Abstract
The increased Salmonella resistance to quinolones and fluoroquinolones is a public health concern in the Southeast Asian region. The objective of this study is to develop a high resolution melt curve (HRM) assay to rapidly screen for mutations in quinolone-resistant determining region (QRDR) of gyrase and topoisomerase IV genes. DNA sequencing was performed on 62 Salmonella strains to identify mutations in the QRDR of gyrA, gyrB, parC, and parE genes. Mutations were detected in QRDR of gyrA (n = 52; S83F, S83Y, S83I, D87G, D87Y, and D87N) and parE (n = 1; M438I). Salmonella strains with mutations within QRDR of gyrA are generally more resistant to nalidixic acid (MIC 16 > 256 μg/mL). Mutations were uncommon within the QRDR of gyrB, parC, and parE genes. In the HRM assay, mutants can be distinguished from the wild-type strains based on the transition of melt curves, which is more prominent when the profiles are displayed in difference plot. In conclusion, HRM analysis allows for rapid screening for mutations at the QRDRs of gyrase and topoisomerase IV genes in Salmonella. This assay markedly reduced the sequencing effort involved in mutational studies of quinolone-resistance genes.
1. Introduction
Salmonella is a human pathogen commonly found in developed and developing countries, causing clinical diseases ranging from mild gastroenteritis to septicaemia [1]. Quinolones are broad spectrum antimicrobial agents that inhibit bacterial DNA from unwinding and duplicating during cell division and are commonly used in treating Salmonella infection. However, the worldwide emergence of quinolone-resistant bacterial strains raises public health concern. Increased incidence of quinolone-resistant Salmonella serovars has been documented in Southeast Asian region [2–5].
Three major mechanisms contribute to quinolone-resistance among Salmonella: altered protein targets for quinolones, decreased uptake of quinolones by bacteria, and DNA gyrase protection via plasmid-derived qnr genes [6]. DNA gyrase and topoisomerase IV are two important enzymes involved in bacterial DNA replication. Quinolones bind to gyrase/topoisomerase IV-DNA complex and inhibit DNA replication. This action is responsible for the bacteriostatic and bactericidal property of quinolones. Mutations in the bacterial genes encoding DNA gyrase and topoisomerase IV may confer resistance to quinolones and it has been shown that altered structures of these enzymes prevent binding of quinolones [6, 7].
Both DNA gyrase and topoisomerase IV are tetrameric enzymes. DNA gyrase is encoded by gyrA and gyrB genes, while topoisomerase IV is encoded by parC and parE genes. Mutation in quinolone-resistant determining region (QRDR) in these genes is associated with quinolone-resistance [6]. Many studies have reported the presence of such mutations in quinolone-resistant Salmonella [8–12]. Indeed, lower fluoroquinolone susceptibility and quinolone-resistance are linked to point mutations in QRDR of the gyrase genes [13, 14]. While most studies revealed that codon 87 of gyrA gene represents the most frequently mutated amino acid in Salmonella [8, 12, 14], point mutations outside of QRDR may also play a role in quinolone-resistance [6]. Nevertheless, QRDR remains the major focus in quinolone-resistance studies since this region exhibits high mutation rate.
DNA sequencing is the gold standard for detecting genetic changes. However, it is relatively costly to sequence all four genes when examining a large sample pool. Furthermore, mutations are rare in parC and parE genes [9]; therefore DNA sequencing will not be an economically viable option when dealing with such conserved regions. The real-time, polymerase chain reaction- (PCR-) based high-resolution melt curve (HRM) analysis is able to detect small genetic variation in PCR amplicons [15–17]. This method is highly sensitive and specific and has been employed in single nucleotide polymorphism (SNP) scanning [18]. During HRM analysis, a DNA intercalating dye is included in the PCR reaction mixture. This compound interacts specifically with double-stranded DNA and emits fluorescence signal but loses fluorescence when released from the DNA during denaturation. Upon completion of PCR, the DNA samples are subject to a temperature gradient, and the loss of fluorescence signal resulting from denaturation gives each DNA sample a unique melting curve that is detected by the real-time PCR system [19]. Hence, different Salmonella strains can be discriminated based on their sequence, length, GC content, or strand composition.
HRM assay has been used for the identification of quinolone-resistance strains in pathogens such as Mycobacterium tuberculosis, Bacillus anthracis, Yersinia pestis, Francisella tularensis, and Salmonella Typhi and Paratyphi A [20–23]. In these studies, only gyrA gene was examined. Although infrequent, mutations in gyrB, parC, and parE genes of Salmonella and their correlation with lower susceptibility to quinolones and fluoroquinolones have been documented too [9]. The objective of the study was to determine the potential of HRM analysis for rapid screening of mutations in QRDR of Salmonella gyrase and topoisomerase IV genes.
2. Materials and Methods
2.1. Bacterial Strains
A total of 195 Salmonella enterica strains were obtained from the culture collection of Laboratory of Biomedical Sciences and Molecular Microbiology, Institute of Graduate Studies, University of Malaya. The susceptibility of the strains towards nalidixic acid and ciprofloxacin was examined and reported in previous studies [24, 25]. Sixty-two Salmonella strains showing resistance or reduced susceptibility towards nalidixic acid were used to develop the HRM assay in this study (Salmonella Typhimurium, n = 12; Salmonella Enteritidis, n = 50). All selected strains were susceptible to ciprofloxacin. The minimum inhibitory concentration (MIC) of nalidixic acid for the strains was determined by using Etest strips (BioMérieux, Marcy l'Etoile, France). The test was performed according to manufacturer's instructions. Genomic DNA for each bacterial strain was extracted by using Wizard genomic DNA purification kit (Promega, Madison, USA). The details of all Salmonella strains used in this study are listed in the Supplementary Table (see Table S1 in the Supplementary Material available online at http://dx.doi.org/10.1155/2014/718084).
2.2. DNA Sequencing
PCR amplification of gyrA, gyrB, parC, and parE QRDRs was performed using previously described primers [26]. The selected primers amplify the QRDRs of the respective genes. PCR amplification was done in a 25 μL monoplex reaction mixture, containing 1x colourless GoTaq Flexi Buffer, 1.5 mmol/L magnesium chloride (MgCl2), 200 μmol/L deoxynucleoside triphosphate (dNTP) mix, 0.3 μmol/L of each primer pair, 1 U Taq DNA polymerase (Promega, Madison, USA), and approximately 100 ng of bacterial genomic DNA. The PCR reaction mixtures were first incubated at 94°C for 2 min, followed by 25 cycles of 94°C for 30 s, 60–63°C (annealing temperature varies for each gene) for 30 s, and 72°C for 45 s, with a final extension step of 72°C for 5 min. The appropriate annealing temperatures for gyrA, gyrB, parC, and parE are 60°C, 60°C, 63°C, and 62°C, respectively. Agarose gel electrophoresis was then carried out to confirm the presence of amplicons prior to DNA sequencing. Next, the PCR products were purified using Wizard SV gel and PCR clean-up system (Promega, Madison, USA), and the purified PCR products were sent to First BASE Laboratories (Selangor, Malaysia) for sequencing. Mutations in the target genes were determined by comparison with the wild-type sequence of the respective genes in Salmonella Typhimurium LT2 (GenBank accession number NC_003197).
2.3. HRM Analysis
A homology search for gyrA, gyrB, parC, and parE gene sequences of Salmonella serovars from the NCBI GenBank database was performed. Sequence alignment was done using the molecular evolutionary genetics analysis software version 5 (MEGA5) [27]. Primers were designed using the online tool Primer3Plus (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi), spanning the QRDR of each gene of interest, and were commercially synthesized (NHK Bioscience Solutions, Kuala Lumpur, Malaysia). Approximately 20 ng of bacterial genomic DNA was added into 1x MeltDoctor HRM master mix (Applied Biosystems, Foster City, CA, USA), which was premixed with 0.3 μM of each primer pair. Sterile deionized distilled water was added to make up a final reaction volume of 20 μL. The reproducibility of the assay was determined by performing all reactions in triplicate. In each assay, the DNA sample from a wild-type reference strain was used as positive control. The selection of the wild-type reference strains for the different target genes was explained in the Results section (Section 3.2). Negative control for the experiment was the HRM reaction mix without the addition of bacterial DNA. The HRM assay was performed on the 7500 Fast Real-Time PCR System (Applied Biosystems). The real-time PCR amplification included a holding stage at 95°C for 10 min, followed by the cycling stage with 40 cycles of 95°C for 15 sec and subsequently 60°C for 1 min. The subsequent melt curve stage consisted of the following steps: 95°C for 15 sec, a melt from 60°C (1 min) to 95°C (30 sec) with a ramp rate of 1%, and 60°C for 15 sec (according to manufacturer's instructions). The amplification plot for each sample was generated by the 7500 software v2.0 (Applied Biosystems) linked to the instrument. The data generated was subsequently imported to the HRM software v2.0.1 (Applied Biosystems) for melting curve analysis. The fluorescence change of each sample was plotted against temperature and was normalized to produce the aligned melt curves. In order to better illustrate the minor fluorescence changes between the wild type and mutants, a difference plot was generated using the HRM software. In a difference plot, one Salmonella strain with wild-type sequence confirmed via sequencing was chosen as a reference. The fluorescence difference between each sample and the reference was plotted against temperature. Mutations in the target region would produce a deviated curve in the difference plot.
3. Results
3.1. Detection of QRDR Mutations by Sequencing
The gyrA, gyrB, parC, and parE genes of 62 Salmonella strains were subjected to sequencing. Of these 62 strains, missense mutations were detected in QRDR of gyrA (n = 52) and parE (n = 1). Silent mutations within and beyond QRDR were observed in gyrB (n = 60), parC (n = 1), and parE (n = 50). In QRDR of gyrA, missense mutations occurred only in codons 83 (conversion of serine to phenylalanine (S83F), tyrosine (S83Y), and isoleucine (S83I)) and 87 (conversion of aspartic acid to glycine (D87G), tyrosine (D87Y), and asparagine (D87N)). In the parE gene, two missense mutations were detected, one within the QRDR (methionine to isoleucine (M438I)) and another outside of the QRDR (valine to phenylalanine (V521F)) (Table 1). The MIC data for all 62 strains are shown in Table 1. In general, mutations at the QRDR of gyrA correlate with increased resistance to nalidixic acid (MIC 16–>256 μg/mL). In the absence of mutations (STM057/05), the strain showed lowest MIC value (MIC 6 μg/mL). Detailed genetic analyses of the QRDRs of each Salmonella strain are listed in the Supplementary Table (Table S1).
Table 1.
Mutations in the gyrase and topoisomerase IV genes as determined by DNA sequencing and the nalidixic acid minimum inhibitory concentration (MIC) of their corresponding Salmonella strains.
| Strain | MIC (μg/mL) | gy rA a | gyrB | parC | pa rE a |
|---|---|---|---|---|---|
| SE 109/07 | >256 | D87Y | Silent mutations | None | V521F |
| SE 005/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 022/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 026/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 027/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 028/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 032/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 033/03 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 047/04 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 054/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 064/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 066/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 067/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 069/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 073/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 075/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 078/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 080/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 081/05 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 086/06 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 101/07 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 104/07 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 105/07 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 106/07 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 107/07 | >256 | D87Y | Silent mutations | None | Silent mutations |
| SE 002/03 | 192 | D87Y | Silent mutations | None | Silent mutations |
| SE 076/05 | 192 | D87Y | Silent mutations | None | Silent mutations |
| SE 004/03 | 128 | D87Y | Silent mutations | None | Silent mutations |
| SE 048/04 | 128 | D87Y | Silent mutations | None | Silent mutations |
| SE 097/06 | 128 | D87Y | Silent mutations | None | Silent mutations |
| SE 074/05 | 96 | D87Y | Silent mutations | None | Silent mutations |
| SE 090/06 | 96 | D87Y | Silent mutations | None | Silent mutations |
| SE 068/05 | 64 | D87Y | Silent mutations | None | Silent mutations |
| SE 102/07 | 64 | D87Y | Silent mutations | None | Silent mutations |
| SE 046/04 | 48 | D87Y | Silent mutations | None | Silent mutations |
| SE 049/04 | 48 | D87Y | Silent mutations | None | Silent mutations |
| SE 084/05 | 24 | D87Y | Silent mutations | None | Silent mutations |
| SE 001/03 | 16 | D87Y | Silent mutations | None | Silent mutations |
| SE 065/05 | 16 | D87Y | Silent mutations | None | Silent mutations |
| STM 032/04 | >256 | D87G | None | None | None |
| SE 089/06 | >256 | D87G | Silent mutations | None | Silent mutations |
| SE 095/06 | >256 | D87G | Silent mutations | None | Silent mutations |
| SE 070/05 | 128 | D87G | Silent mutations | None | Silent mutations |
| SE 050/04 | 64 | D87G | Silent mutations | None | Silent mutations |
| SE 012/03 | >256 | D87N | Silent mutations | None | Silent mutations |
| SE 015/03 | >256 | D87N | Silent mutations | None | Silent mutations |
| SE 013/03 | 96 | D87N | Silent mutations | None | Silent mutations |
| STM 043/05 | >256 | S83F | Silent mutations | Silent mutations | Silent mutations |
| SE 055/05 | >256 | S83F | Silent mutations | None | Silent mutations |
| SE 051/04 | >256 | S83I | Silent mutations | None | Silent mutations |
| SE 052/04 | >256 | S83I | Silent mutations | None | Silent mutations |
| SE 099/06 | >256 | S83Y | Silent mutations | None | Silent mutations |
| STM 018/03 | 24 | None | Silent mutations | None | M438I |
| STM 071/07 | 64 | None | Silent mutations | None | None |
| STM 055/05 | 32 | None | Silent mutations | None | None |
| STM 006/02 | 16 | None | Silent mutations | None | None |
| STM 033/04 | 16 | None | Silent mutations | None | None |
| STM 053/05 | 16 | None | Silent mutations | None | None |
| STM 056/05 | 16 | None | Silent mutations | None | None |
| STM 034/04 | 12 | None | Silent mutations | None | None |
| STM 048/05 | 8 | None | Silent mutations | None | None |
| STM 057/05 | 6 | None | None | None | None |
D, aspartic acid; F, phenylalanine; G, glycine; I, isoleucine; M, methionine; N, asparagine; S, serine; V, valine; Y, tyrosine; “silent mutations” denotes synonymous base substitutions in the QRDR of the target genes and thus does not result in amino acid change; “none” denotes wild-type sequence in the QRDR; that is, base substitution did not take place.
aThe first letter indicates the original amino acid, followed by numbers that indicate the position of the amino acid in respective gene, and the last letter indicates the substituting amino acid. For example, D87Y means tyrosine replaced aspartic acid in position 87 in GyrA subunit.
3.2. HRM Analysis for Rapid Mutation Screening of Gyrase and Topoisomerase IV Genes
HRM primers were designed to span the QRDRs of gyrA (Ala67-Gln106), gyrB (Asp426-Lys447), parC (Ala64-Gln103), and parE (Asp420-Lys441) genes. The primer pairs produce amplicons ranging from 150 to 250 bp (Table 2), with a melting temperature of approximately 60°C. HRM analysis showed distinct melt curves for wild-type versus mutant strains, which were confirmed by sequencing. Using sequencing as a guideline, two QRDR mutation-free strains were selected as references to generate difference plots (STM006/02 for gyrA, parC, and parE genes; STM032/04 for gyrB gene). The QRDRs of gyrA, parC, and parE genes in the reference strain STM006/02 and the gyrB QRDR in the reference strain STM032/04 have matching sequences with the Salmonella Typhimurium reference genome LT2 (NC_003197). Detailed information for these two reference strains is listed in the Supplementary Table (Table S1).
Table 2.
PCR primer sequences for HRM analysis of gyrase and topoisomerase IV genes.
| Primer | Sequence (5′-3′)b | Amplicon size (bp) |
|---|---|---|
| gyrA-F | CAATGACTGGAACAAAGCCTA | 164 |
| gyrA-R | AACCGAAGTTACCCTGACCA | |
| gyrB-F | TGTCCGAACTGTACCTGGTG | 198 |
| gyrB-R | ACTCGTCGCGACCGATAC | |
| parC-F | CGTCTATGCGATGTCAGAGC | 219 |
| parC-R | ATCGCCGCGAATGACTTC | |
| parE-F | TACCGCGCAGGATCTTAATC | 193 |
| parE-R | GATCGCCACGGAAATATCAT |
bPrimers designed in this study.
The melting temperature of wild-type gyrA allele is 82.5–82.8°C, while that of the mutants (D87Y, D87N, S83F, and S83Y) is at 82.0–82.4°C. Interestingly, Salmonella strains with gyrA D87G mutation produced a distinct melting curve compared to other mutants, at slightly higher melting temperature (82.9–83.5°C). The differences in the melting curves of the wild type versus mutants were distinguishable in both aligned and difference plot (Figure 1). The melting temperature for gyrB wild-type allele is 86.1°C–86.2°C, while that of the mutants is 86.2–86.7°C. One mutant (STM043/05) with three mutations in gyrB has a slightly lower melting temperature at 86.0°C. The aligned plot did not show any difference when comparing the melt curves of wild-type versus mutant gyrB alleles. However, they can be distinguished in difference plot (Figure 2). For parC gene, the melting temperature for wild-type allele is at 86.1–86.5°C (98% melted below 86.4°C) and the mutant at 86.4°C. Although the differences in the melting temperature were small, the mutant produced a unique melting curve in the difference plot when compared to the wild type (Figure 3). For parE gene, the melting temperature for wild-type allele is at 84.6–84.9°C, whereas that of the mutants containing multiple mutations is at 84.9–85.2°C. One mutant (STM018/03), which harbors a single mutation in parE QRDR, has a lower melting temperature at 83.9°C. The differences in wild-type versus mutant melt curves were apparent in both aligned and difference plot (Figure 4).
Figure 1.
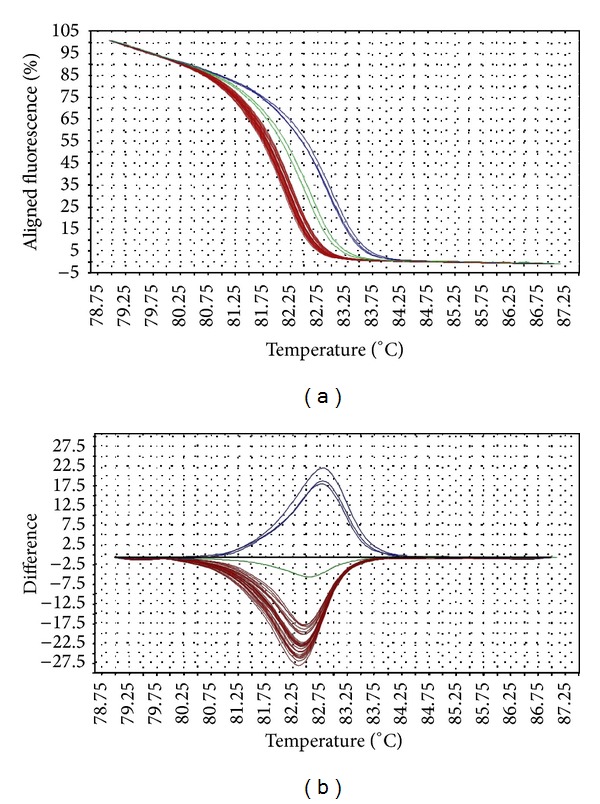
Representative HRM aligned melting curves (a) and difference plot (b) for mutations in gyrA QRDR. The aligned melting curves were set at 100% at the beginning and 0% at the end of melting process. In the difference plot, the melting curves represent the temperature at which the amplicons were completely denatured. The difference of the melting temperature among the samples is clearly illustrated in the difference plot. The reference strain is indicated by the horizontal black line in the difference plot; and the wild-type samples are indicated as green line. The blue curves derived from mutants with the missense mutation D87G, whereas the red curves belong to other mutants (D87Y, D87N, S83F, and S83Y). Mutants with missense mutations D87Y, D87N, S83F, and S83Y were denatured at similar melting temperature and therefore form a tight cluster.
Figure 2.
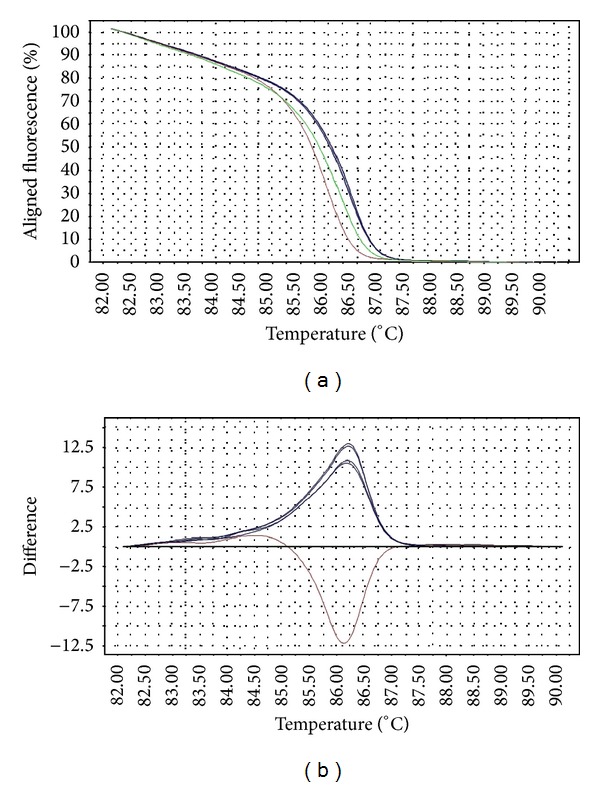
Representative HRM aligned melting curves (a) and difference plot (b) for mutations in gyrB QRDR. The reference strain is indicated by the horizontal black line in the difference plot, while mutants are represented by red (6 mutations within the HRM target region) or blue curves (3 mutations within the HRM target region). All gyrB mutants are silent mutations.
Figure 3.
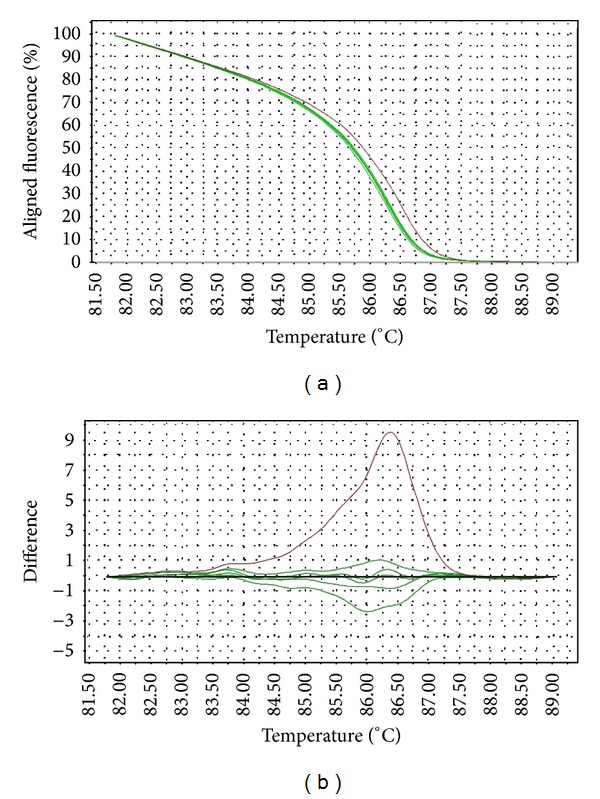
Representative HRM aligned melting curves (a) and difference plot (b) for mutations in parC QRDR. The reference strain is indicated by the horizontal black line in difference plot. Wild-type samples are indicated by green curves and the single mutant is indicated by red curve.
Figure 4.
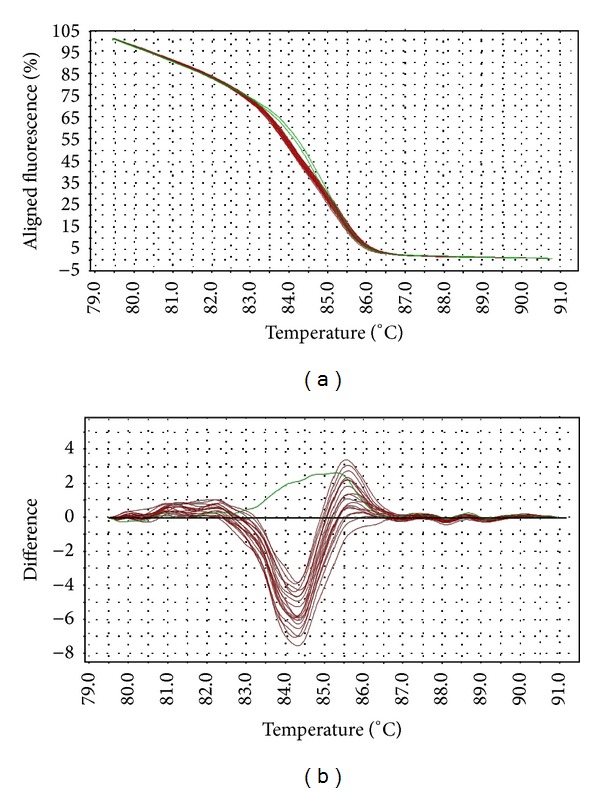
Representative HRM aligned melting curves (a) and difference plot (b) for mutations in parE QRDR. The reference strain is indicated by the horizontal black line in difference plot. Wild-type samples are indicated by green curves and mutants are indicated by red curves. All mutant strains contain similar nucleotide changes in the QRDR, resulting in tightly clustered melting curves.
4. Discussion
The HRM primers designed in this study spanned the entire QRDR of all four target genes. The QRDRs are short regions of DNA, making them suitable targets for HRM analysis (optimal amplicon length 150–350 bp). The transition of melt curves in HRM analysis of all target regions successfully discriminated the wild types from mutants. Although the melting transitions were not very distinct in aligned melt curves, especially for gyrB, the melting transitions of the mutants became apparent when displayed in a difference plot. These transitions are direct consequences of the mutations in the target gene QRDRs, since they are the only genetic variation within the PCR region.
Point mutations were identified in QRDRs of gyrA, gyrB, parC, and parE. Consistent with the literature [9, 14], our study identified that mutations in gyrA QRDR most commonly take place at codons 83 and 87, often resulting in amino acid change, and are associated with the nalidixic acid resistance in Salmonella strains. Gyrase A-DNA complex is the primary target for quinolones in Salmonella [7]. Hence, prolonged usage of quinolones often selects for development of resistant strains with mutations in gyrA, since such mutations confer better resistance to quinolone compared to that of gyrB mutants [9]. HRM analysis of the gyrA successfully resolved the mutant strains into three distinct melting profiles. The melting profiles of the D87G mutants were highly divergent from those of the other mutants. The D87G variants consisted of a single base substitution from adenosine to guanine. This type of mutation represents class I SNP, which often results in a higher melting temperature shift compared to other classes of SNP.
Mutation was not common in the QRDR of gyrB among Salmonella strains in this study. Furthermore, most studies did not identify mutations in gyrB of quinolone-resistant Salmonella [9, 14, 28]. The HRM primers designed in this study covered a region larger than the QRDR, with an additional 132 bp. Hence, the HRM variants detected in this study consisted of three point mutations outside of QRDR regions. However, all nucleotide changes resulted in silent mutations. One strain (STM043/05) consisted of a silent mutation in the QRDR and an additional three mutations compared to other mutants. This strain consisted of class I SNPs at two sites, thus producing a melting profile different from other variants. The different variants can be resolved by HRM as distinct difference plots.
Topoisomerase is the secondary target of quinolones in Gram-negative bacteria. Mutations within QRDRs of parC and parE genes are uncommon among Salmonella [9, 14, 28]. In this study, amino acid changes were not detected in parC genes and were identified in parE gene of only two Salmonella strains. The strains used in this study showed classical phenotype, that is, resistant to nalidixic acid but not to ciprofloxacin. These observations are in agreement with previous reports, which suggested that the mutations of these genes are most probably infrequently detected because they either are not important in quinolone-resistance or are only required for higher level of resistance, that is, resistance to fluoroquinolones [9]. Multiple mutations were detected both within and beyond the QRDRs of parC and parE genes. However, these were mostly silent mutations. HRM assay successfully differentiated the mutants from the wild type. The melting profiles of the mutants were highly divergent from those of the wild type, as clearly shown in both aligned melt curves and difference plots.
Despite the high efficiency of HRM analysis in mutation screening, this assay could only detect the presence of base substitutions. The melting profiles should not be used directly for variants classification without first validating with DNA sequencing. False positive may occur due to synonymous mutations, while false negative occurs due to mutations outside targeted regions or resistance caused by other mechanisms. To date, sequencing remains the ultimate proof for mutations that lead to amino acid changes and subsequently an alteration of enzyme structure [29]. Nevertheless, melting profiles are useful parameters to distinguish mutants with different characteristic and number of mutations, since individual mutant presents a distinctive melt curve when viewed in the difference plot. Hence, HRM analysis can be considered as an efficient and cost-effective preliminary step in the process of identifying QRDR mutations in Salmonella.
In conclusion, HRM analysis allows for detection of mutations in Salmonella gyrase and topoisomerase IV genes with sufficient sensitivity. The adoption of this assay for mutation screening prior to DNA sequencing is an attractive option to reduce the cost for mutational studies related to quinolone resistance. Furthermore, HRM analysis is time-saving, allows for high throughput screening, and is easy to perform since there is no post-PCR processing or purification steps. Nevertheless, sequencing shall be used for final validation and should not be completely replaced by HRM analysis.
Supplementary Material
The background of the Salmonella strains used to develop the HRM assay is tabulated in the supplementary table (Table S1). Additional information include the susceptibility of each Salmonella strain to nalidixic acid and the corresponding minimum inhibitory concentration value, type of base substitution and the corresponding amino acid change in each strain, and the melting temperature of the HRM amplicons.
Acknowledgments
The authors acknowledge University of Malaya for the facilities and financial support. The study was financially supported by Ministry of Science, Technology and Innovation (MOSTI) Science Fund (GA013-2013), High Impact Research (HIR) Grant (UM.C/625/HIR/MOHE/02), and Post Graduate Research Fund (PPP) (PS319/2010B). They thank K. H. Chua and C. S. J. Teh for advice in the initial setup of HRM. Soo Tein Ngoi was a recipient of the UM fellowship.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunology and Cell Biology. 2007;85(2):112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 2.Ling ML, Wang GCY. Epidemiological analysis of Salmonella enteritidis isolates in Singapore. Journal of Infection. 2001;43(3):169–172. doi: 10.1053/jinf.2001.0877. [DOI] [PubMed] [Google Scholar]
- 3.Padungtod P, Kaneene JB. Salmonella in food animals and humans in northern Thailand. International Journal of Food Microbiology. 2006;108(3):346–354. doi: 10.1016/j.ijfoodmicro.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Benacer D, Thong KL, Watanabe H, Puthucheary SD. Characterization of drug-resistant Salmonella enterica serotype typhimurium by antibiograms, plasmids, integrons, resistance genes, and PFGE. Journal of Microbiology and Biotechnology. 2010;20(6):1042–1052. doi: 10.4014/jmb.0910.10028. [DOI] [PubMed] [Google Scholar]
- 5.Thong KL, Modarressi S. Antimicrobial resistant genes associated with Salmonella from retail meats and street foods. Food Research International. 2011;44(9):2641–2646. [Google Scholar]
- 6.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. Journal of Antimicrobial Chemotherapy. 2003;51(5):1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 7.Michael GB, Butaye P, Cloeckaert A, Schwarz S. Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes and Infection. 2006;8(7):1898–1914. doi: 10.1016/j.micinf.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Eaves DJ, Randall L, Gray DT, et al. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica . Antimicrobial Agents and Chemotherapy. 2004;48(10):4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. International Journal of Antimicrobial Agents. 2005;25(5):358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Preisler A, Mraheil MA, Heisig P. Role of novel gyrA mutations in the suppression of the fluoroquinolone resistance genotype of vaccine strain Salmonella Typhimurium vacT (gyrA D87G) Journal of Antimicrobial Chemotherapy. 2006;57(3):430–436. doi: 10.1093/jac/dki475. [DOI] [PubMed] [Google Scholar]
- 11.Weill F-X, Bertrand S, Guesnier F, Baucheron S, Grimont PAD, Cloeckaert A. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerging Infectious Diseases. 2006;12(10):1611–1612. doi: 10.3201/eid1210.060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Cui S, McDermott PF, et al. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrobial Agents and Chemotherapy. 2007;51(2):535–542. doi: 10.1128/AAC.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehrenberg C, de Jong A, Friederichs S, Cloeckaert A, Schwarz S. Molecular mechanisms of decreased susceptibility to fluoroquinolones in avian Salmonella serovars and their mutants selected during the determination of mutant prevention concentrations. Journal of Antimicrobial Chemotherapy. 2007;59(5):886–892. doi: 10.1093/jac/dkm072. [DOI] [PubMed] [Google Scholar]
- 14.Lunn AD, Fàbrega A, Sánchez-Céspedes J, Vila J. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. International Microbiology. 2010;13(1):15–20. doi: 10.2436/20.1501.01.107. [DOI] [PubMed] [Google Scholar]
- 15.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clinical Chemistry. 2003;49(6):853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 16.Liew M, Pryor R, Palais R, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clinical Chemistry. 2004;50(7):1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 17.Pietzka AT, Stöger A, Huhulescu S, Allerberger F, Ruppitsch W. Gene scanning of an internalin B gene fragment using high-resolution melting curve analysis as a tool for rapid typing of Listeria monocytogenes . The Journal of Molecular Diagnostics. 2011;13(1):57–63. doi: 10.1016/j.jmoldx.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clinical Chemistry. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 19.White H, Potts G. Mutation Scanning by High Resolution Melt Analysis. Evaluation of Rotor-Gene 6000 (Corbett Life Science), HR-1 and 384-Well Lightscanner (Idaho Technology) Wessex, UK: National Genetics Reference Laboratory; 2006. [Google Scholar]
- 20.Slinger R, Bellfoy D, Desjardins M, Chan F. High-resolution melting assay for the detection of gyrA mutations causing quinolone resistance in Salmonella enterica serovars Typhi and Paratyphi. Diagnostic Microbiology and Infectious Disease. 2007;57(4):455–458. doi: 10.1016/j.diagmicrobio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Loveless BM, Yermakova A, Christensen DR, et al. Identification of ciprofloxacin resistance by SimpleProbe, High Resolution Melt and Pyrosequencing nucleic acid analysis in biothreat agents: Bacillus anthracis, Yersinia pestis and Francisella tularensis . Molecular and Cellular Probes. 2010;24(3):154–160. doi: 10.1016/j.mcp.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL. Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. Journal of Clinical Microbiology. 2011;49(10):3450–3457. doi: 10.1128/JCM.01068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee ASG, Ong DCT, Wong JCL, Siu GKH, Yam W-C. High-resolution melting analysis for the rapid detection of fluoroquinolone and streptomycin resistance in Mycobacterium tuberculosis . PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0031934.e31934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngoi ST, Lindstedt B-A, Watanabe H, Thong KL. Molecular characterization of Salmonella enterica serovar Typhimurium isolated from human, food, and animal sources in Malaysia. Japanese Journal of Infectious Diseases. 2013;66(3):180–188. doi: 10.7883/yoken.66.180. [DOI] [PubMed] [Google Scholar]
- 25.Ngoi ST, Thong KL. Molecular characterization showed limited genetic diversity among Salmonella Enteritidis isolated from humans and animals in Malaysia. Diagnostic Microbiology and Infectious Disease. 2013;77(4):304–311. doi: 10.1016/j.diagmicrobio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Fàbrega A, du Merle L, Le Bouguénec C, de Anta MTJ, Vila J. Repression of invasion genes and decreased invasion in a high-level fluoroquinolone-resistant Salmonella typhimurium mutant. PLoS ONE. 2009;4(11) doi: 10.1371/journal.pone.0008029.e8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K-Y, Park J-H, Kwak H-S, Woo G-J. Characterization of the quinolone resistance mechanism in foodborne Salmonella isolates with high nalidixic acid resistance. International Journal of Food Microbiology. 2011;146(1):52–56. doi: 10.1016/j.ijfoodmicro.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 29.Tindall EA, Petersen DC, Woodbridge P, Schipany K, Hayes VM. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Human Mutation. 2009;30(6):876–883. doi: 10.1002/humu.20919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The background of the Salmonella strains used to develop the HRM assay is tabulated in the supplementary table (Table S1). Additional information include the susceptibility of each Salmonella strain to nalidixic acid and the corresponding minimum inhibitory concentration value, type of base substitution and the corresponding amino acid change in each strain, and the melting temperature of the HRM amplicons.


