google Scholar]
- International Consortium for Systemic Lupus Erythematosus, G., et al (2008) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet . 40(2):204–10 [DOI] [PMC free article] [PubMed]
- Saruhan-Direskeneli G, et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am J Hum Genet. 2013;93(2):298–305. doi: 10.1016/j.ajhg.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41(12):1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42(4):332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. 2009;41(12):1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- Lessard CJ, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren’s syndrome. Nat Genet. 2013;45(11):1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku C, et al. Fcgamma receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: contribution of FCGR2B to genetic susceptibility. Arthritis Rheum. 2002;46(5):1242–1254. doi: 10.1002/art.10257. [DOI] [PubMed] [Google Scholar]
- Li X, et al. A novel polymorphism in the Fcgamma receptor IIB (CD32B) transmembrane region alters receptor signaling. Arthritis Rheum. 2003;48(11):3242–3252. doi: 10.1002/art.11313. [DOI] [PubMed] [Google Scholar]
- Kono H, et al. FcgammaRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum Mol Genet. 2005;14(19):2881–2892. doi: 10.1093/hmg/ddi320. [DOI] [PubMed] [Google Scholar]
- Floto RA, et al. Loss of function of a lupus-associated FcgammaRIIb polymorphism through exclusion from lipid rafts. Nat Med. 2005;11(10):1056–1058. doi: 10.1038/nm1288. [DOI] [PubMed] [Google Scholar]
- Su K et al (2004a) A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcgammaRIIb alters receptor expression and associates with autoimmunity. I. Regulatory FCGR2B polymorphisms and their association with systemic lupus erythematosus. J Immunol 172(11):7186–7191 [DOI] [PubMed]
- Su K et al (2004b) A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcgammaRIIb alters receptor expression and associates with autoimmunity. II. Differential binding of GATA4 and Yin-Yang1 transcription factors and correlated receptor expression and function. J Immunol 172(11):7192–7199 [DOI] [PubMed]
- Blank MC, et al. Decreased transcription of the human FCGR2B gene mediated by the -343 G/C promoter polymorphism and association with systemic lupus erythematosus. Hum Genet. 2005;117(2–3):220–227. doi: 10.1007/s00439-005-1302-3. [DOI] [PubMed] [Google Scholar]
- Metes D, et al. Ligand binding specificities and signal transduction pathways of Fc gamma receptor IIc isoforms: the CD32 isoforms expressed by human NK cells. Eur J Immunol. 1999;29(9):2842–2852. doi: 10.1002/(SICI)1521-4141(199909)29:09<2842::AID-IMMU2842>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Stewart-Akers AM, et al. Fc gamma R expression on NK cells influences disease severity in rheumatoid arthritis. Genes Immun. 2004;5(7):521–529. doi: 10.1038/sj.gene.6364121. [DOI] [PubMed] [Google Scholar]
- Metes D, et al. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood. 1998;91(7):2369–2380. [PubMed] [Google Scholar]
- Li X et al (2013) Allelic-dependent expression of an activating fc receptor on B cells enhances humoral immune responses. Sci Transl Med 5(216):216ra175 [DOI] [PMC free article] [PubMed]
- Ernst LK, et al. Allelic polymorphisms in the FcgammaRIIC gene can influence its function on normal human natural killer cells. J Mol Med (Berl) 2002;80(4):248–257. doi: 10.1007/s00109-001-0294-2. [DOI] [PubMed] [Google Scholar]
- Breunis WB, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111(3):1029–1038. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest. 1997;100(5):1059–1070. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koene HR, et al. Fc gammaRIIIa-158 V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood. 1997;90(3):1109–1114. [PubMed] [Google Scholar]
- Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med. 1989;170(2):481–497. doi: 10.1084/jem.170.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon JE, Edberg JC, Kimberly RP. Fc gamma receptor III on human neutrophils. Allelic variants have functionally distinct capacities. J Clin Invest. 1990;85(4):1287–1295. doi: 10.1172/JCI114566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bux J, et al. Characterization of a new alloantigen (SH) on the human neutrophil Fc gamma receptor IIIb. Blood. 1997;89(3):1027–1034. [PubMed] [Google Scholar]
- Jasek M, et al. A novel polymorphism in the cytoplasmic region of the human immunoglobulin A Fc receptor gene. Eur J Immunogenet. 2004;31(2):59–62. doi: 10.1111/j.1365-2370.2004.00445.x. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. FcalphaRI (CD89) alleles determine the proinflammatory potential of serum IgA. J Immunol. 2007;178(6):3973–3982. doi: 10.4049/jimmunol.178.6.3973. [DOI] [PubMed] [Google Scholar]
- Shikanai T et al (1985, 2002) Sequence variants in the FcepsilonRI alpha chain gene. J Appl Physiol 93(1):37–41 [DOI] [PubMed]
- Hasegawa M, et al. A novel -66T/C polymorphism in Fc epsilon RI alpha-chain promoter affecting the transcription activity: possible relationship to allergic diseases. J Immunol. 2003;171(4):1927–1933. doi: 10.4049/jimmunol.171.4.1927. [DOI] [PubMed] [Google Scholar]
- Potaczek DP, et al. The alpha-chain of high-affinity receptor for IgE (FcepsilonRIalpha) gene polymorphisms and serum IgE levels. Allergy. 2006;61(10):1230–1233. doi: 10.1111/j.1398-9995.2006.01195.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama C. Molecular mechanism of allergy-related gene regulation and hematopoietic cell development by transcription factors. Biosci Biotechnol Biochem. 2006;70(1):1–9. doi: 10.1271/bbb.70.1. [DOI] [PubMed] [Google Scholar]
- Kim SH, et al. Genetic mechanism of aspirin-induced urticaria/angioedema. Curr Opin Allergy Clin Immunol. 2006;6(4):266–270. doi: 10.1097/01.all.0000235899.57182.d4. [DOI] [PubMed] [Google Scholar]
- Bae JS, et al. Significant association of FcepsilonRIalpha promoter polymorphisms with aspirin-intolerant chronic urticaria. J Allergy Clin Immunol. 2007;119(2):449–456. doi: 10.1016/j.jaci.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Kanada S, et al. Two different transcription factors discriminate the -315C > T polymorphism of the Fc epsilon RI alpha gene: binding of Sp1 to -315C and of a high mobility group-related molecule to -315T. J Immunol. 2008;180(12):8204–8210. doi: 10.4049/jimmunol.180.12.8204. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. Conservation of FcepsilonRI gamma chain coding region in normals and in SLE patients. Lupus. 2002;11(1):42–45. doi: 10.1191/0961203302lu145oa. [DOI] [PubMed] [Google Scholar]
- Nishiyama C, et al. Polymorphisms in the Fc epsilon RI beta promoter region affecting transcription activity: a possible promoter-dependent mechanism for association between Fc epsilon RI beta and atopy. J Immunol. 2004;173(10):6458–6464. doi: 10.4049/jimmunol.173.10.6458. [DOI] [PubMed] [Google Scholar]
- Meng JF, McFall C, Rosenwasser LJ. Polymorphism R62 W results in resistance of CD23 to enzymatic cleavage in cultured cells. Genes Immun. 2007;8(3):215–223. doi: 10.1038/sj.gene.6364376. [DOI] [PubMed] [Google Scholar]
- Liu YJ, et al. Recombinant 25-kDa CD23 and interleukin 1 alpha promote the survival of germinal center B cells: evidence for bifurcation in the development of centrocytes rescued from apoptosis. Eur J Immunol. 1991;21(5):1107–1114. doi: 10.1002/eji.1830210504. [DOI] [PubMed] [Google Scholar]
- Chan MA, et al. FCER2 (CD23) Asthma-Related Single Nucleotide Polymorphisms Yields Increased IgE Binding and Egr-1 Expression in Human B Cells. Am J Respir Cell Mol Biol. 2014;50(2):263–269. doi: 10.1165/rcmb.2013-0112OC. [DOI] [PubMed] [Google Scholar]
- Davis RS, et al. Fc receptor homologs: newest members of a remarkably diverse Fc receptor gene family. Immunol Rev. 2002;190:123–136. doi: 10.1034/j.1600-065x.2002.19009.x. [DOI] [PubMed] [Google Scholar]
- Kochi Y, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37(5):478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AW, et al. The FCRL3-169CT promoter single-nucleotide polymorphism, which is associated with systemic lupus erythematosus in a Japanese population, predicts expression of receptor protein on CD19 + B cells. Arthritis Rheum. 2009;60(11):3510–3512. doi: 10.1002/art.24915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat Genet. 2011;43(9):897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Chistiakov AP. Is FCRL3 a new general autoimmunity gene? Hum Immunol. 2007;68(5):375–383. doi: 10.1016/j.humimm.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Sachs UJ, et al. A variable number of tandem repeats polymorphism influences the transcriptional activity of the neonatal Fc receptor alpha-chain promoter. Immunology. 2006;119(1):83–89. doi: 10.1111/j.1365-2567.2006.02408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, et al. An abnormality of the gene that encodes neutrophil Fc receptor III in a patient with systemic lupus erythematosus. J Clin Invest. 1990;86(1):341–346. doi: 10.1172/JCI114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga TW, et al. Maternal genomic neutrophil FcRIII deficiency leading to neonatal isoimmune neutropenia. Blood. 1990;76(10):1927–1932. [PubMed] [Google Scholar]
- Koene HR, et al. Fc gamma RIIIB gene duplication: evidence for presence and expression of three distinct Fc gamma RIIIB genes in NA(1 + ,2 +)SH(+) individuals. Blood. 1998;91(2):673–679. [PubMed] [Google Scholar]
- Willcocks LC, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205(7):1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas M, et al. Neutrophil Fc gamma RIIIb deficiency, nature, and clinical consequences: a study of 21 individuals from 14 families. Blood. 1995;86(6):2403–2413. [PubMed] [Google Scholar]
- Reilly AF, et al. Variation in human FCGR2C gene copy number. Immunogenetics. 1994;40(6):456. doi: 10.1007/BF00177829. [DOI] [PubMed] [Google Scholar]
- Breunis WB, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat. 2009;30(5):E640–E650. doi: 10.1002/humu.20997. [DOI] [PubMed] [Google Scholar]
- Li X, et al. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes Immun. 2009;10(5):380–389. doi: 10.1038/gene.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R, Kumararatne DS. Selective IgG subclass deficiency: quantification and clinical relevance. Clin Exp Immunol. 1990;81(3):357–367. doi: 10.1111/j.1365-2249.1990.tb05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endeman H, et al. The Fcgamma receptor IIA-R/R131 genotype is associated with severe sepsis in community-acquired pneumonia. Clin Vaccine Immunol. 2009;16(7):1087–1090. doi: 10.1128/CVI.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platonov AE, et al. Association of human Fc gamma RIIa (CD32) polymorphism with susceptibility to and severity of meningococcal disease. Clin Infect Dis. 1998;27(4):746–750. doi: 10.1086/514935. [DOI] [PubMed] [Google Scholar]
- Jansen WT, et al. Fcgamma receptor polymorphisms determine the magnitude of in vitro phagocytosis of Streptococcus pneumoniae mediated by pneumococcal conjugate sera. J Infect Dis. 1999;180(3):888–891. doi: 10.1086/314920. [DOI] [PubMed] [Google Scholar]
- Yee AM, et al. Association between FcgammaRIIa-R131 allotype and bacteremic pneumococcal pneumonia. Clin Infect Dis. 2000;30(1):25–28. doi: 10.1086/313588. [DOI] [PubMed] [Google Scholar]
- Sanders LA, et al. Fc gamma receptor IIa (CD32) heterogeneity in patients with recurrent bacterial respiratory tract infections. J Infect Dis. 1994;170(4):854–861. doi: 10.1093/infdis/170.4.854. [DOI] [PubMed] [Google Scholar]
- Bredius RG, et al. Fc gamma receptor IIa (CD32) polymorphism in fulminant meningococcal septic shock in children. J Infect Dis. 1994;170(4):848–853. doi: 10.1093/infdis/170.4.848. [DOI] [PubMed] [Google Scholar]
- Yuan FF, et al. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66(4):291–296. doi: 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MP, et al. C-reactive protein binding to FcgammaRIIa on human monocytes and neutrophils is allele-specific. J Clin Invest. 2000;105(3):369–376. doi: 10.1172/JCI7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser JN, et al. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp Med. 1998;187(4):631–640. doi: 10.1084/jem.187.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZN, et al. Topical distribution of Fc gammaRI, Fc gammaRII and Fc gammaRIII in inflamed human gingiva. J Clin Periodontol. 1999;26(7):441–447. doi: 10.1034/j.1600-051x.1999.260705.x. [DOI] [PubMed] [Google Scholar]
- Nicu EA, et al. Hyper-reactive PMNs in FcgammaRIIa 131 H/H genotype periodontitis patients. J Clin Periodontol. 2007;34(11):938–945. doi: 10.1111/j.1600-051X.2007.01136.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, et al. Relevance of IgG receptor IIIb (CD16) polymorphism to handling of Porphyromonas gingivalis: implications for the pathogenesis of adult periodontitis. J Periodontal Res. 2000;35(2):65–73. doi: 10.1034/j.1600-0765.2000.035002065.x. [DOI] [PubMed] [Google Scholar]
- Song GG, Lee YH. Associations between FCGR2A rs1801274, FCGR3A rs396991, FCGR3B NA1/NA2 polymorphisms and periodontitis: a meta-analysis. Mol Biol Rep. 2013;40(8):4985–4993. doi: 10.1007/s11033-013-2599-y. [DOI] [PubMed] [Google Scholar]
- Yasuda K, et al. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003;4(8):541–546. doi: 10.1038/sj.gene.6364021. [DOI] [PubMed] [Google Scholar]
- Horino K, et al. Effects of anti-Bacteroides gingivalis (Bg), and anti-Actinobacillus actinomycetemcomitans (Aa) antibodies on Bg and Aa. Nihon Shishubyo Gakkai Kaishi. 1989;31(1):1–12. doi: 10.2329/perio.31.1. [DOI] [PubMed] [Google Scholar]
- Sugita N, et al. Association of the FcgammaRIIB-nt645 + 25A/G polymorphism with the expression level of the FcgammaRIIb receptor, the antibody response to Porphyromonas gingivalis and the severity of periodontitis. J Periodontal Res. 2012;47(1):105–113. doi: 10.1111/j.1600-0765.2011.01411.x. [DOI] [PubMed] [Google Scholar]
- Chai L, et al. SNPs of Fc-gamma receptor genes and chronic periodontitis. J Dent Res. 2010;89(7):705–710. doi: 10.1177/0022034510365444. [DOI] [PubMed] [Google Scholar]
- Moi ML, et al. Involvement of the Fc gamma receptor IIA cytoplasmic domain in antibody-dependent enhancement of dengue virus infection. J Gen Virol. 2010;91(Pt 1):103–111. doi: 10.1099/vir.0.014829-0. [DOI] [PubMed] [Google Scholar]
- Littaua R, Kurane I, Ennis FA. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990;144(8):3183–3186. [PubMed] [Google Scholar]
- Garcia G, et al. Long-term persistence of clinical symptoms in dengue-infected persons and its association with immunological disorders. Int J Infect Dis. 2011;15(1):e38–e43. doi: 10.1016/j.ijid.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Loke H, et al. Susceptibility to dengue hemorrhagic fever in vietnam: evidence of an association with variation in the vitamin d receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67(1):102–106. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- Garcia G, et al. Asymptomatic dengue infection in a Cuban population confirms the protective role of the RR variant of the FcgammaRIIa polymorphism. Am J Trop Med Hyg. 2010;82(6):1153–1156. doi: 10.4269/ajtmh.2010.09-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga J, et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J. 2012;39(3):604–610. doi: 10.1183/09031936.00020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantopoulos PT, et al. Correlation of Fc-gamma RIIA polymorphisms with latent Epstein-Barr virus infection and latent membrane protein 1 expression in patients with low grade B-cell lymphomas. Leuk Lymphoma. 2013;54(9):2030–2034. doi: 10.3109/10428194.2012.762512. [DOI] [PubMed] [Google Scholar]
- Forthal DN, et al. FcgammaRIIa genotype predicts progression of HIV infection. J Immunol. 2007;179(11):7916–7923. doi: 10.4049/jimmunol.179.11.7916. [DOI] [PubMed] [Google Scholar]
- Lehrnbecher TL, et al. Variant genotypes of FcgammaRIIIA influence the development of Kaposi’s sarcoma in HIV-infected men. Blood. 2000;95(7):2386–2390. [PubMed] [Google Scholar]
- Nolle B, et al. Anticytoplasmic autoantibodies: their immunodiagnostic value in Wegener granulomatosis. Ann Intern Med. 1989;111(1):28–40. doi: 10.7326/0003-4819-111-1-28. [DOI] [PubMed] [Google Scholar]
- Ralston DR, et al. Antineutrophil cytoplasmic antibodies induce monocyte IL-8 release. Role of surface proteinase-3, alpha1-antitrypsin, and Fcgamma receptors. J Clin Invest. 1997;100(6):1416–1424. doi: 10.1172/JCI119662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porges AJ, et al. Anti-neutrophil cytoplasmic antibodies engage and activate human neutrophils via Fc gamma RIIa. J Immunol. 1994;153(3):1271–1280. [PubMed] [Google Scholar]
- Kessenbrock K, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15(6):623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaletti S, et al. Neutrophil extracellular traps mediate transfer of cytoplasmic neutrophil antigens to myeloid dendritic cells toward ANCA induction and associated autoimmunity. Blood. 2012;120(15):3007–3018. doi: 10.1182/blood-2012-03-416156. [DOI] [PubMed] [Google Scholar]
- Edberg JC, et al. Analysis of FcgammaRII gene polymorphisms in Wegener’s granulomatosis. Exp Clin Immunogenet. 1997;14(3):183–195. [PubMed] [Google Scholar]
- Tse WY, et al. Neutrophil FcgammaRIIIb allelic polymorphism in anti-neutrophil cytoplasmic antibody (ANCA)-positive systemic vasculitis. Clin Exp Immunol. 2000;119(3):574–577. doi: 10.1046/j.1365-2249.2000.01182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse WY, et al. No association between neutrophil FcgammaRIIa allelic polymorphism and anti-neutrophil cytoplasmic antibody (ANCA)-positive systemic vasculitis. Clin Exp Immunol. 1999;117(1):198–205. doi: 10.1046/j.1365-2249.1999.00960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher M, et al. Antineutrophil cytoplasmic antibodies preferentially engage Fc gammaRIIIb on human neutrophils. J Immunol. 1998;161(12):6909–6914. [PubMed] [Google Scholar]
- Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39(6):721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira O, et al. Lyme disease in Chile. Prevalence study in selected groups. Rev Med Chil. 1996;124(5):537–544. [PubMed] [Google Scholar]
- Kelley JM, et al. IgA and IgG antineutrophil cytoplasmic antibody engagement of Fc receptor genetic variants influences granulomatosis with polyangiitis. Proc Natl Acad Sci U S A. 2011;108(51):20736–20741. doi: 10.1073/pnas.1109227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, et al. Role of activating FcgammaR gene polymorphisms in Kawasaki disease susceptibility and intravenous immunoglobulin response. Circ Cardiovasc Genet. 2012;5(3):309–316. doi: 10.1161/CIRCGENETICS.111.962464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi Y, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. 2012;44(5):517–521. doi: 10.1038/ng.2220. [DOI] [PubMed] [Google Scholar]
- Taniuchi S, et al. Polymorphism of Fc gamma RIIa may affect the efficacy of gamma-globulin therapy in Kawasaki disease. J Clin Immunol. 2005;25(4):309–313. doi: 10.1007/s10875-005-4697-7. [DOI] [PubMed] [Google Scholar]
- Breunis WBH, Long T, Eileen Png, Geissler J, Nagelkerke S, Ellis J, Davila S, Chiea Chuen K, Levin M, Burgner D, Shimizu C, Burns JC.; Hibberd ML, Kuijpers TW (2013) Fc-Gamma Receptor Genetic Variation In Kawasaki Disease. In: American college of rheumatology2013, San Diego
- Asano K, et al. Impact of allele copy number of polymorphisms in FCGR3A and FCGR3B genes on susceptibility to ulcerative colitis. Inflamm Bowel Dis. 2013;19(10):2061–2068. doi: 10.1097/MIB.0b013e318298118e. [DOI] [PubMed] [Google Scholar]
- Harley IT, et al. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10(5):285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon JE, et al. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest. 1996;97(5):1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edberg JC, et al. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum. 2002;46(8):2132–2140. doi: 10.1002/art.10438. [DOI] [PubMed] [Google Scholar]
- Manger K, et al. Fcgamma receptor IIa, IIIa, and IIIb polymorphisms in German patients with systemic lupus erythematosus: association with clinical symptoms. Ann Rheum Dis. 2002;61(9):786–792. doi: 10.1136/ard.61.9.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karassa FB, et al. Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum. 2002;46(6):1563–1571. doi: 10.1002/art.10306. [DOI] [PubMed] [Google Scholar]
- Magnusson V, et al. Polymorphisms of the Fc gamma receptor type IIB gene are not associated with systemic lupus erythematosus in the Swedish population. Arthritis Rheum. 2004;50(4):1348–1350. doi: 10.1002/art.20151. [DOI] [PubMed] [Google Scholar]
- Kyogoku C, et al. Association of Fcgamma receptor IIA, but not IIB and IIIA, polymorphisms with systemic lupus erythematosus: A family-based association study in Caucasians. Arthritis Rheum. 2004;50(2):671–673. doi: 10.1002/art.20029. [DOI] [PubMed] [Google Scholar]
- Siriboonrit U, et al. Association of Fcgamma receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens. 2003;61(5):374–383. doi: 10.1034/j.1399-0039.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- Lee EB, et al. Fcgamma receptor IIIA polymorphism in Korean patients with systemic lupus erythematosus. Rheumatol Int. 2002;21(6):222–226. doi: 10.1007/s00296-001-0171-x. [DOI] [PubMed] [Google Scholar]
- Chu ZT, et al. Association of Fcgamma receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue Antigens. 2004;63(1):21–27. doi: 10.1111/j.1399-0039.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- Weersma RK, et al. Association of FcgR2a, but not FcgR3a, with inflammatory bowel diseases across three Caucasian populations. Inflamm Bowel Dis. 2010;16(12):2080–2089. doi: 10.1002/ibd.21342. [DOI] [PubMed] [Google Scholar]
- van der Pol WL, et al. Association of the Fc gamma receptor IIA-R/R131 genotype with myasthenia gravis in Dutch patients. J Neuroimmunol. 2003;144(1–2):143–147. doi: 10.1016/j.jneuroim.2003.08.043. [DOI] [PubMed] [Google Scholar]
- Zhou XJ, et al. FCGR2B gene polymorphism rather than FCGR2A, FCGR3A and FCGR3B is associated with anti-GBM disease in Chinese. Nephrol Dial Transplant. 2010;25(1):97–101. doi: 10.1093/ndt/gfp374. [DOI] [PubMed] [Google Scholar]
- Jonsen A, et al. Association between SLE nephritis and polymorphic variants of the CRP and FcgammaRIIIa genes. Rheumatology (Oxford) 2007;46(9):1417–1421. doi: 10.1093/rheumatology/kem167. [DOI] [PubMed] [Google Scholar]
- Dong C et al (2013) FcgammaRIIIa SNPs and haplotypes affect human IgG binding and association with lupus nephritis in African Americans. Arthritis Rheum, doi: 10.1002/art.38337 [DOI] [PMC free article] [PubMed]
- Koene HR, et al. The Fc gammaRIIIA-158F allele is a risk factor for systemic lupus erythematosus. Arthritis Rheum. 1998;41(10):1813–1818. doi: 10.1002/1529-0131(199810)41:10<1813::AID-ART13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kyogoku C, et al. Studies on the association of Fc gamma receptor IIA, IIB, IIIA and IIIB polymorphisms with rheumatoid arthritis in the Japanese: evidence for a genetic interaction between HLA-DRB1 and FCGR3A. Genes Immun. 2002;3(8):488–493. doi: 10.1038/sj.gene.6363921. [DOI] [PubMed] [Google Scholar]
- Alarcon GS, et al. Time to renal disease and end-stage renal disease in PROFILE: a multiethnic lupus cohort. PLoS Med. 2006;3(10):e396. doi: 10.1371/journal.pmed.0030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta Y, et al. Association of Fc gamma receptor IIIB, but not of Fc gamma receptor IIA and IIIA polymorphisms with systemic lupus erythematosus in Japanese. Genes Immun. 1999;1(1):53–60. doi: 10.1038/sj.gene.6363639. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escribano MF, et al. FcgammaRIIA, FcgammaRIIIA and FcgammaRIIIB polymorphisms in Spanish patients with systemic lupus erythematosus. Eur J Immunogenet. 2002;29(4):301–306. doi: 10.1046/j.1365-2370.2002.00324.x. [DOI] [PubMed] [Google Scholar]
- You Y, et al. Lack of association between Fc receptor-like 3 gene polymorphisms and systemic lupus erythematosus in Chinese population. J Dermatol Sci. 2008;52(2):118–122. doi: 10.1016/j.jdermsci.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Choi CB, et al. The -169C/T polymorphism in FCRL3 is not associated with susceptibility to rheumatoid arthritis or systemic lupus erythematosus in a case-control study of Koreans. Arthritis Rheum. 2006;54(12):3838–3841. doi: 10.1002/art.22248. [DOI] [PubMed] [Google Scholar]
- Ramirez-Bello J, et al. Juvenile rheumatoid arthritis and asthma, but not childhood-onset systemic lupus erythematosus are associated with FCRL3 polymorphisms in Mexicans. Mol Immunol. 2013;53(4):374–378. doi: 10.1016/j.molimm.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Piotrowski P et al (2013) The FCRL3 -169T > C polymorphism might be associated with some autoantibody presence in patients with SLE in a Polish population. Mod Rheumatol 24(2):296–299 [DOI] [PubMed]
- Mao C, et al. Association between Fc receptor-like 3 C169T polymorphism and risk of systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. 2010;37(1):191–196. doi: 10.1007/s11033-009-9591-6. [DOI] [PubMed] [Google Scholar]
- Song GG, et al. Fc receptor-like 3 (FCRL3) -169 C/T polymorphism and systemic lupus erythematosus: a meta-analysis. Rheumatol Int. 2013;33(9):2323–2329. doi: 10.1007/s00296-013-2722-3. [DOI] [PubMed] [Google Scholar]
- Morgan AW, et al. Fcgamma receptor type IIIA is associated with rheumatoid arthritis in two distinct ethnic groups. Arthritis Rheum. 2000;43(10):2328–2334. doi: 10.1002/1529-0131(200010)43:10<2328::AID-ANR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Morgan AW, et al. FcgammaRIIIA-158 V and rheumatoid arthritis: a confirmation study. Rheumatology (Oxford) 2003;42(4):528–533. doi: 10.1093/rheumatology/keg169. [DOI] [PubMed] [Google Scholar]
- Thabet MM, et al. FCRL3 promoter 169 CC homozygosity is associated with susceptibility to rheumatoid arthritis in Dutch Caucasians. Ann Rheum Dis. 2007;66(6):803–806. doi: 10.1136/ard.2006.064949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eike MC, et al. The FCRL3 -169T > C polymorphism is associated with rheumatoid arthritis and shows suggestive evidence of involvement with juvenile idiopathic arthritis in a Scandinavian panel of autoimmune diseases. Ann Rheum Dis. 2008;67(9):1287–1291. doi: 10.1136/ard.2007.077826. [DOI] [PubMed] [Google Scholar]
- Maehlen MT, et al. FCRL3 -169C/C genotype is associated with anti-citrullinated protein antibody-positive rheumatoid arthritis and with radiographic progression. J Rheumatol. 2011;38(11):2329–2335. doi: 10.3899/jrheum.110489. [DOI] [PubMed] [Google Scholar]
- Wu H, et al. Fc receptor-like 3 gene polymorphisms confer susceptibility to rheumatoid arthritis in a Chinese population. Hum Immunol. 2010;71(12):1203–1208. doi: 10.1016/j.humimm.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Bajpai UD, et al. A functional variant in FCRL3 is associated with higher Fc receptor-like 3 expression on T cell subsets and rheumatoid arthritis disease activity. Arthritis Rheum. 2012;64(8):2451–2459. doi: 10.1002/art.34457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, et al. FCRL3 gene polymorphisms contribute to the radiographic severity rather than susceptibility of rheumatoid arthritis. Hum Immunol. 2012;73(5):537–542. doi: 10.1016/j.humimm.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Zeng Z, et al. Association of FCRL4 polymorphisms on disease susceptibility and severity of ankylosing spondylitis in Chinese Han population. Clin Rheumatol. 2012;31(10):1449–1454. doi: 10.1007/s10067-012-2028-y. [DOI] [PubMed] [Google Scholar]
- Tang X, et al. A single-nucleotide polymorphism marker within the FCRL5 gene and HLA-B27 positive Han Chinese ankylosing spondylitis patients. Tissue Antigens. 2009;74(4):314–316. doi: 10.1111/j.1399-0039.2009.01335.x. [DOI] [PubMed] [Google Scholar]
- Owen CJ, et al. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab. 2007;92(3):1106–1111. doi: 10.1210/jc.2006-2183. [DOI] [PubMed] [Google Scholar]
- Duchatelet S, et al. FCRL3 -169CT functional polymorphism in type 1 diabetes and autoimmunity traits. Biomed Pharmacother. 2008;62(3):153–157. doi: 10.1016/j.biopha.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Inoue N, et al. Associations between autoimmune thyroid disease prognosis and functional polymorphisms of susceptibility genes, CTLA4, PTPN22, CD40, FCRL3, and ZFAT, previously revealed in genome-wide association studies. J Clin Immunol. 2012;32(6):1243–1252. doi: 10.1007/s10875-012-9721-0. [DOI] [PubMed] [Google Scholar]
- Gu LQ, et al. Clinical associations of the genetic variants of CTLA-4, Tg, TSHR, PTPN22, PTPN12 and FCRL3 in patients with Graves’ disease. Clin Endocrinol (Oxf) 2010;72(2):248–255. doi: 10.1111/j.1365-2265.2009.03617.x. [DOI] [PubMed] [Google Scholar]
- Plagnol V, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7(8):e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson JM, et al. Genetic association of zinc transporter 8 (ZnT8) autoantibodies in type 1 diabetes cases. Diabetologia. 2012;55(7):1978–1984. doi: 10.1007/s00125-012-2540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, et al. FcRL3 and multiple sclerosis pathogenesis: role in autoimmunity? J Neuroimmunol. 2007;189(1–2):132–136. doi: 10.1016/j.jneuroim.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Matesanz F, et al. The high producer variant of the Fc-receptor like-3 (FCRL3) gene is involved in protection against multiple sclerosis. J Neuroimmunol. 2008;195(1–2):146–150. doi: 10.1016/j.jneuroim.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Martinez A, et al. Epistatic interaction between FCRL3 and MHC in Spanish patients with IBD. Tissue Antigens. 2007;69(4):313–317. doi: 10.1111/j.1399-0039.2007.00816.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, et al. Association of polymorphisms in the promoter region of FCER1A gene with atopic dermatitis, chronic uticaria, asthma, and serum immunoglobulin E levels in a Han Chinese population. Hum Immunol. 2012;73(3):301–305. doi: 10.1016/j.humimm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Niwa Y, et al. FcepsilonRIalpha gene (FCER1A) promoter polymorphisms and total serum IgE levels in Japanese atopic dermatitis patients. Int J Immunogenet. 2010;37(2):139–141. doi: 10.1111/j.1744-313X.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- Weidinger S, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8):e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granada M et al (2012) A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol 129(3):840–845 e21 [DOI] [PMC free article] [PubMed]
- MacGlashan D., Jr IgE and FcepsilonRI regulation. Clin Rev Allergy Immunol. 2005;29(1):49–60. doi: 10.1385/criai:29:1:049. [DOI] [PubMed] [Google Scholar]
- Kim YK, et al. Association and functional relevance of E237G, a polymorphism of the high-affinity immunoglobulin E-receptor beta chain gene, to airway hyper-responsiveness. Clin Exp Allergy. 2007;37(4):592–598. doi: 10.1111/j.1365-2222.2007.02680.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. The E237G polymorphism of the high-affinity IgE receptor beta chain and asthma. Ann Allergy Asthma Immunol. 2004;93(5):499–503. doi: 10.1016/s1081-1206(10)61419-6. [DOI] [PubMed] [Google Scholar]
- Laprise C, et al. Evidence for association and linkage between atopy, airway hyper-responsiveness, and the beta subunit Glu237Gly variant of the high-affinity receptor for immunoglobulin E in the French-Canadian population. Immunogenetics. 2000;51(8–9):695–702. doi: 10.1007/s002510000185. [DOI] [PubMed] [Google Scholar]
- Nagata H, et al. Association between nasal allergy and a coding variant of the Fc epsilon RI beta gene Glu237Gly in a Japanese population. Hum Genet. 2001;109(3):262–266. doi: 10.1007/s004390100561. [DOI] [PubMed] [Google Scholar]
- Li A, Hopkin JM. Atopy phenotype in subjects with variants of the beta subunit of the high affinity IgE receptor. Thorax. 1997;52(7):654–655. doi: 10.1136/thx.52.7.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizawa N, et al. A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med. 2000;161(3 Pt 1):906–909. doi: 10.1164/ajrccm.161.3.9903128. [DOI] [PubMed] [Google Scholar]
- Kim SH, et al. A polymorphism of MS4A2 (- 109T > C) encoding the beta-chain of the high-affinity immunoglobulin E receptor (FcepsilonR1beta) is associated with a susceptibility to aspirin-intolerant asthma. Clin Exp Allergy. 2006;36(7):877–883. doi: 10.1111/j.1365-2222.2006.02443.x. [DOI] [PubMed] [Google Scholar]
- Yang HJ et al (2014) Association of the MS4A2 gene promoter C-109T or the 7th exon E237G polymorphisms with asthma risk: A meta-analysis. Clin Biochem 47(7-8):605–611 [DOI] [PubMed]
- Laitinen T, et al. Association study of the chromosomal region containing the FCER2 gene suggests it has a regulatory role in atopic disorders. Am J Respir Crit Care Med. 2000;161(3 Pt 1):700–706. doi: 10.1164/ajrccm.161.3.9810056. [DOI] [PubMed] [Google Scholar]
- Koster ES, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy. 2011;66(12):1546–1552. doi: 10.1111/j.1398-9995.2011.02701.x. [DOI] [PubMed] [Google Scholar]
- Tantisira KG, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120(6):1285–1291. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Sharma V, et al. A role of FCER1A and FCER2 polymorphisms in IgE regulation. Allergy. 2014;69(2):231–236. doi: 10.1111/all.12336. [DOI] [PubMed] [Google Scholar]
- Potaczek DP, et al. Interaction of functional FCER2 promoter polymorphism and phenotype-associated haplotypes. Tissue Antigens. 2009;74(6):534–538. doi: 10.1111/j.1399-0039.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- Kaneko M, et al. Allergen-specific IgG1 and IgG3 through Fc gamma RII induce eosinophil degranulation. J Clin Invest. 1995;95(6):2813–2821. doi: 10.1172/JCI117986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson F, et al. Human FcgammaRIIA induces anaphylactic and allergic reactions. Blood. 2012;119(11):2533–2544. doi: 10.1182/blood-2011-07-367334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Sperling AI. The contribution of allergen-specific IgG to the development of th2-mediated airway inflammation. J Allergy (Cairo) 2012;2012:236075. doi: 10.1155/2012/236075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, et al. Longitudinal study on the relationship between cat allergen and endotoxin exposure, sensitization, cat-specific IgG and development of asthma in childhood–report of the German Multicentre Allergy Study (MAS 90) Allergy. 2005;60(6):766–773. doi: 10.1111/j.1398-9995.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- Bruhns P, Fremont S, Daeron M. Regulation of allergy by Fc receptors. Curr Opin Immunol. 2005;17(6):662–669. doi: 10.1016/j.coi.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Wu et al (2014) Functional Fc gamma receptor polymorphisms are associated with human allergy. PLOS one 9(2):e89196 [DOI] [PMC free article] [PubMed]
- Robledo G, et al. Association of the FCGR3A-158F/V gene polymorphism with the response to rituximab treatment in Spanish systemic autoimmune disease patients. DNA Cell Biol. 2012;31(12):1671–1677. doi: 10.1089/dna.2012.1799. [DOI] [PubMed] [Google Scholar]
- Cooper N, et al. Platelet-associated antibodies, cellular immunity and FCGR3a genotype influence the response to rituximab in immune thrombocytopenia. Br J Haematol. 2012;158(4):539–547. doi: 10.1111/j.1365-2141.2012.09184.x. [DOI] [PubMed] [Google Scholar]
- Dijstelbloem HM, et al. Fcgamma receptor polymorphisms in Wegener’s granulomatosis: risk factors for disease relapse. Arthritis Rheum. 1999;42(9):1823–1827. doi: 10.1002/1529-0131(199909)42:9<1823::AID-ANR5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Norsworthy P, et al. Overrepresentation of the Fcgamma receptor type IIA R131/R131 genotype in caucasoid systemic lupus erythematosus patients with autoantibodies to C1q and glomerulonephritis. Arthritis Rheum. 1999;42(9):1828–1832. doi: 10.1002/1529-0131(199909)42:9<1828::AID-ANR6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Song YW, et al. Abnormal distribution of Fc gamma receptor type IIa polymorphisms in Korean patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41(3):421–426. doi: 10.1002/1529-0131(199803)41:3<421::AID-ART7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Dijstelbloem HM, et al. Fcgamma receptor polymorphisms in systemic lupus erythematosus: association with disease and in vivo clearance of immune complexes. Arthritis Rheum. 2000;43(12):2793–2800. doi: 10.1002/1529-0131(200012)43:12<2793::AID-ANR20>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Morgan AW, et al. Association of FCGR2A and FCGR2A-FCGR3A haplotypes with susceptibility to giant cell arteritis. Arthritis Res Ther. 2006;8(4):R109. doi: 10.1186/ar1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol WL, et al. IgG receptor IIa alleles determine susceptibility and severity of Guillain-Barre syndrome. Neurology. 2000;54(8):1661–1665. doi: 10.1212/wnl.54.8.1661. [DOI] [PubMed] [Google Scholar]
- Khor CC, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. 2011;43(12):1241–1246. doi: 10.1038/ng.981. [DOI] [PubMed] [Google Scholar]
- Orru V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155(1):242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, et al. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 2006;54(12):3908–3917. doi: 10.1002/art.22220. [DOI] [PubMed] [Google Scholar]
- Olferiev M, et al. The role of activating protein 1 in the transcriptional regulation of the human FCGR2B promoter mediated by the -343 G - > C polymorphism associated with systemic lupus erythematosus. J Biol Chem. 2007;282(3):1738–1746. doi: 10.1074/jbc.M605808200. [DOI] [PubMed] [Google Scholar]
- Morgan AW, et al. Analysis of Fcgamma receptor haplotypes in rheumatoid arthritis: FCGR3A remains a major susceptibility gene at this locus, with an additional contribution from FCGR3B. Arthritis Res Ther. 2006;8(1):R5. doi: 10.1186/ar1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XJ, et al. Copy number variation of FCGR3A rather than FCGR3B and FCGR2B is associated with susceptibility to anti-GBM disease. Int Immunol. 2010;22(1):45–51. doi: 10.1093/intimm/dxp113. [DOI] [PubMed] [Google Scholar]
- Foster CB, et al. Polymorphisms in inflammatory cytokines and Fcgamma receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol. 2001;113(3):596–599. doi: 10.1046/j.1365-2141.2001.02807.x. [DOI] [PubMed] [Google Scholar]
- Niederer HA, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. 2010;19(16):3282–3294. doi: 10.1093/hmg/ddq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossent JC, Rischmueller M, Lester S. Low copy number of the Fc-gamma receptor 3B gene FCGR3B is a risk factor for primary Sjogren’s syndrome. J Rheumatol. 2012;39(11):2142–2147. doi: 10.3899/jrheum.120294. [DOI] [PubMed] [Google Scholar]
- Osuga Y, et al. Lymphocytes in endometriosis. Am J Reprod Immunol. 2011;65(1):1–10. doi: 10.1111/j.1600-0897.2010.00887.x. [DOI] [PubMed] [Google Scholar]
- Teles JS, et al. Association of FCRL3 C-169T promoter single-nucleotide polymorphism with idiopathic infertility and infertility-related endometriosis. J Reprod Immunol. 2011;89(2):212–215. doi: 10.1016/j.jri.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Szczepanska M, et al. The FCRL3 -169T > C polymorphism and the risk of endometriosis-related infertility in a Polish population. Arch Gynecol Obstet. 2013;288(4):799–804. doi: 10.1007/s00404-013-2829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


