Abstract
Functionalized proline residues have diverse applications. Herein we describe a practical approach, proline editing, for the synthesis of peptides with stereospecifically modified proline residues. Peptides are synthesized by standard solid-phase-peptide-synthesis to incorporate Fmoc-Hydroxyproline (4R-Hyp). In an automated manner, the Hyp hydroxyl is protected and the remainder of the peptide synthesized. After peptide synthesis, the Hyp protecting group is orthogonally removed and Hyp selectively modified to generate substituted proline amino acids, with the peptide main chain functioning to “protect” the proline amino and carboxyl groups. In a model tetrapeptide (Ac-TYPN-NH2), 4R-Hyp was stereospecifically converted to 122 different 4-substituted prolyl amino acids, with 4R or 4S stereochemistry, via Mitsunobu, oxidation, reduction, acylation, and substitution reactions. 4-Substituted prolines synthesized via proline editing include incorporated structured amino acid mimetics (Cys, Asp/Glu, Phe, Lys, Arg, pSer/pThr), recognition motifs (biotin, RGD), electron-withdrawing groups to induce stereoelectronic effects (fluoro, nitrobenzoate), handles for heteronuclear NMR (19F:fluoro; pentafluorophenyl or perfluoro-tert-butyl ether; 4,4-difluoro; 77SePh) and other spectroscopies (fluorescence, IR: cyanophenyl ether), leaving groups (sulfonate, halide, NHS, bromoacetate), and other reactive handles (amine, thiol, thioester, ketone, hydroxylamine, maleimide, acrylate, azide, alkene, alkyne, aryl halide, tetrazine, 1,2-aminothiol). Proline editing provides access to these proline derivatives with no solution phase synthesis. All peptides were analyzed by NMR to identify stereoelectronic and steric effects on conformation. Proline derivatives were synthesized to permit bioorthogonal conjugation reactions, including azide-alkyne, tetrazinetrans-cyclooctene, oxime, reductive amination, native chemical ligation, Suzuki, Sonogashira, cross-metathesis, and Diels-Alder reactions. These proline derivatives allowed three parallel bioorthogonal reactions to be conducted in one solution.
Introduction
Protein activity, defined broadly, results from the geometrically controlled three-dimensional arrangement of a series of functional groups. In folded proteins, protein activity relies on secondary structure formation, the hydrophobic effect, and other non-covalent interactions to organize functional groups in a manner that allows specific recognition of substrates and binding partners and catalysis. Because functions are effected through specific structures, there has been broad interest in developing ways to stabilize protein structures. In addition, nature is limited by the genetically encoded amino acids, combined with post-translational modifications, plus metals and organic co-factors that bind these amino acids. Enhanced activity and novel functions may be incorporated into proteins by expanding beyond the array of natural amino acids, for example by introducing conformationally restrained amino acids, novel non-native functionalities, or amino acids with an enhanced hydrophobic effect.1
The field of protein design and engineering aims to achieve the functions of proteins in novel structures and/or the development of novel functions in proteins. The goals include achieving protein function within smaller structures, developing new structural topologies, de novo catalytic activity, and the development of hyperstable proteins.2 The use of conformationally restricted amino acids has had particular application in achieving enhanced protein stability and in stabilizing protein recognition epitopes within small peptides, especially in medicinal chemistry applications of peptides.3
Proline residues are unique among the canonical amino acids, due to the conformational restraint of backbone cyclization and the presence of a tertiary amide bond.4 These structural characteristics limit the available conformations for proline residues, with the φ torsion angle restricted to −65° ± 25°. The conformational restriction and absence of a hydrogen bond donor results in proline being preferentially observed in certain structural contexts, including secondary structure termination, loops, turns, and polyproline helices (PPII). Proline residues are also preferentially employed in biomolecular recognition based on their distinction from other canonical amino acids, the hydrophobicity of the pyrrolidine ring, and the possibility of favorable aromatic-proline interactions.5
Proline has two key conformational equilibria: endo versus exo ring pucker, and trans versus cis amide bond (Figure 1).4 Proline ring pucker correlates with protein φ and ψ main chain conformation, with an exo ring pucker favoring more compact conformations (PPII, αR) and an endo ring pucker favoring more extended conformations.6 Trans versus cis amide bond conformation defines the ω torsion angle, with the cis conformation strongly preferring the endo ring pucker. Thus, control of proline ring pucker permits control of all protein backbone torsion angles (φ, ψ, ω).7
Figure 1.
Proline conformational equilibria. (a) Cis-trans isomerism of the prolyl amide bond. In proteins, 95% of prolyl amide bonds are trans, 5% are cis. The trans and cis amide bond are in slow exchange on the NMR timescale, so the relative populations (Ktrans/cis = [peptide with trans amide bond]/[peptide with cis amide bond]) are determined via quantification of their distinct NMR resonances. (b) Exo and endo ring puckers of proline. Exo and endo ring puckers are in rapid equilibrium. (c) Definition of main chain torsion angles in proline.
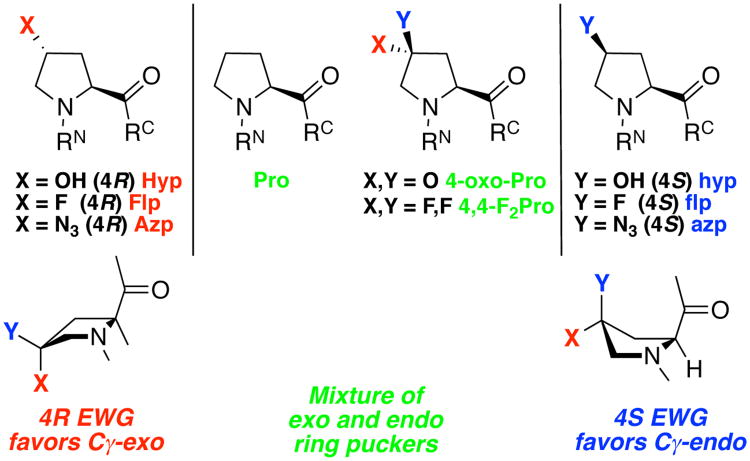
The most abundant human protein, collagen, exhibits obligatory hydroxylation of proline at certain positions (Yaa of the XaaYaaGly collagen repeat, consensus sequence ProHypGly) to 4R-hydroxyproline (Hyp).8 Proline hydroxylation results in a substantial enhancement of the stability of collagen, via the induction of a stereoelectronic effect.9 This stereoelectronic effect is manifested in a preferred gauche relationship of the 4-hydroxyl substituent and the carbon-amide bond (Figure 2, Figure 3, Figure 4). The sterically disfavored gauche conformation is preferred due to favorable hyperconjugative interactions from overlap of electron-rich σC-H orbitals and electron-deficient σ*C-X orbitals (X = amide or hydroxyl in Hyp).10 This favorable gauche interaction leads to a preference for 4R-Hyp to adopt the exo ring pucker. Analogously, the non-native 4S-hydroxyproline (hyp) diastereomer leads to a preference for the endo ring pucker. The strength of this interaction depends on how electron-withdrawing the 4-substitutent is, with a more electron-withdrawing group leading to a greater stereoelectronic effect and a greater conformational preference for the sterically disfavored conformation. Thus, the incorporation of 4R-fluoroproline (Flp) in place of Hyp in collagen mimetic peptides leads to enhanced collagen triple helix stability due to the more electron-withdrawing nature of fluorine versus hydroxyl, and thus a greater preference for the exo conformation required at the Yaa position of collagen. In contrast, incorporation of non-electron-withdrawing 4R-methyl substituent, with a steric preference for an anti conformation, or an electron-withdrawing 4S-fluoro (flp) substituent, leads to a bias toward the endo ring pucker. The endo ring pucker is destabilizing at the Yaa position of collagen but stabilizing at the Xaa position, which is typically a Pro residue with an endo ring pucker and a more extended conformation.11
Figure 2.
Stereoelectronic effects in 4-substituted prolines lead to a preference in the Cγ-CS bond for a gauche relationship between the amide and electron-withdrawing 4-substituents. (a) In 4R-fluoroproline (Flp) and 4S-fluoroproline (flp), two major hyperconjugative interactions stabilize the gauche conformation: orbital overlap between the electron-rich C-Hδ bond (σ) and the electron-deficient C-F bond (σ*) (left), and orbital overlap between the electron-rich C-H, bond and the electron-deficient C-N bond (middle). When N and F are anti, neither stabilizing interaction is possible (right). Additional hyperconjugative stabilization is provided by an anti arrangement between a beta proton and the fluorine when Flp is exo or when flp is endo. (b) Manifestation of gauche effects on ring pucker. With electron-withdrawing substituents, a gauche conformation with 4R-substitution leads to a strong preference for exo ring pucker, while a gauche conformation with 4S-substitution leads to a strong preference for endo ring pucker. The exo/endo preference thus depends on both the stereochemistry of the substitution and how electron-withdrawing the substituent is. For sterically demanding and/or non-electron-withdrawing substituents, the reverse preferences will be observed due to a steric preference for anti over gauche conformation. Newman projections are simplified to show idealized torsion angles.
Figure 3.
Effects of proline ring pucker on main chain φ, ψ, and ω torsion angles. (a) 4-Substituted proline residues can bias proline ring pucker by stereoelectronic effects or by steric effects. (b) An n→π* interaction between the carbonyl lone pair (n) of residue i and the π* orbital of the carbonyl on the subsequent residue (i+1) provides local organization of the protein main chain, including stabilization of the trans amide bond and the αR and PPII conformations. This interaction is preferential with the Cγ-exo ring pucker of proline and is only possible with a trans amide bond.
Figure 4.
Most-studied 4-substituted proline derivatives, three letter codes, and their conformational preferences. Red color and upper case 3-letter code indicates trans relative stereochemistry (4R substitution) of the 4-substituent and the carbonyl. Blue color and lower case 3-letter code indicates cis relative stereochemistry (4S) of the 4-substitutent and the carbonyl. Green color indicates 4,4-disubstituted prolines. EWG = electron-withdrawing group. Non-electron-withdrawing or sterically demanding substituents have opposite conformational preferences: 4R-substituted methylproline (X = CH3) and 4R-mercaptoproline (X = SH) favor Cγ-endo ring pucker, while 4S-substituted-methylproline and 4S-mercaptoproline favor Cγ-exoring pucker.7a, 11, 39
The stereospecific effect of proline 4-substitution on ring pucker is also manifested in amide cis-trans isomerism (ω torsion angle). The exo ring pucker stabilizes the trans amide bond, whereas an endo ring pucker is strongly favored in a cis amide bond. In proteins, proline amide conformation at specific residues is conserved evolutionarily.12 Moreover, proper amide conformation is necessary for protein function. Cis-trans isomerism is often a rate-determining step in protein folding, with prolyl isomerases critical enzymes for rapid folding of proteins and for prevention of protein misfolding.13 In addition, there are a growing number of examples of proteins with two alternating functions, one of which is mediated by having a prolyl cis amide and another with a trans amide.14 Slow or incorrect prolyl cis-trans isomerism has also been implicated pathologically in protein misfolding. For example, overexpression of the prolyl isomerase Pin1 slows tau misfolding and neurofibrillary tangle formation in cellular models of Alzheimer's disease.15 Strategies to stabilize cis or trans amide bonds thus can directly impact protein structure, stability, and function.
In addition to the stereoelectronic (gauche) effect on controlling ring pucker, and thus protein main chain conformation, conformations in peptides containing proline residues are affected by a favorable n→π* interaction between adjacent carbonyls (Figure 3).9b, 16 In the n→π* interaction, which is also observed in non-proline residues,16c there is orbital overlap between a lone pair (n) on the carbonyl oxygen of the i residue with the π* orbital of the carbonyl at the carbon of the i+1 residue, with a near-ideal Bürgi-Dunitz trajectory internuclear angle. This n→π* interaction is strongest with an exo ring pucker on the proline residue, and fundamentally impacts the conformational preferences of proline residues, thus connecting proline ring pucker with protein secondary structure.
The ability to control protein structure through the incorporation of stereodefined substituted proline residues, either by favoring particular conformations or through enhanced hydrophobic effect, is an emerging strategy to increase protein stability and/or activity. Proline derivatives have been incorporated into collagen mimetics and other advanced materials, in neurotransmitter receptor proteins, in enzymes, in synthetic polyproline scaffolds, and in globular proteins, in addition to many peptides with diverse applications.9, 14d, 16a, 17 Certain proline derivatives (particularly fluoroprolines) have been incorporated into expressed proteins, either site-specifically through amber suppression/orthogonal aminoacyl tRNA synthetases or by global incorporation at all proline amino acids in proline auxotrophic bacterial strains.14d, 17c, 17i, j, 17n, 18 In every case, protein stability, activity, and/or function is dependent on both the identity and the stereochemistry of the substitution, allowing modulation of function in a manner that is predictable based on the known conformational preferences of proline derivatives.
The presence of proline in critical biological recognition and structural motifs has inspired the synthesis of a wide range of proline derivatives.19 Interest in the synthesis of modified proline residues has also risen due to the reemergence and substantial advances in proline-catalyzed organic reactions.20 There is an extensive history of development of novel proline derivatives, particularly for applications in medicinal chemistry, which has been the stimulus for the synthesis of most proline derivatives.5d, 19, 21 Proline derivatives have also been significantly applied to the field of collagen mimetics, where multiple copies of proline derivatives are incorporated in the collagen triple helix, allowing modulation of stability of synthetic collagens.9,11,17a-f,h,p-q
A range of creative strategies has been developed for the synthesis of proline derivatives, both based on cyclization strategies and based on modification reactions on the commercially available and inexpensive Hyp. The structural effects of proline modification have been extensively measured and categorized based on a combination of steric effects and stereoelectronic effects. In addition to synthetic proline derivatives, it has also been observed that hydroxyproline may be natively phosphorylated (observed in rat crystallin protein and in diatom biosilica) and glycosylated (observed in plant cell walls).22 Moreover, within bacteria, fungi, sponges, algae, and higher plants, an even wider range of proline analogues has been identified, in compounds with antibiotic, antifungal, and other activities.19a, 19c
While a significant number of proline derivatives have been synthesized, the application of these novel proline amino acids is restricted by the substantial solution phase synthetic effort to make each derivative as an Fmoc or Boc amino acid suitable for solid phase peptide synthesis, with typically 5-7 steps required per amino acid, including protection and deprotection strategies for the amine and carboxylic acids. For the preparation of a series of peptides, each with a different modified proline amino acid, for example to test structure-activity relationships, there is the requirement to synthesize each protected amino acid in solution, then incorporate each into a peptide, repeating all amide coupling steps N-terminal to the site of modification. While there has been an increase in the commercial availability of proline derivatives in recent years, highlighting the interest in their applications, these amino acids are generally quite expensive, other than the naturally derived 4R-Hyp.23
We recently communicated an approach, termed proline editing, to address the synthetic challenges of preparing peptides with proline derivatives.24 In proline editing, the commercially available and inexpensive amino acid Fmoc-4R-Hyp is incorporated in a peptide and orthogonally protected in an automated manner. After the peptide synthesis is completed, the hydroxyproline trityl protecting group is removed and the hydroxyl group selectively modified in an otherwise fully protected peptide on solid phase. This approach was applied in a model peptide, Ac-TYPN-NH2, and was used to synthesize 4R- and 4S- fluoro, hydroxyl, and O-nitrobenzoate proline-containing peptides, as well as the 4-oxoproline and 4,4-difluoroproline derivatives. That work was subsequently applied to incorporate proline derivatives in a more complex peptide, Andersen's trp cage miniprotein, and used to modulate the stability of the trp cage in a manner that was predictable based on stabilization or destabilization of the observed exo ring pucker at residue 12 of the trp cage.17r This strategy fundamentally employs the peptide main chain as a “protecting group,” abrogating the need to protect the amine and carboxylic acid functional groups. It is also a fundamentally diversity-oriented strategy, since a single fully synthesized peptide can be converted into a series of peptides exhibiting diverse functional groups and/or structural effects. Herein we provide a full description of the proline editing approach, considerably expanding the scope of the proline derivatives that may be incorporated into a peptide by the method and broadly examining the effects of incorporated proline derivatives on structure and in applications.
Results
Design of the model peptide
Proline editing was examined within a model tetrapeptide context24-25 that was chosen to contain a central tyrosine-proline sequence (residues i+1 and i+2) that promotes cis amide bonds via a favorable interaction between the aromatic and proline rings (Figure 5).5a, b, 5d, e, 5h, 26 The i (Thr) and i+3 (Asn) residues were selected to further promote cis amide bond formation.25-26, 26c, 27 This context permits the ready examination of the structural effects of proline modification by 1H NMR. The parent peptide Ac-TYPN-NH2 exhibits a Ktrans/cis = 2.7 in aqueous solution at room temperature, with all amide protons resolved and 3JαN values measurable to allow identification of the effects of proline modification on peptide main chain conformation. This model peptide has also been applied to examine aromatic effects on peptide structure, replacing tyrosine with electon-rich and electron-deficient aromatic amino acids as well as control peptides containing Ala and cyclyhexylalanine (Cha).5h, 24, 28 In this context, it was observed that aromatic electronics correlated with cis-trans isomerism (Ktrans/cis), with electron-rich aromatics favoring cis amide bond via a CH/π interaction between the aromatic ring and the prolyl ring, while electron-deficient aromatics, as well as Cha and Ala, relatively favored a trans amide bond due to the absence of a favorable CH/π interaction. By combining electronic and stereoelectronic effects, it was demonstrated that peptides containing aromatic-proline sequences could be designed to strongly promote cis (Ac-TWflpN-NH2, Ktrans/cis = 0.65) or trans (Ac-T(4-NO2-Phe)HypN-NH2, Ktrans/cis = 20.1) amide bonds. In the current work, a series of peptides was synthesized via proline editing to examine the effects of proline substitution on cis-trans isomerism, via a combination of stereoelectronic and steric effects on proline conformation and thus consequently on peptide main chain conformation.
Figure 5.

Aromatic-proline CH/π interactions between Hα (partially positive due to adjacent electron-withdrawing groups) and the negatively charged aromatic π face leads to an increased preference for cis amide bond in aromatic-proline sequences such as the model peptide sequence Ac-TYPN-NH2.
Synthesis of protected Hyp peptides
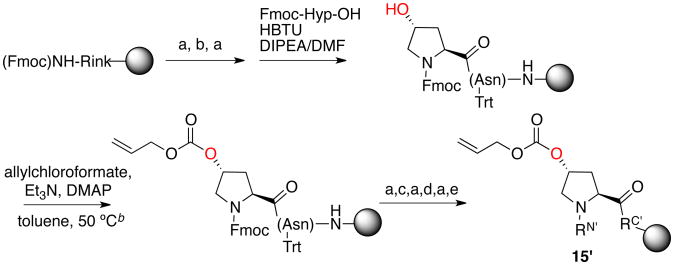
The central concept of proline editing is to incorporate a Hyp residue within a peptide and to subsequently modify the Hyp using stereospecific reactions at the γ-hydroxyl. Hyp is commercially available as an Fmoc amino acid either with a free hydroxyl group or protected as the t-butyl ether. Since t-butyl is a common protecting group in Fmoc solid phase peptide synthesis, and is not readily removed selectively on solid phase, proline editing was accomplished via coupling of the unprotected Fmoc-Hyp-OH via standard solid phase peptide synthesis (Scheme 1). The Hyp hydroxyl group was then trityl-Protected using trityl chloride in an automated manner, via programming as a double coupling with trityl chloride (and optionally imidazole) in place of an amino acid in the second coupling position. The trityl-Protected peptide was then subjected to standard solid phase peptide synthesis to complete the synthesis of the peptide, which proceeded cleanly. The effectiveness of this approach was demonstrated both within the model peptide and within the trp cage miniprotein.17r, 24, 28a After peptide synthesis was completed, the trityl group could be selectively removed with 2% TFA and subjected to reaction chemistry on the free hydroxyl.
Scheme 1.
Proline editing general approach: automated synthesis of the peptide Ac-TYHypN-NH2(1) via trityl hydroxyl protection.a
a (a) 20% piperidine/DMF; (b) Fmoc-Asn(Trt)-OH, HBTU, DIPEA/DMF; (c) Fmoc-Tyr(OtBu)-OH, HBTU, DIPEA/DMF; (d) Fmoc-Thr(OtBu)-OH, HBTU, DIPEA/DMF; (e) 10% Ac2O/pyridine. RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2. For all peptides in all schemes, the prime designation (e.g. 1′, RN′, RC′) indicates the protected peptide on solid phase, whereas the absence of a prime designation indicates free peptide in solution.
In order to potentially allow the incorporation of multiple different proline derivatives within a peptide, we also examined alternative and orthogonal protecting group strategies for the Hyp hydroxyl.29 Using an analogous approach, we were able to readily protect the Hyp as a silyl (TBS) ether using TBSCl with subsequent deprotection with TBAF on solid phase (Scheme 2). We also were able to protect the hydroxyl as an Alloc carbonate, although this method required elevated temperature and was not conducted as part of an automated synthetic procedure (Scheme 3). In addition, we also found that direct acylation of the Hyp hydroxyl after coupling proceeded readily as an alternative, using the orthogonally removable (NaN3/MeOH)30 nitrobenzoate ester or via acylation and alkylation to generate the bromoacetyl and subsequently azidoacetyl derivatives (Scheme 4). The direct acylation of Hyp approach was also applied in the synthesis of some modified trp cage miniproteins via proline editing.17r
Scheme 2.
Proline editing alternative approach: automated synthesis of the peptide Ac-TYHypN-NH2(1) via TBS hydroxyl protection.a
a (a) 20% piperidine/DMF; (b) Fmoc-Asn(Trt)-OH, HBTU, DIPEA/DMF; (c) Fmoc-Tyr(OtBu)-OH, HBTU, DIPEA/DMF; (d) Fmoc-Thr(OtBu)-OH, HBTU, DIPEA/DMF; (e) 10% Ac2O/pyridine. RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Scheme 3.
Proline editing alternative approach: synthesis of the peptide Ac-TYP(4R-O-Alloc)N-NH2(15) via Alloc protection and direct acylation. The Alloc group was stable to TFA cleavage/deprotection. a
a (a) 20% piperidine/DMF; (b) Fmoc-Asn(Trt)-OH, HBTU, DIPEA/DMF; (c) Fmoc-Tyr(OtBu)-OH, HBTU, DIPEA/DMF; (d) Fmoc-Thr(OtBu)-OH, HBTU, DIPEA/DMF; (e) 10% Ac2O/pyridine. RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2
b Reaction was performed in a glass vial manually.
Scheme 4.
Proline editing alternative approach: direct modification immediately after coupling of Fmoc-Hyp-OH. Synthesis of the peptides Ac-TYP(4R-OBzNO2)N-NH2(3) (top) and Ac-TYP(4R-azidoacetate)N-NH2(83) via direct acylation.a
a (a) 20% piperidine/DMF; (b) Fmoc-Asn(Trt)-OH, HBTU, DIPEA/DMF; (c) Fmoc-Tyr(OtBu)-OH, HBTU, DIPEA/DMF; (d) Fmoc-Thr(OtBu)-OH, HBTU, DIPEA/DMF; (e) 10% Ac2O/pyridine. RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2
b Reaction was performed manually in a disposable fritted tube.
Selective and stereospecific modification of Hyp residues in peptides on solid phase
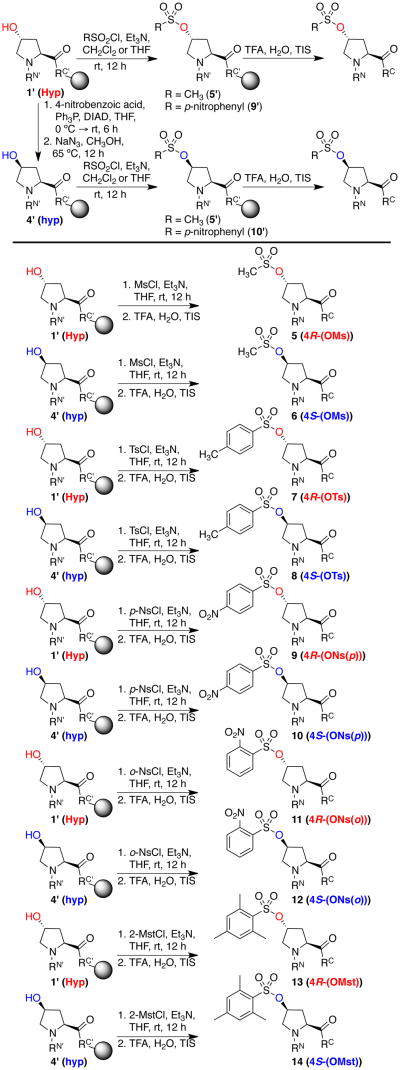
After peptide synthesis and Hyp deprotection, the free Hyp-containing peptide could potentially be subjected to a series of modification reactions to control stereochemistry, to promote specific structures, and to introduce novel functional groups into peptides (Scheme 5). The reactions examined include the generation of sulfonates and other leaving groups for substitution reactions; Mitsunobu and related stereospecific inversion reactions; acylation; and oxidation. Because we sought to synthesize peptides containing any given functional group with defined stereochemistry at the 4-position, we examined the conduct of these reactions on either the 4R (trans relative stereochemistry, indicated with upper case in 3-letter codes and red lettering) or 4S (cis relative stereochemistry, indicated with lower case and blue lettering) hydroxyprolines. The coupled 4R-Hyp (1) was readily converted on solid phase to 4S-hyp (4) via Mitsunobu reaction with 4-nitrobenzoic acid and subsequent selective removal of the nitrobenzoate with NaN3 in MeOH (Scheme 6, Figure 6), providing access to both 2S-hydroxyproline diastereomers (Hyp (4R) and hyp (4S)) on solid phase.30
Scheme 5.
Proline editing to introduce functional groups with defined stereochemistry into peptides synthesized with commercially available and inexpensive Fmoc-Hyp-OH.a
a LG = leaving group. RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin.
Scheme 6.
Mitsunobu inversion on solid phase to convert Hyp (4R) (1′) to hyp (4S) (4′). The nitrobenzoate may be deprotected selectively on solid phase with NaN3/MeOH30 or non-selectively with LiOH or K2CO3/MeOH.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Figure 6.
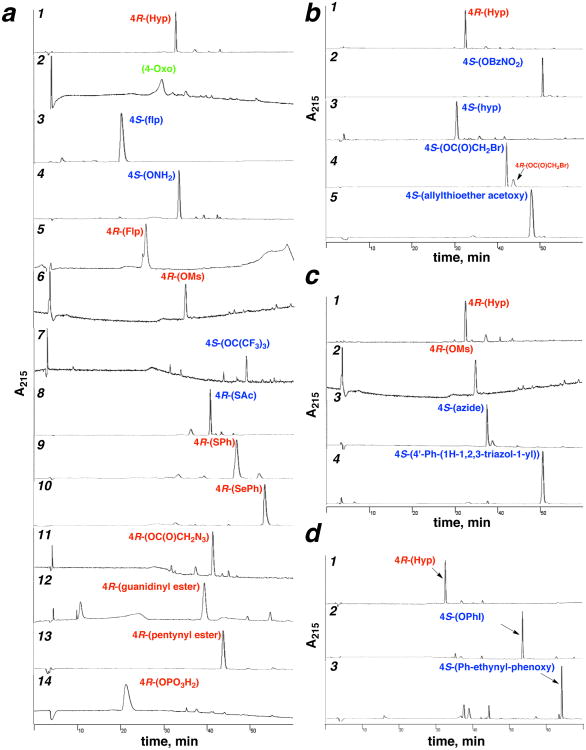
Representative crude HPLC chromatograms of proline editing reactions on solid phase, including inversion of hydroxyproline stereochemistry, sulfonylation, SN2, Mitsunobu, acylation, oxidation, and solid phase organometallic reactions. HPLC chromatograms for the synthesis of all peptides are in the Supporting Information (Part 2). (a) Representative reaction products from Hyp (1), including oxidation (entry 2 (91)), DAST reaction (entries 3 (26) and 5 (27) (via hyp)), Mitsunobu reaction (entries 4 (52), 7 (56)), sulfonation (entry 6 (5)), 3-step sequences via Mitsunobu reaction on hyp (entries 8 (43), 9 (16), 10 (41)), acylation plus SN2 (entry 11 (83)), acylation (entries 12 (64), 13 (79)), and phosphorylation (entry 14 (97)). Entry 5 (Flp) includes flp as a minor impurity in the chromatogram due to incomplete Mitsunobu reaction to form the hyp starting material for the DAST reaction. (b) Synthesis of 4S-acetoxy allylthioether (90) from Hyp (1) via Mitsunobu inversion to the hyp nitrobenzoate (2), azidemediated deesterification to hyp (4), acetylation to the bromoacetate (75), and SN2 reaction with allyl mercaptan. (c) Synthesis of the 4S-azide-alkyne cycloaddition product (109) from Hyp (1) via mesylation (5), SN2 reaction with sodium azide (21), and copper-mediated Huisgen cycloaddition. (d) Synthesis of the 4S-Sonogashira product (107) from Hyp (1) via Mitsunobu reaction with 4-iodophenol (37) and Pd-mediated cross-coupling with phenylacetylene.
Synthesis of hydoxyproline sulfonates and SN2 reactions
We examined the conversion of hydroxyproline-containing peptides to incorporate sulfonate leaving groups, for modification on solid phase or in solution (Scheme 7).19a, 21b, 21e, 21j, k, 31 Hydroxyproline residues were readily converted to include leaving groups including mesylates, tosylates, para-nosylates, and ortho-nosylates. These derivatives were stable to standard TFA cleavage conditions and isolable as the sulfonates (see below for structural analysis of the sulfonates and all derivatives). Attempts to synthesize the more reactive triflates and tresylates for solid phase SN2 reactions did not succeed and resulted in formation of multiple side products (data not shown).
Scheme 7.
Sulfonylation reactions on solid phase. Sulfonates were subsequently modified on solid phase (Scheme 8) or were subjected to TFA cleavage/deprotection and purified and analyzed in aqueous solution.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
The application of sulfonates for the synthesis of diverse 4-substituted prolines via SN2 reactions was examined with a series of nucleophiles (Scheme 8), including thiolate, azide, and iodide. These reactions were applied to generate the respective 4-substituted prolines, including the highly versatile 4-azidoprolines (4R (18) and 4S (21)) with good conversion.17o, p, 21f, 32 In addition, the azidoproline-containing peptides were converted to the ionizable aminoprolines (19, 22) and guanidinoprolines (20, 23) via reduction of the azide to the amine and guanylation.21l, 31d, 33 In general, the proline mesylates (5, 6) and para-nosylates (9, 10) were similarly effective for SN2 reactions on solid phase (see HPLC chromatograms in the Supporting Information), whereas the tosylates (7, 8) were less effective, resulting in poorer conversion and/or more side products.21j, k
Scheme 8.
Solid phase SN2 reactions on sulfonates and subsequent reactions on SN2 products.a
a RN = Ac-Thr(O tBu)-Tyr(O tBu)-, RC = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn- NH2.
Synthesis of 4S-substituted prolines via SN2 reaction involved a straightforward 2-step protocol of conversion of Hyp to a sulfonate followed by SN2 reaction on the sulfonate (Scheme 7 and Scheme 8). In contrast, synthesis of 4R-substituted prolines via SN2 chemistry as described above involved Mitsunobu inversion with nitrobenzoic acid, azide-mediated release of the nitrobenzoate ester, sulfonylation to generate a leaving group, and then SN2 reaction. While these 4-step sequences proceeded with high conversion in several cases, a shorter protocol to access 4R-subsituted prolines via SN2 chemistry would be preferable. Alcohols can be converted to halide leaving groups with inversion via Mitsunobu-like reactions with appropriate halide sources. Therefore, as proof of principle, we investigated the synthesis of the 4R-thiophenyl proline-containing peptide (16) via direct conversion of Hyp to 4S-bromoproline (25), followed by SN2 reaction with the thiophenolate (Scheme 9).31d, 34 Both reactions proceeded with good overall conversion, providing simpler two-step access to 4R-substituted prolines via SN2 reaction.
Scheme 9.

Alternative solid phase synthesis of 4R-substituted proline via an SN2 reaction on 4S- bromoproline.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Fluorination reactions
Fluoroprolines have been the most broadly employed 4-substituted proline derivatives after hydroxyproline, due to the strong stereoelectronic effect induced stereospecifically by fluorine substitution, which results in strong structural preferences (Figure 2).1l, m, 9, 16a, 17a-f, 17h-m, 35 Using the appropriate hydroxyprolines, both the 4R (Flp, 27) and 4S (flp, 26) fluoroproline-containing peptides were synthesized using conditions analogous to those developed for the solution phase synthesis of these amino acids (Scheme 10).36 The flp-containing peptide is readily accessible directly in one step from Hyp (Figure 6a, entry 3). While the Flp peptide requires a three-step peptide modification from Hyp, including two stereoinversions, it nonetheless proceeds with good conversion (Figure 6a, entry 5). Comparison of the NMR of the Flp-containing peptide synthesized via proline editing and the peptide synthesized using commercially available ($265/1 g) Fmoc-Flp-OH revealed identical NMR spectra, confirming that these reactions proceeded stereospecifically and without alpha-epimerization (see the Supporting Information for details).
Scheme 10.
Solid phase fluorination of Hyp and hyp to generate flp and Flp. a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Mitsunobu reactions
Mitsunobu reactions are among the most broadly employed reactions for the modification of alcohols, occurring with stereospecific inversion of stereochemistry.37 Mitsunobu reactions require a relatively acidic proton (pKa < 11) on the nucleophile, and thus were examined with carboxylic acids, phenols, thiophenols, selenophenol, phthalimide, hydroxyphthalimide, and thiolacetic acid (Scheme 11). Mitsunobu conditions were applied for the solid phase modification of both 4R- and 4S-hydroxyproline-containing peptides to generate a series of esters, ethers, and thioethers.
Scheme 11.
Mitsunobu reactions on solid phase.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Mitsunobu reactions of phenol derivatives to yield aryl prolyl ethers proceeded readily, including with the modestly sterically hindered 2,6-dimethylphenol and with the electron-deficient pentafluorophenol (Scheme 11, 29-38).21b, 21i, 38 Other aryl prolyl ethers synthesized included cyanophenyl, which could be used as a spectroscopic probe (IR or fluorescence, see below), and aryl halides (I, Br) for palladium-mediated cross-coupling reactions. These phenyl ethers could also be employed to tune the recognition properties of proline-containing peptides.21b, 21i, 38 While all other Mitsunobu reactions herein were conducted with DIAD, the Mitsunobu reactions with iodophenol and bromophenol both proceeded to higher conversion using the more reactive ADDP than they did with DIAD.
Thiophenol and selenophenol are larger chalcogen analogues of phenol. In addition to longer C-S and C-Se bonds and larger electron clouds, and thus greater sterics than oxygen, the thiophenyl and selenophenyl ethers are subject to modification by oxidation, and thus their properties tunable by oxidation. Mitsunobu reactions on solid phase with Hyp and hyp proceeded readily for 4-nitrothiophenol and selenophenol (derived from in situ reduction of diphenyl diselenide with sodium borohydride) (Scheme 11, 39-42).31b In contrast, Mitsunobu reaction with thiophenol only proceeded to high conversion for hyp, to generate the 4R phenyl thioether (16). The 4S phenyl thioether (17) was instead synthesized by SN2 reaction (Scheme 8), which was also alternatively used to synthesize the 4R phenyl thioether (Scheme 8 and Scheme 9).21b
4-Thioproline (4-mercaptoproline, Mpc (4R)/mpc (4S)) has recently been employed to prepare collagen mimetics that are readily modified by alkylation, oxidation, or pH-dependent structural switching.39 4R-thioproline has also been demonstrated to be highly effective in native chemical ligation reactions at proline.40 Given the range of structural, redox, nucleophilic, and metal-binding properties of cysteines and the emerging role of cysteine oxidation (disulfide, glutathionylation, sulfenic acid, sulfinic acid, S-nitrosylation) in intracellular post-translational modifications, 4-thioproline could potentially have broad applications if there were increased accessibility.21g, 28b, 41 4-Thioproline-containing peptides were readily synthesized via Mitsunobu reaction with thiolacetic acid, to generate the thioesters (43, 45), followed by deesterification to generate the free thiols (44, 46), which was conducted on solid phase and can also be effected in solution after TFA cleavage.21g, 28b
Mitsunobu reactions in general proceed poorly with amines. However, amines can be prepared via Mitsunobu reaction with phthalimide, followed by hydrazine deprotection. In addition, Mitsunobu reactions with phthalimide-protected hydroxylamines allow the introduction of aminoxy substitutents into molecules, with the possibility after hydrazine deprotection of a functional group capable of rapid chemoselective oxime formation.42 The phthalimide and hydroxyphthalimide Mitsunobu reactions proceeded readily on solid phase on both Hyp- and hyp- containing peptides to generate the desired products (Scheme 11, 47, 48, 49, 51).21h The hydroxyphthalimides were also deprotected on solid phase with hydrazine to generate after TFA cleavage the aminoxy peptides (50, 52). Notably, a previous synthesis of the protected Fmoc 4R-aminoxyproline required 9 steps of solution phase synthesis.42b
Halides are excellent leaving groups for substitution chemistry and also allow modulation of steric and stereoelectronic effects as a function of halogen size and electronegativity. The 4S-chloro, -bromo, and -iodo prolines (Scheme 11, 53, 25, 24) were all synthesized cleanly under Mitsunobu conditions and were stable to TFA cleavage and under aqueous conditions (Scheme 11).19a, 32a, 33b, 43 4S-Bromoproline was employed directly on solid phase for the SN2 reaction with thiophenol (Scheme 9, see above).31d, 34a 4S-Iodoproline was generated more cleanly under Mitsunobu conditions than by SN2 reaction with sodium iodide and 4R-hydroxyproline nosylate (above, Scheme 8).
Of particular interest was the application of the Mitsunobu reaction with perfluoro-tert-butanol (pKa 5) to incorporate a perfluoro-tert-butyl ether with either stereochemistry (55, 56).44 tert-Butyl groups have broad applications in medicinal chemistry and catalysis due to their sterics, hydrophobicity, and symmetry, which permits target binding with a reduced cost in conformational entropy. tert-Butyl groups have similar advantages in amino acids and peptides, with tert-leucine observed in pharmaceuticals and catalysts due to a strong conformational bias and steric effect.45 A perfluoro-tert-butyl group would be expected to have enhanced steric and hydrophobic effects over a tert-butyl group, while also introducing nine equivalent fluorines that would provide a strong singlet signal in 19F NMR, suggesting its possible use as a functional probe.1e, 1l, m, 44b, 46 Mitsunobu reactions of perfluoro-tert-butanol proceeded with good conversion on solid phase to generate both 4R- and 4S-perfluoro-tert-butylhydroxyproline ethers (55, 56) within peptides (Scheme 11). These reactions were more synthetically demanding than other Mitsunobu reactions, and proceeded substantially more effectively on lower loading resin (0.3-0.4 mmol/g) than on standard or higher loading resin (0.6-0.9 mmol/g). Other reactions that benefited from lower loading resin are specifically indicated as such in the Supporting Information.
Acylation reactions and reactions on acylates
Acylation of alcohols is a facile reaction that, combined with the large number of available carboxylic acids, could provide ready access to a wide range of substituted proline derivatives, including those that might induce significant stereoelectronic effects (due to the electron-withdrawing effects of the substituent) and those incorporated for the functional or recognition properties of the conjugated substituent. Hydroxyproline-containing peptides were readily acylated on solid phase using DIC/DMAP, providing access to peptides containing diverse functional groups. (Scheme 12). Acylation could be conducted on both Hyp and hyp peptides; alternatively, the formation of the 4S esters of hyp can be more directly conducted by Mitsunobu reaction on Hyp with the relevant carboxylic acids (Scheme 9).
Scheme 12.
Acylation reactions on solid phase. a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Acylation was applied to incorporate a broad range of functionalities, including modified proline residues with enhanced hydrophobic effect, proteinogenic side chain functional groups, reactive groups, and affinity ligands (Scheme 12). Hydrophobic groups added to proline include benzoates, acetates, and pivaloylate, which differ in hydrophobicity and in alkyl versus aryl functionality, and which could be used to optimize target binding (3, 57-61).47 Acylation was particularly effective for the incorporation of polar amino acid side chain functional groups. Functionalities added include ammonium (lysine mimetics (62, 63), via β-alanine), guanidinium (arginine mimetics (64, 65), via the guanidino acid33c, 33k, 48 of β-alanine), and carboxylates (aspartic/glutamic acid mimetics (66-70), via maleic anhydride, succinic anhydride, or glutaric anhydride).21e, 49 In addition, amino acids or peptides could be directly incorporated at the site of the hydroxyproline. This approach, via a β-alanine linker, allowed the incorporation of a cysteine residue (1,2 aminothiol functionality (73)), for native chemical ligation, and of an RGD peptide (72), for cell surface recognition.1h, 50
Acylation also permitted the incorporation of other reactive functional groups for subsequent further modification. Functionalities thus introduced via acylation reactions included α-bromo acetates (74, 75) for SN2 reactions, biotin (76, 77) for affinity recognition, maleimide (78) for Michael reaction with thiols or for Diels-Alder reactions, alkyne (79) for Huisgen [3+2] cycloaddition, NHS ester (80) for reaction with amine nucleophiles, acrylate (81) for polymerization, and tetrazine (82) for tetrazine-trans-cyclooctene ligation.51 These reactions were all conducted on solid phase, and the peptides containing reactive functionalities isolated in good yield after standard TFA cleavage/deprotection (typically using TFA/TIS/H2O and avoiding thiol additives such as ethanedithiol that could react with these functional groups, particularly maleimide, fumarate, alkyne, and NHS esters). The acrylate ester was particularly sensitive to reaction and cleavage/deprotection conditions, requiring oxygen and hydroquinone as inhibitors of polymerization; even under these conditions the acrylate exhibited evidence of polymerization and substantially lower overall desired product formation than other reactions, though the peptide was still isolable and characterized by NMR.
SN2 reactions are among the most versatile in their capability to stereospecifically incorporate a range of functional groups into molecules. The mesylate, tosylate, p-nosylate, o-nosylate, iodo, bromo, chloro, and bromoactetyl-substituted prolines thus represent a versatile range of derivatives for subsequent peptide modification. Bromoacetates exhibit enhanced SN2 reactivity due to the adjacent carbonyl. In addition, substitution at the primary carbon is faster and provides a spacer element between the proline and potential conjugates. As proof of principle for the application of the hydroxyproline bromoacetate as an alternative site of modification, we examined a series of SN2 reactions on this peptide on solid phase (Scheme 13). These reactions proceeded to high conversion with azide, alkyne, primary amine, aniline, and thiol nucleophiles (83-90). These reactions proceeded under milder conditions than those on proline 4-sulfonates (Scheme 8), with all reactions proceeding at room temperature, in contrast to elevated temperature and/or the presence of a crown ether in reactions on the corresponding proline-4-sulfonates. In addition, some nucleophiles that did not react cleanly or at all on the proline 4-sulfonates (phenyl acetylene, allyl mercaptan) proceeded with high conversion on the bromoacetates. The azide and allyl thioether acetates also subsequently reacted more rapidly in bioorthogonal ligation reactions than the equivalent derivatives directly substituted on proline (see below), presumably due to reduced steric hindrance. In sum, these data indicate that hydroxyproline bromoacetates are an effective alternative site for modification of proline residues with nucleophiles.
Scheme 13.
Substitution reactions on 4R- or 4S-hydroxyproline bromoacetate on solid phase. a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Oxidation reactions
Alcohols may be oxidized under a variety of conditions to the corresponding carbonyls. Following established solution conditions for oxidation of protected hydroxyprolines,36c the Hyp-containing peptide was readily oxidized to 4-oxoproline (91) (Scheme 14).31a 4-Oxoproline contains an interesting ketone electrophilic handle that is not normally present in proteins. Notably, ketone-containing acetophenone and benzophenone derivatives of phenylalanine have been exploited as electrophiles in ligation reactions in proteins.52
Scheme 14.
Oxidation reactions on 4-substituted prolines in TYXN peptides.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Oxidation reactions could also modulate the structure of 4-substituted proline derivatives, through either steric or stereoelectronic effects. The proline thiophenyl ethers could be selectively oxidized to either the sulfoxide or sulfone derivatives (Scheme 14, 92-95).21b The sulfoxides were obtained as an inseparable mixture of diastereomeric sulfoxides. The 4R-diastereomeric sulfoxides (92) were produced in an approximately 1:1 ratio and had apparently similar effects on cis/trans isomerism, though the NMR spectra exhibited distinct resonances in the amide region for the sulfoxide diastereomeric species. In contrast, the 4S-substituted thioether appeared to produce one sulfoxide diastereomer preferentially over the other (93), presumably due to the greater steric restraints present in the 4S diastereomer (Figure 3). Notably, the 4S-selenophenyl ether could be oxidized to generate the 3,4-dehydroproline (96) elimination product,31b which modulates the proline ring and main chain conformations and introduces an internal alkene to the peptide.19a, 53
Phosphorylation is a major intracellular post-translational modification of proteins.54 Phosphorylation of proteins can lead to protein-protein interactions, or alternatively or additionally may lead to protein structural changes, particularly within natively disordered regions of proteins.15a, 45g, 55 The TYPN peptide model system provides a context for potentially understanding the inherent stereoelectronic effects of phosphorylation, to provide a basis for understanding structural effects of protein phosphorylation. In addition, recent work has found that proteins may be phosphorylated and glycosylated on hydroxyproline residues, suggesting a richer range of post-translational modifications accessible in this residue.22 In order to address these questions, both 4R and 4S diastereomers of hydroxyproline were modified to the phosphorylated and to the diethylphosphate triester forms (Scheme 14, 97-100). Notably, phosphorylation ionization state changes as a function of pH, and therefore the structure of peptides with phosphorylated hydroxyproline could change as a function of pH.
Subjection of the 4-oxoproline-containing peptide to the fluorinating reagent DAST generated the 4,4-difluoroproline-containing peptide (101) (Scheme 15).17b, 21d, 36c 4,4-Difluoroproline is a noteworthy amino acid because it is conformationally similar to Pro, exhibiting a mixture of exo and endo ring puckers, while reducing the activation barrier for cistrans isomerism.17c Thus, given that cis-trans isomerism is commonly a rate-determining step in protein folding, peptides or proteins containing 4,4-difluoroproline could exhibit enhanced rates of protein folding. 4,4-Difluoroproline also could be exploited for a potentially enhanced hydrophobic effect in proteins without significantly changing the inherent conformational preferences of the protein.1e, 1l, m 4,4-Difluoroproline also introduces two 19F atoms as potential NMR probes of protein folding and function (see below). Notably, 4,4-difluoroproline has been incorporated in expressed proteins in proline auxotrophs, although the 19F NMR spectra of these proteins have not been reported.17c, 18b
Scheme 15.
Synthesis of disubstituted prolines.a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
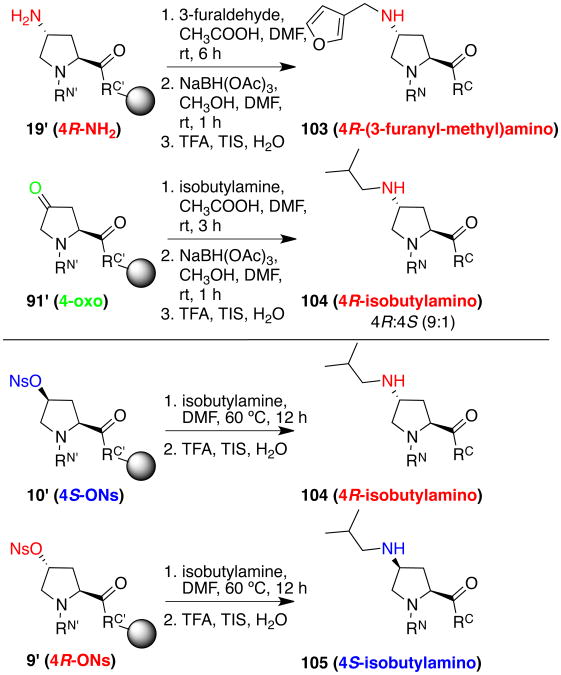
Ketones are otherwise exploitable as chemical handles in peptides. Peptide cleavage/deprotection of the 4-oxoproline-containing peptide in the presence of the typical thiol scavenger ethanedithiol resulted in isolation in high yield of the 4,4-dithiolane-containing peptide (102) (Scheme 16).21b, 21d Notably, the proline 4,4-dithiolane amino acid is part of the ACE inhibitor drug Spirapril.21c This reaction could also potentially be exploited to conjugate other thiols into 4-oxoproline-containing peptides. Alternatively, the 4-oxoproline-containing peptide (91) was cleanly obtained using TIS and water as scavengers.
Scheme 16.
Reductive amination on 4R-aminoproline and 4-oxoproline. The stereochemistry of the oxoproline reductive amination products, suggested by 1H NMR spectra to indicate 4R (major) versus 4S (minor) stereochemistry, was confirmed by SN2 reactions of isobutyl amine on the respective nosylates. a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
Ketones are readily converted to amines using reductive amination. Reductive amination was accomplished on the 4-oxoproline-containing peptide on solid phase using isobutyl amine and sodium triacetoxyborohydride (Scheme 16), generating the substituted 4-aminoproline as a mixture of diastereomers (104, 105), with the 4R diastereomer the major diastereomer (9:1 4R:4S). The stereochemistry of the major and minor diastereomers obtained by reductive amination was confirmed by the stereospecific synthesis of both diastereomers via SN2 reactions. Alternatively, 4-aminoproline could react with an aldehyde on the solid phase to generate a substituted 4-aminoproline (103). Reductive amination provides a handle to incorporate both amines and the functional groups associated with the amine.
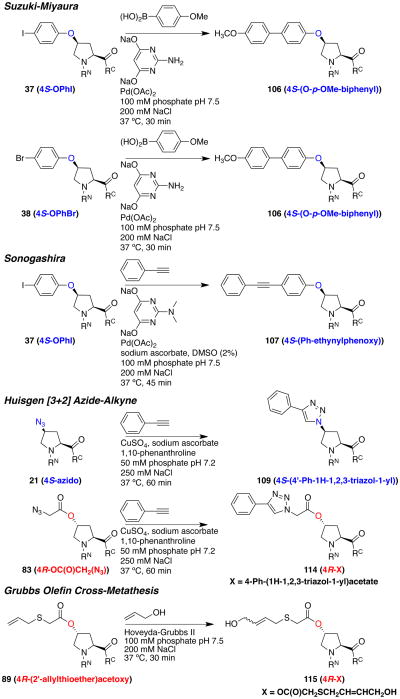
Organometallic reactions on solid phase
The derivatives described above include a range of chemical handles capable of application in organometallic reactions, including azides, alkenes, alkynes, and aryl halides. We initially examined the exploitation of these groups for peptide modification on solid phase (Scheme 17). The 4S-hydroxyproline bromophenyl ether was subjected to standard conditions for the Suzuki reaction, resulting in facile generation of the cross-coupled product (106) of reaction with the boronic acid of p-anisole. Similarly, the 4S-hydroxyproline iodophenyl ether readily underwent a Sonogashira reaction with phenyl acetylene on solid phase to generate the disubstituted alkyne (107). The azidoproline-containing peptides also reacted on solid phase with phenyl acetylene to form either the 4- or 5-substituted triazoles (108, 109, 112, 113), depending on the use of copper or ruthenium catalyst.51e, 56 The azidoproline peptides also readily underwent copper-mediated azide-alkyne coupling with 1-ocytne (110, 111). These approaches may be useful for solid phase diversification of peptide ligands in medicinal chemistry and other applications.38
Scheme 17.
Organometallic reactions on solid phase. Peptides were subjected to TFA cleavage/deprotection, purified, and characterized. Chromatograms for the multistep synthesis of 107 are in Figure 6d. Chromatograms for the multistep synthesis of 109 are in Figure 6ca
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Solution phase and bioorthogonal modification of peptides using proline derivatives
Unnatural amino acids containing unique functional groups can introduce reactivity not present within canonical amino acids.57 Aryl halides, azides, alkynes, alkenes, 1,2-aminothiols, maleimides, tetrazines, and hydroxylamines incorporated via proline editing thus provide a diverse and complementary toolkit for the site-specific modification of peptides and proteins within defined structural contexts. In order to examine the possible application of these 4-substituted proline residues in bioorthogonal ligations, these derivatives were subjected to established solution reaction conditions.
Organometallic reactions provide unique selectivity in modification at distinct functional groups. In particular, palladium-mediated cross-coupling reactions have achieved preeminent status due to high chemoselectivity under highly diverse reaction conditions. Peptides containing proline iodophenyl and bromophenyl ethers were examined under conditions developed by Davis and Lin for aqueous Suzuki and Sonogashira cross-coupling reactions (Scheme 18).58 These palladium-catalyzed reactions proceeded rapidly (30-45 minutes reaction at 37 °C) and cleanly to generate products (106, 107) with high conversion in mild, neutral aqueous conditions. Suzuki-Miyaura reactions proceeded effectively with both the aryl bromide and the aryl iodide under these conditions, as did the palladium-catalyzed Sonogashira reaction with the aryl iodide, using the alternative dimethylamino ligand.
Scheme 18.
Organometallic reactions on peptides in aqueous solution. Bottom: HPLC chromatograms of reactions. For each, top panel: starting material used for that reaction, bottom panel: crude HPLC of reaction products. a
a RN′ = Ac-Thr(OtBu)-Tyr(OtBu)-, RC′ = -Asn(Trt)-NHRink-Resin. RN = Ac-Thr-Tyr-, RC = - Asn-NH2.
As expected, both the 4-azidoproline and the 4-azidoacetate of hydroxyproline reacted readily with the model alkyne phenyl acetylene in the [3+2] Huisgen azide-alkyne coupling under copper-mediated conditions (109, 114).17o, p, 32c, 59 In general, the azidoacetate reacted more rapidly than the azidoproline, likely due to reduced steric restriction, though both were effective in azide-alkyne coupling reactions.
Alkenes provide an alternative, orthogonal handle for chemical reactivity. Allylic groups substituted with chalcogens provide enhanced reactivity in cross-metathesis reactions, with reactivity Se > S > O.60 The allyl thioether acetate of hydroxyproline reacted rapidly (30 minutes, 37 °C) in cross-metathesis with allyl alcohol in water (115), in a manner similar to reactions previously described with S-allyl cysteine, O-allyl tyrosine, and S-allyl-thiophenylalanine.28b, 60
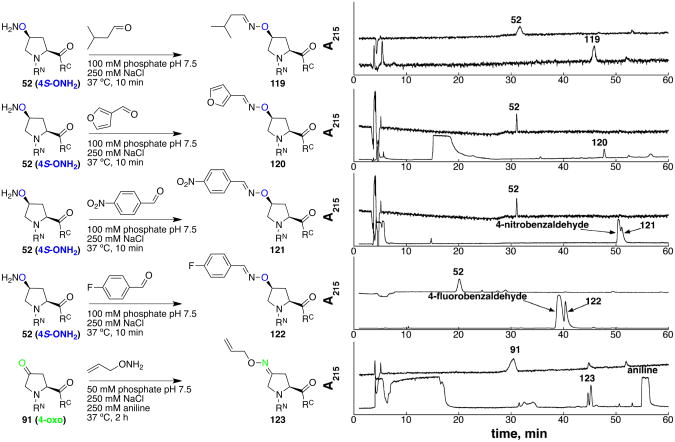
Bioorthogonal reactions that do not require a metal catalyst have particular value in intracellular and extracellular labeling and in situations where removal of metal catalysts would be problematic.57, 61 Four types of reactions were examined for peptide modification at proline under metal-free conditions: native chemical ligation reaction via 1,2-aminothiol (116), Diels-Alder reaction via maleimide (117), inverse electron demand tetrazine-trans-cyclooctene cycloaddition via conjugated tetrazine (118) (Scheme 19), and oxime reactions (Scheme 20) via 4-aminoxyproline (119-122) or via 4-oxoproline (123).42, 51a-d, 62 All model reactions proceeded cleanly in high conversion. The oxime reaction with 4-oxoproline proceeded more slowly and required aniline as a nucleophilic catalyst to achieve high conversion (123).42a The fastest reactions in this group were tetrazine-trans-cyclooctene ligations (118) and oxime reactions between aldehydes and 4S-aminoxyproline (119-122). The 4S-aminoxyproline reactions proceeded without added nucleophilic catalyst and allowed the rapid incorporation of diverse functional substituents, including 4-fluoro-benzaldehyde (122), which has been synthesized in one step in 18F-labeled form and subsequently incorporated via oxime chemistry in RGD peptides, hormone peptides, the hormone protein leptin, and a ligand of the CXCR4 chemokine receptor for application in 18F-positron emission tomography (PET).63
Scheme 19.
Bioorthogonal ligation reactions in aqueous solution. Bottom: HPLC chromatograms of reactions. For each, top panel: starting material used for that reaction, bottom panel: crude HPLC of reaction products. For 116 the amide product was distinguished from the thioester product based on the presence a new amide proton in the NMR spectrum. See the Supporting Information for details. Compound 117 could be the Diels-Alder endo and/or exo diastereomeric products (Diels-Alder stereochemistry not determined). Compound 118 results from isomerization of an initial 1,3-diene product.51a RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Scheme 20.
Oxime reactions in aqueous solution. Right: HPLC chromatograms of reactions. For each, top panel: starting material used for that reaction, bottom panel: crude HPLC of reaction products. The reaction with oxoproline to synthesize 123 generates two products that were indistinguishable by NMR and mass spectrometry, presumably the E and Z oximes. RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
The range of functionalities incorporated in 4-substituted prolines provides the possibility of performing multiple bioorthogonal ligations in parallel, with potential future applications of incorporating multiple modifications within a single protein.62b, 64 As proof of principle, peptides containing tetrazine, hydroxylamine, and maleimide functionalities were incubated in a single solution with the reactive partners trans-cyclooctene, isovaleraldehyde, and cyclopentadiene. These reactive functionalities ligated selectively and rapidly (30 minutes) in aqueous solution in good yield with their reactive partners in the presence of the alternative reactive groups. Notably, all of these reactions are metal-free. These data suggest that substituted prolines could be broadly applied as sites of multiple selective ligations within peptides and proteins. In this context, it is noteworthy that proline residues commonly occur in loops and turns and in natively disordered sequences, and are thus often at or near the surface or proteins.5g, 65
Structural analysis of the stereospecific effects of proline substitution in model peptides
Stereospecific substitutions on proline residues can significantly impact peptide and protein conformation (Figures 2, 3, 4). 4R-Hydroxylation (Hyp) of collagen at the Yaa position is obligatory for collagen stability and function, and 4R-fluorination (Flp) provides enhanced stability in collagen model peptides. In contrast, 4S-hydroxylation (hyp) or 4S-fluorination (flp) at the same position dramatically destabilizes collagen model peptides, due to the stereoelectronic preference for an endo ring pucker in these derivatives.8-9, 17a, 17d-f, 17h The magnitude of stereoelectronic effects on conformation allows modulation of the stability of collagen and other proline-containing proteins. However, stereoelectronic effects can be counteracted by steric effects. For example, 4S-methyl proline may substitute effectively at the Yaa position of collagen model peptides because this substitution leads to a steric preference for an anti relationship between the methyl substituent and the amide about the Cγ-Cδ bond, and thus a preference for an exo ring pucker (Figure 2).7a, 11, 66 Thus, the interplay between steric and stereoelectronic effects provides alternative approaches to tune proline side chain and main chain conformation.
The derivatives described above represent a continuum of proline substitutions, representing both steric and stereoelectronic effects modulated by substitution, with both relative stereochemistries. Numerous proline derivatives have been analyzed for their conformational effects as the methyl esters of acetylated amino acids.8-9,11,17a-c,o,39,43,47,66,70 Many of these substituted prolines have also been examined in collagen model peptides, where multiple copies of the proline derivative are incorporated in the collagen triple helix (3-30 proline substitutions, depending on single site (host-guest) modification (three copies via one copy in each strand of the triple helix) or global substitution at the Xaa or Yaa site in collagen model peptides ((ProHypGly)n, n = 7-10) (3 × n = 21-30 substituted prolines)). The Ac-TYPN-NH2 peptide is an intermediate structural context, in which substitutions can impact the conformation of adjacent amino acids without impacting tertiary structure, and thus complements the limiting cases of isolated amino acids (Ac-Prox-OMe) and the tertiary structure of a collagen triple helix (e.g. (ProProxGly)7-10 or (ProxProGly)7-10, where Prox = 4-substituted proline). These peptides were analyzed by NMR spectroscopy (Figure 7, Table 1, Table 2, Table 3, and Supporting Information) to provide a readout of stereoelectronic versus steric effects for proline substituents, revealing additional context on the conformational effects of substitution for established proline derivatives and new data on the conformational effects of proline substitution for a wide range of previously unsynthesized or unstudied proline derivatives.
Figure 7.
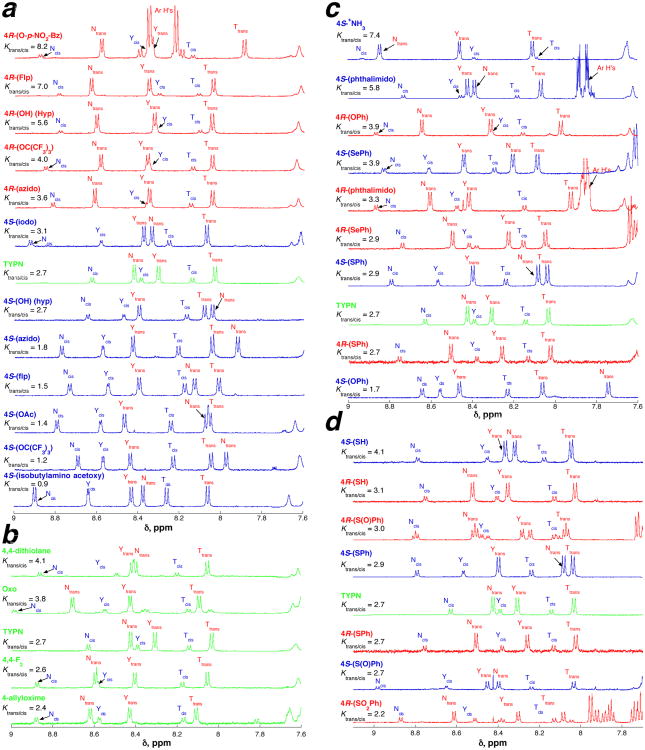
Representative 1H NMR spectra (5 mM phosphate (pH 4 unless an ionizable proton is present), 25 mM NaCl, 90% H2O/10% D2O) of Ac-TYP(4-substituted)N-NH2 peptides with 4-substituted prolines, with species with trans and cis amide bonds and Ktrans/cis indicated. The largest differences between 4R- versus 4S-substituted peptides are observed in Ktrans/cis, the δ of Tyrcis and Asntrans, and the 3JαN of Tyrcis. These effects are modulated as a function of the nature of the substituent with the 4R or 4S series. (a) Stereoelectronic effects of 4-substituents. (b) Disubstituted prolines. The NMR spectrum of 4-oxo-proline also includes the hydrate, which is in equilibrium with the ketone (Khydrate/ketone = 0.15).67b (c) Steric effects of 4-substituted prolines and the interplay of steric versus stereoelectronic effects in chalcogen-substituted prolines. (d) Steric effects versus stereoelectronic effects as a result of sulfur oxidation. NMR spectra for all peptides are in the Supporting Information (part 1: amide regions of the NMR spectra for all peptides; part 3: full NMR spectra for all peptides).
Table 1.
Ktrans/cis and NMR data for Ac-TYXN-NH2 peptides in aqueous solution (25 mM NaCl, 5 mM phosphate (pH 4 unless otherwise indicated), 90% H2O/10% D2O) at 25 °C. Ktrans/cis = ratio of peptide with trans amide bond to peptide with cis amide bond as determined by NMR. ΔGtrans/cis = −RT ln Ktrans/cis. ΔΔGtrans/cis = ΔGtrans/cis (peptide) − ΔGtrans/cis (Pro). n.d. = not determined due to spectral overlap. 3JαN = coupling constant between HN and Hα, which can be correlated to φ via a parametrized Karplus relationship.69 All NMR spectra are in the Supporting Information (amide region, Part 1; full NMR spectra, Part 3).
| Ac-TYP(X)N-NH2 X = | Ktrans/cis | ΔGtrans/cis | ΔΔGtrans/cis | 3JαN | δ, HN | δ, HN |
|---|---|---|---|---|---|---|
| kcal mol-1 | kcal mol-1 | Tyrcis | Tyrcis | Asntrans | ||
|
|
8.2 | -1.25 | -0.66 | n.d. | n.d. | 8.57 |
|
|
8.2 | -1.25 | -0.66 | n.d. | n.d. | 8.58 |
|
|
7.5 | -1.19 | -0.61 | 6.0 | 8.43 | 8.92 |
|
|
7.0 | -1.15 | -0.56 | 7.7 | 8.26 | 8.73 |
|
|
7.0 | -1.15 | -0.56 | 7.7 | 8.30 | 8.63 |
|
|
6.7 | -1.13 | -0.54 | n.d. | 8.41 | 8.58 |
|
|
6.6 | -1.12 | -0.53 | 7.6 | 8.36 | 8.07 |
|
|
6.5 | -1.11 | -0.52 | 6.6 | 8.45 | 8.59 |
|
|
6.4 | -1.10 | -0.51 | n.d. | 8.33 | 8.61 |
|
|
6.3 | -1.09 | -0.50 | n.d. | 8.31 | 8.67 |
|
|
5.8 | -1.04 | -0.45 | 6.5 | 8.45 | 8.42 |
|
|
5.8 | -1.04 | -0.45 | 7.7 | 8.33 | 8.60 |
|
|
5.8 | -1.04 | -0.45 | 7.8 | 8.38 | 8.66 |
|
|
5.8 | -1.04 | -0.45 | n.d. | 8.30 | 8.54 |
|
|
5.7 | -1.03 | -0.44 | n.d. | 8.35 | 8.54 |
|
|
5.7 | -1.03 | -0.44 | n.d. | 8.32 | 8.72 |
|
|
5.6 | -1.02 | -0.43 | 7.5 | 8.34 | 8.59 |
|
|
5.3 | -0.99 | -0.40 | 7.3 | 8.35 | 8.59 |
|
|
5.1 | -0.96 | -0.38 | n.d. | 8.32 | 8.80 |
|
|
5.1 | -0.96 | -0.38 | 7.4 | 8.47 | 8.60 |
|
|
5.1 | -0.96 | -0.38 | n.d. | 8.38 | 8.66 |
|
|
5.1 | -0.96 | -0.38 | n.d. | 8.35 | 8.64 |
|
|
5.1 | -0.96 | -0.38 | 7.2 | 8.34 | 8.61 |
|
|
5.0 | -0.95 | -0.36 | 7.4 | 8.38 | 8.62 |
|
|
4.9 | -0.94 | -0.35 | n.d. | 8.31 | 8.71 |
|
|
4.7 | -0.92 | -0.33 | 6.0 | 8.30 | 8.73 |
|
|
4.7 | -0.92 | -0.33 | 7.3 | 8.33 | 8.59 |
|
|
4.6 | -0.90 | -0.32 | 7.2 | 8.34 | 8.59 |
|
|
4.5 | -0.89 | -0.30 | n.d. | 8.38 | 8.61 |
|
|
4.5 | -0.89 | -0.30 | 7.2 | 8.33 | 8.67 |
|
|
4.4 | -0.88 | -0.29 | 7.4 | 8.31 | 8.57 |
|
|
4.3 | -0.86 | -0.28 | 6.0 | 8.47 | 8.63 |
|
|
4.3 | -0.86 | -0.28 | n.d. | 8.25 | 8.57 |
|
|
4.2 | -0.85 | -0.26 | n.d. | 8.34 | 8.61 |
|
|
4.2 | -0.85 | -0.26 | 7.9 | 8.56 | 8.77 |
|
|
4.1 | -0.84 | -0.25 | 6.0 | 8.46 | 8.33 |
|
|
4.1 | -0.84 | -0.25 | 4.9 | 8.96 | 8.26 |
|
|
4.1 | -0.84 | -0.25 | 7.2 | 8.31 | 8.61 |
|
|
4.1 | -0.84 | -0.25 | n.d. | 8.36 | 8.66 |
|
|
4.1 | -0.84 | -0.25 | 6.1 | 8.48 | 8.41 |
|
|
4.0 | -0.82 | -0.23 | n.d. | 8.35 | 8.58 |
|
|
3.9 | -0.81 | -0.22 | 8.2 | 8.15 | 8.64 |
|
|
3.9 | -0.81 | -0.22 | 4.9 | 8.61 | 8.20 |
|
|
3.9 | -0.81 | -0.22 | 5.2 | 8.55 | 8.14 |
|
|
3.8 | -0.79 | -0.20 | 5.7 | 8.56 | 8.72 |
|
|
3.8 | -0.79 | -0.20 | n.d. | 8.33 | 8.67 |
|
|
3.7 | -0.77 | -0.19 | 6.6 | 8.33 | 8.61 |
|
|
3.6 | -0.76 | -0.17 | n.d. | 8.32 | 8.60 |
|
|
3.6 | -0.76 | -0.17 | 5.6 | 8.53 | 8.19 |
|
|
3.5 | -0.74 | -0.15 | 7.8 | 8.33 | 8.30 |
|
|
3.5 | -0.74 | -0.15 | n.d. | 8.32 | 8.62 |
|
|
3.4 | -0.72 | -0.14 | n.d. | 8.32 | 8.64 |
|
|
3.4 | -0.72 | -0.14 | n.d. | 8.38 | 8.66 |
|
|
3.3 | -0.71 | -0.12 | 7.6 | 8.44 | 8.60 |
|
|
3.2 | -0.69 | -0.10 | n.d. | 8.30 | 8.58 |
|
|
3.2 | -0.69 | -0.10 | n.d. | 8.31 | 8.61 |
|
|
3.2 | -0.69 | -0.10 | 5.2 | 8.54 | 8.39 |
|
|
3.1 | -0.67 | -0.08 | 6.8 | 8.40 | 8.52 |
|
|
3.1 | -0.67 | -0.08 | 4.2 | 8.62 | 7.98 |
|
|
3.1 | -0.67 | -0.08 | 4.2 | 8.58 | 8.32 |
|
|
3.0 | -0.65 | -0.06 | 5.0 | 8.54 | 8.08 |
|
|
3.0 | -0.65 | -0.06 | n.d. | 8.53 | 8.37 |
|
|
2.9 | -0.63 | -0.04 | 6.2 | 8.40 | 8.48 |
|
|
2.9 | -0.63 | -0.04 | 4.0 | 8.56 | 8.03 |
|
|
2.8 | -0.61 | -0.02 | 6.6 | 8.40 | 8.57 |
|
|
2.8 | -0.61 | -0.02 | 5.3 | 8.56 | 8.10 |
|
|
2.8 | -0.61 | -0.02 | 6.9 | 8.33 | 8.68 |
| H (Pro) | 2.7 | -0.59 | 0.00 | 6.1 | 8.37 | 8.40 |
|
|
2.7 | -0.59 | 0.00 | n.d. | 8.36 | 8.67 |
|
|
2.7 | -0.59 | 0.00 | 6.4 | 8.42 | 8.52 |
|
|
2.7 | -0.59 | 0.00 | 6.2 | 8.40 | 8.52 |
|
|
2.7 | -0.59 | 0.00 | 4.2 | 8.65 | 8.40 |
|
|
2.7 | -0.59 | 0.00 | 4.9 | 8.49 | 8.08 |
|
|
2.6 | -0.57 | 0.02 | n.d. | 8.56 | 8.72 |
|
|
2.4 | -0.52 | 0.07 | 7.2 | 8.42 | 8.57 |
|
|
2.4 | -0.52 | 0.07 | 5.6 | 8.51 | 8.10 |
|
|
2.4 | -0.52 | 0.07 | 4.1 | 8.55 | 7.74 |
|
|
2.3 | -0.49 | 0.09 | 5.3 | 8.49 | 8.08 |
|
|
2.2 | -0.47 | 0.12 | 5.4 | 8.56 | 8.01 |
|
|
2.2 | -0.47 | 0.12 | 5.5 | 8.52 | 8.61 |
|
|
2.1 | -0.44 | 0.15 | 4.6 | 8.60 | 8.04 |
|
|
2.1 | -0.44 | 0.15 | 5.4 | 8.58 | 7.72 |
|
|
2.1 | -0.44 | 0.15 | 5.6 | 8.48 | 8.02 |
|
|
2.0 | -0.41 | 0.18 | 4.2 | 8.60 | 8.06 |
|
|
2.0 | -0.41 | 0.18 | 4.5 | 8.58 | 7.97 |
|
|
2.0 | -0.41 | 0.18 | 5.4 | 8.58 | 8.27 |
|
|
1.9 | -0.38 | 0.21 | 4.8 | 8.54 | 7.72 |
|
|
1.9 | -0.38 | 0.21 | 4.4 | 8.56 | 7.70 |
|
|
1.9 | -0.38 | 0.21 | 5.4 | 8.53 | 8.18 |
|
|
1.9 | -0.38 | 0.21 | 5.4 | 8.54 | 8.09 |
|
|
1.8 | -0.35 | 0.24 | 4.8 | 8.57 | 7.77 |
|
|
1.8 | -0.35 | 0.24 | 4.8 | 8.58 | 7.73 |
|
|
1.8 | -0.35 | 0.24 | 5.4 | 8.56 | 8.05 |
|
|
1.8 | -0.35 | 0.24 | 5.3 | 8.56 | 8.17 |
|
|
1.8 | -0.35 | 0.24 | 4.2 | 8.57 | 7.91 |
|
|
1.8 | -0.35 | 0.24 | 4.1 | 8.57 | 8.07 |
|
|
1.7 | -0.31 | 0.27 | 4.7 | 8.60 | 7.75 |
|
|
1.7 | -0.31 | 0.27 | 4.6 | 8.56 | 8.10 |
|
|
1.5 | -0.24 | 0.35 | 4.8 | 8.56 | 7.72 |
|
|
1.5 | -0.24 | 0.35 | 4.9 | 8.58 | 8.16 |
|
|
1.5 | -0.24 | 0.35 | 4.8 | 8.62 | 8.07 |
|
|
1.4 | -0.20 | 0.39 | 4.2 | 8.59 | 8.07 |
|
|
1.4 | -0.20 | 0.39 | 5.4 | 8.49 | 7.91 |
|
|
1.4 | -0.20 | 0.39 | 4.8 | 8.59 | 8.06 |
|
|
1.4 | -0.20 | 0.39 | 5.2 | 8.57 | 8.31 |
|
|
1.3 | -0.16 | 0.43 | 3.7 | 8.58 | 8.06 |
|
|
1.3 | -0.16 | 0.43 | 4.5 | 8.60 | 8.06 |
|
|
1.3 | -0.16 | 0.43 | 4.3 | 8.60 | 8.21 |
|
|
1.3 | -0.16 | 0.43 | 4.4 | 8.53 | 8.18 |
|
|
1.2 | -0.11 | 0.48 | 4.7 | 8.59 | 8.24 |
|
|
1.0 | 0.00 | 0.59 | 6.0 | 8.47 | 8.05 |
|
|
1.0 | 0.00 | 0.59 | 4.4 | 8.57 | 8.05 |
|
|
0.9 | 0.06 | 0.65 | 4.2 | 8.62 | 8.24 |
|
| ||||||
|
|
all trans | n.d. | n.d. | 2.5 | 8.34 | 8.33 |
|
|
7.2 | -1.17 | -0.58 | 8.4 | 8.53 | 8.67 |
Table 2.
Overall functional group effects on cis-trans isomerism in Ac-TYXN-NH2 peptides. ΔΔG = ΔGtrans/cis(4R stereoisomer) − ΔGtrans/cis(4S stereoisomer), with negative ΔG consistent with the magnitude of stereoelectronic effects and positive ΔG indicating steric effects dominating over stereoelectronic effects. By definition, for Pro (4R = 4S = H), ΔΔG = 0.00.
| Ac-TYP(4-X)N-NH2 X = | ΔΔG(4R-4S) kcal mol-1 |
|---|---|
| ONH3+ | -1.13 |
| isobutylaminoacetoxy | -0.93 |
| β-Alanyl ester | -0.92 |
| F | -0.91 |
| OBzNO2 | -0.90 |
| OC(O)CH2Br | -0.88 |
| OBzCF3 | -0.87 |
| hydroxyphthalimide | -0.81 |
| OAc | -0.76 |
| O-pivaloyl | -0.76 |
| OC(O)CH2N3 | -0.74 |
| O-(2,6-(CH3)2Ph) | -0.71 |
| OC(CF3)3 | -0.71 |
| succinyl ester | -0.68 |
| allylthioetheracetoxy | -0.64 |
| OPO3H- | -0.60 |
| OMs | -0.60 |
| OC6F5 | -0.56 |
| OPh | -0.50 |
| maleinyl ester | -0.49 |
| β-Ala-guanidinyl ester | -0.45 |
| OPO3Et2 | -0.44 |
| OH | -0.43 |
| azido | -0.41 |
| OPhCN | -0.41 |
| OMst | -0.39 |
| ONs | -0.33 |
| guanidinyl | -0.31 |
| 5′-Ph-(1H-1,2,3-triazol-1-yl) | -0.28 |
| OTs | -0.17 |
| S(O)Ph | -0.06 |
| 4′-Ph-(1H-1,2,3-triazol-1-yl) | -0.04 |
| SPh | 0.04 |
| SPhNO2 | 0.06 |
| SAc | 0.06 |
| 4′-hexyl-(1H-1,2,3-triazol-1-yl) | 0.15 |
| SH | 0.17 |
| SePh | 0.18 |
| SO2Ph | 0.22 |
| phthalimide | 0.33 |
| +NH3 | 0.33 |
Table 3.
NMR data for ionizable derivatives as a function of pH and protonation state. Expected major protonation state at the given pH is indicated.
| Ac-TYP(X)N-NH2 X = | Ktrans/cis | ΔG | ΔΔG | 3J αN | δ, HN | δ, HN | pH |
|---|---|---|---|---|---|---|---|
| kcal mol-1 | kcal mol-1 | Tyrcis | Tyrcis | Asntrans | |||
|
|
4.3 | -0.86 | -0.28 | 7.0 | 8.46 | 8.62 | 2.0 |
|
|
7.5 | -1.19 | -0.60 | 6.0 | 8.43 | 8.92 | 2.0 |
|
|
4.2 | -0.85 | -0.26 | 5.4 | 8.47 | 8.63 | 4.0 |
|
|
7.4 | -1.18 | -0.60 | 6.6 | 8.41 | 8.91 | 4.0 |
|
|
4.0 | -0.82 | -0.23 | 6.6 | 8.44 | 8.61 | 6.3 |
|
|
7.3 | -1.18 | -0.59 | 6.1 | 8.45 | 8.93 | 6.3 |
|
|
3.9 | -0.81 | -0.22 | n.d. | n.d. | n.d. | 7.6 |
|
|
6.8 | -1.13 | -0.55 | n.d. | n.d. | n.d. | 7.6 |
|
|
4.1 | -0.84 | -0.25 | n.d. | n.d. | n.d. | 8.5 |
|
|
6.2 | -1.08 | -0.49 | n.d. | n.d. | n.d. | 8.5 |
|
| |||||||
| Ac-TYP(X)N-NH2 X = | Ktrans/cis | ΔG | ΔΔG | 3J αN | δ, HN | δ, HN | pH |
| kcal mol-1 | kcal mol-1 | Tyrcis | Tyrcis | Asntrans | |||
|
| |||||||
|
|
6.4 | -1.10 | -0.51 | n.d. | 8.32 | 8.57 | 2.0 |
|
|
1.0 | 0.00 | 0.59 | 4.4 | 8.57 | 8.05 | 2.0 |
|
|
5.9 | -1.05 | -0.46 | n.d. | 8.32 | 8.61 | 4.0 |
|
|
1.4 | -0.20 | 0.39 | 4.4 | 8.58 | 7.83 | 4.0 |
|
|
5.5 | -1.01 | -0.42 | n.d. | 8.33 | 8.61 | 5.6 |
|
|
1.4 | -0.20 | 0.39 | 4.4 | 8.58 | 7.83 | 5.6 |
|
|
3.2 | -0.69 | -0.10 | n.d. | n.d. | n.d. | 7.6 |
|
|
1.5 | -0.24 | 0.35 | n.d. | n.d. | n.d. | 7.6 |
|
|
2.7 | -0.59 | 0.00 | n.d. | n.d. | n.d. | 10.0 |
|
|
1.4 | -0.20 | 0.39 | n.d. | n.d. | n.d. | 10.0 |
|
| |||||||
| Ac-TYP(X)N-NH2 X = | Ktrans/cis | ΔG | ΔΔG | 3J αN | δ, HN | δ, HN | pH |
| kcal mol-1 | kcal mol-1 | Tyrcis | Tyrcis | Asntrans | |||
|
| |||||||
|
|
6.3 | -1.09 | -0.50 | n.d. | 8.31 | 8.67 | 4.0 |
|
|
2.3 | -0.49 | 0.10 | 5.3 | 8.49 | 8.08 | 4.0 |
|
|
6.0 | -1.06 | -0.47 | n.d. | 8.30 | 8.66 | 5.6 |
|
|
2.2 | -0.47 | 0.12 | 6.0 | 8.47 | 8.14 | 5.6 |
|
|
5.9 | -1.05 | -0.46 | 7.9 | 8.20 | 8.68 | 6.8 |
|
|
2.9 | -0.63 | -0.04 | 6.6 | 8.45 | 8.21 | 6.8 |
|
|
6.7 | -1.13 | -0.54 | n.d. | n.d. | n.d. | 8.5 |
|
|
2.9 | -0.63 | -0.04 | n.d. | n.d. | n.d. | 8.5 |
|
| |||||||
| Ac-TYP(X)N-NH2 X = | Ktrans/cis | ΔG | ΔΔG | 3J αN | δ, HN | δ, HN | pH |
| kcal mol-1 | kcal mol-1 | Tyrcis | Tyrcis | Asntrans | |||
|
| |||||||
|
|
3.1 | -0.67 | -0.08 | 6.8 | 8.40 | 8.52 | 4.0 |
|
|
4.1 | -0.84 | -0.25 | 6.0 | 8.46 | 8.33 | 4.0 |
|
|
3.2 | -0.69 | -0.10 | 7.1 | 8.41 | 8.52 | 6.8 |
|
|
4.6 | -0.90 | -0.32 | 7.0 | 8.46 | 8.33 | 6.8 |
|
|
2.8 | -0.61 | -0.02 | - | - | - | 7.6 |
|
|
4.4 | -0.88 | -0.29 | - | - | - | 7.6 |
|
|
3.0 | -0.65 | -0.06 | - | - | - | 8.5 |
|
|
4.4 | -0.88 | -0.29 | - | - | - | 8.5 |
In addition to providing data on the conformational effects of proline substitution, the NMR spectroscopy also confirms that the reactions proceeded stereospecifically, as expected. As noted above, one method used to confirm that sequences of multiple reactions proceeded stereospecifically was the conversion of Ac-TYHypN-NH2 to Ac-TYhypN-NH2 (stereospecific Mitsunobu inversion) and the subsequent modification of the hyp-containing peptide with DAST on solid phase to generate Flp (Scheme 6, Scheme 10). The NMR spectrum of the peptide containing Flp synthesized via proline editing was identical to that of the peptide containing Flp synthesized using commercial Fmoc-Flp-OH, confirming that the two sequential stereospecific inversions had proceeded with expected stereochemistry and more generally indicating that all peptides derived from both Hyp and hyp were originating from the expected stereochemistry.
In general, the NMR spectra of peptides with the same stereochemistry and similar electronics were relatively comparable in global appearance, while peptides with the same substituent but opposite stereochemistry were divergent in appearance, indicating that the main effects of modification were determined by the stereochemistry and electronics of the 4-substitutent (Figure 7 and Supporting Information). However, one potential alternative side reaction of proline modification chemistry is epimerization at the alpha carbon. Thus, in addition to the 2S,4R (Hyp) and 2S,4S (hyp) hydroxyproline derivatives, the peptides with 2R,4S (d-hyp) and 2R,4R (d-hyp) hydroxyprolines were synthesized and analyzed by NMR spectroscopy, in order to identify the NMR signatures of peptides with alpha epimerization. These peptides were synthesized via coupling of Fmoc-d-hyp-OH and Fmoc-d-hyp-OH, which were prepared by solution phase synthesis using controlled epimerization of Hyp, subsequent modification by standard solution phase synthetic methods, and full characterization of the amino acids.17g, 67 Comparison of the Ac-TYProxN-NH2 peptides containing all four hydroxyproline stereoisomers indicates distinct NMR spectral signatures for all four stereoisomers (Figure 8). In particular, the NMR signatures of peptides with 2R stereochemistry (d-hyp (125) and d-hyp (126)) are easily distinguished from those containing 2S stereochemistry, as would be expected based on the substantial literature describing the large conformational effects of replacement of l- with d- amino acids in peptides. For proline these conformational effects are particularly significant because of the obligatory switch of proline from the left side of the Ramachandran plot to the right side of the Ramachandran plot. Interestingly, the peptide containing d-hyp exhibited expected cis-trans isomerism about the Tyr-d-hyp bond, whereas the peptide containing d-hyp exhibited no evidence of cis amide bond, with the peptides exhibiting very different NMR spectra from each other. Given the number of peptides employing d-Pro-Gly, d-Pro-l-Pro, and other d-Pro sequences in turns and in cyclic peptides, these data suggest substantial potential applications in the use of stereoelectronic effects to control conformation in heterochiral peptides.1k, 2h, 3d, 20i, j, 68
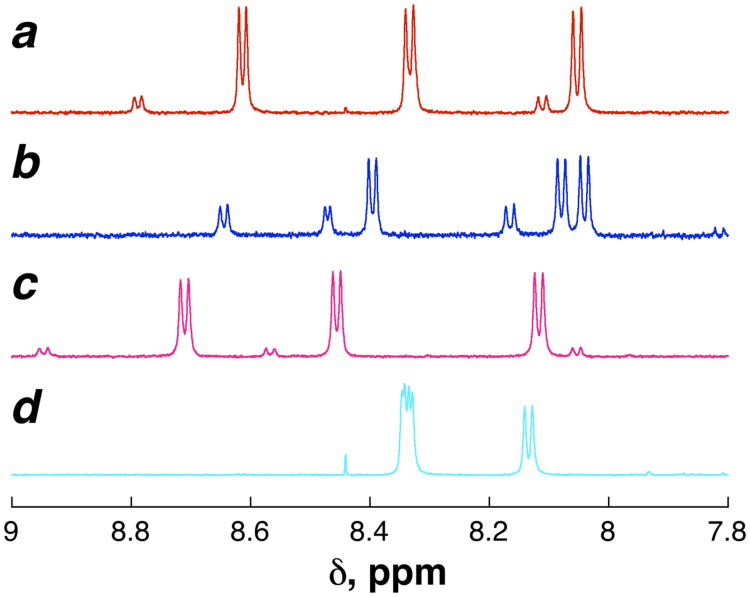
Figure 8.
1H NMR spectra (amide region) of the four 4-hydroxyproline Ac-TY-4-hydroxyproline-N-NH2 diastereomers. (a) 2S,4R-hydroxyproline (trans-hydroxyproline, Hyp) (1) (red). (b) 2S,4S-hydroxyproline (cis-hydroxyproline, hyp) (4) (blue); (c) 2R,4S-hydroxyproline (d-trans-hydroxyproline, d-hyp) (125) (magenta); (d) 2R,4R-hydroxyproline (d-cis-hydroxyproline, d-hyp) (126) (cyan). Full NMR spectra and TOCSY-derived amide resonance assignments are in the Supporting Information.
Global analysis of the peptides allows the comparison of the effects of proline substitution on cis-trans isomerism in Ac-TYProxN-NH2 peptides as a function of the stereochemistry and the nature of the substituent (Figure 7, Table 1, Table 2, Table 3). Three key parameters were examined initially: Ktrans/cis, the equilibrium constant of trans to cis prolyl amide bond (Figure 1); the free energy of proline modification on cis-trans isomerism relative to proline (ΔΔGtrans/cis = ΔGtrans/cis (peptide) − ΔGtrans/cis (Pro)); and the effect of proline modification on the backbone conformation at Tyr in the cis conformation (3JαN = coupling constant between HN and Hα, which can be correlated to φ via a parametrized Karplus relationship, with smaller values indicating more compact conformations).69 We also noted specific changes in Thr Hγ-methyl and Asn HN chemical shifts in peptides as a function of proline substituent electronics and stereochemistry, indicating that proline 4-substitution changed the conformation of the entire peptide, and that the effects of proline modification should be considered within a broader structural context than cis-trans isomerism.7a
In total, 109 different 4-substituted prolines were analyzed by NMR within this peptide context (Figure 7, Table 1, Table 2, Table 3, and Supporting Information). For 41 substituents, data were collected on both the 4R- and the 4S-substituted variants of the functional group, and the free energy difference of conformational effects of the stereoisomeric substitutions on cistrans isomerism calculated (Table 2). Comparison of the NMR data of the peptides revealed three general trends. First, for electron-withdrawing 4-substitutents, the Ac-TYProxN-NH2 model peptide context provides a basis for the comparison of stereoelectronic versus steric effects. Based on these data, benzoates, substituted acetates, and fluorine had the largest stereoelectronic effects, with aryl ethers, hydroxyl, azide, and sulfonates displaying relatively smaller overall effects on cis-trans isomerism. Second, increasing the size of the atom connected to the proline resulted in reduction or elimination of the stereoelectronic effect as a function of steric size and/or reduced electronegativity, suggesting that steric effects were becoming dominant in determining conformation. Third, the effects of a substitution on conformation could be modulated by changes in protonation state, oxidation, or other chemical reactions (e.g. cycloaddition, oxime formation, or cross-coupling reaction).
Protonated-aminoxy, phthalimide-protected aminoxy, isobutylaminoacetoxy, benzoates, and fluoro were the most potent substitutions to change peptide conformation. In all of these cases, either a highly electron-withdrawing atom is directly connected to the proline ring, or an electron-withdrawing atom bound to additional electron-withdrawing atoms (e.g. nitrogen, carbonyl) was attached to the ring, enhancing the stabilization that could be provided by hyperconjugation (Figure 2) and thus enhancing the conformational bias induced by the substituent. Lesser effects were observed with hydroxyl, aryl ethers, sulfonates, and azide, with the effects of azide modification similar to those of hydroxyl. These differences could be due to reduced inherent stereoelectronic effect, and/or due to steric interaction of these functional groups with the adjacent peptide backbone or side chains leading to an increased preference for the less sterically congested anti conformation, effects that would not be observed within acetylated amino acid methyl esters lacking this extended context.
Effects of proline substitution within the Ac-TYProxN-NH2 context could also potentially be influenced by interaction of the proline substituent with the tyrosine aromatic ring. This prospective interaction is more likely with the 4R substituent, which is on the same face of proline as that with which tyrosine interacts via the proline Hα in an aromatic-proline interaction (Figure 5). Therefore, in order to more thoroughly quantify stereoelectronic effects of peptides, for a representative subset of substitutions the Ac-TAProxN-NH2 peptides (127-149) were also synthesized and analyzed by NMR (Table 4, Table 5, and Supporting Information).67b These peptides lack a tyrosine ring that can interact with the proline. In general, proline editing reactions on Ac-TAProxN-NH2 peptides proceeded more readily (shorter reaction times, lower temperatures, and/or higher conversions) than those on Ac-TYProxN-NH2 peptides, presumably due to aromatic-proline interactions suppressing proline accessibility, suggesting that Ac-TYProxN-NH2 peptides, with sterically demanding groups both N-terminal (TyrOtBu) and C-terminal (Asn(Trt)) to proline, are an appropriate test of the generality of proline editing.
Table 4.
Ktrans/cis for Ac-TAXN-NH2 peptides. ΔG = −RT ln Ktrans/cis. ΔΔGtrans/cis = ΔGtrans/cis (peptide) − ΔGtrans/cis (Pro). n.d. = not determined due to spectral overlap. 3JαN = coupling constant between HN and Hα, which can be correlated to φ via a parametrized Karplus relationship.
| Ac-TAP(4-X)N-NH2X = | Ktrans/cis | ΔGtrans/cis, | ΔΔGtrans/cis, | 3JαN | δ, HN | δ, HN |
|---|---|---|---|---|---|---|
| kcal mol-1 | kcal mol-1 | Alacis | Alacis | Asntrans | ||
|
|
23.8 | -1.88 | -0.48 | 7.2 | 8.31 | 8.68 |
|
|
20.8 | -1.80 | -0.40 | 6.6 | 8.60 | 8.65 |
|
|
19.0 | -1.74 | -0.34 | n.d. | n.d. | 8.72 |
|
|
18.2 | -1.72 | -0.32 | 5.9 | 8.32 | 8.69 |
|
|
16.3 | -1.65 | -0.25 | n.d. | n.d. | 8.67 |
|
|
15.1 | -1.61 | -0.21 | n.d. | 8.89 | 8.68 |
|
|
11.6 | -1.45 | -0.05 | 6.6 | 8.30 | 8.68 |
|
|
11.6 | -1.45 | -0.05 | n.d. | n.d. | 8.89 |
|
|
11.1 | -1.42 | -0.02 | 5.7 | 8.36 | 8.71 |
| H (Pro) | 10.7 | -1.40 | 0.00 | 5.4 | 8.32 | 8.45 |
|
|
10.1 | -1.37 | 0.03 | 6.0 | 8.50 | 8.61 |
|
|
9.2 | -1.31 | 0.09 | 6.8 | 8.31 | 8.67 |
|
|
8.5 | -1.27 | 0.13 | n.d. | 8.53 | 8.69 |
|
|
8.5 | -1.27 | 0.13 | 4.8 | 8.42 | 8.17 |
|
|
8.2 | -1.24 | 0.16 | 5.4 | 8.44 | 8.18 |
|
|
6.5 | -1.11 | 0.29 | 6.2 | 8.41 | 8.28 |
|
|
5.8 | -1.04 | 0.36 | 4.7 | 8.49 | 8.22 |
|
|
5.5 | -1.01 | 0.39 | 4.8 | 8.53 | 8.34 |
|
|
5.1 | -0.96 | 0.44 | 4.4 | 8.49 | 8.33 |
|
|
4.7 | -0.92 | 0.48 | 4.7 | 8.46 | 8.19 |
|
|
4.7 | -0.92 | 0.48 | n.d. | n.d. | 8.26 |
|
|
4.7 | -0.92 | 0.48 | 4.8 | 8.49 | 8.37 |
|
|
4.4 | -0.88 | 0.52 | n.d. | n.d. | 8.21 |
|
|
4.3 | -0.86 | 0.54 | n.d. | 8.73 | 8.44 |
Table 5.
Overall functional group effects on cis-trans isomerism in Ac-TAXN-NH2 peptides. ΔΔG = ΔGtrans/cis(4R stereoisomer) − ΔGtrans/cis(4S stereoisomer), with negative ΔG consistent with the magnitude of stereoelectronic effects and positive indicating steric effects dominating over stereoelectronic effects. By definition, for Pro (4R = 4S = H), ΔΔG = 0.00.
| Ac-TAP(4-X)N-NH2 X = | ΔΔG(4R-4S) kcal mol-1 |
|---|---|
| F | -0.87 |
| OBzNO2 | -0.82 |
| OBzCF3 | -0.65 |
| OMs | -0.59 |
| OH | -0.56 |
| azido | -0.54 |
| ONs | -0.53 |
| OPO3H– | -0.45 |
| OTs | -0.39 |
| OPO3Et2 | -0.38 |
The data on Ac-TAProxN-NH2 peptides broadly corroborated that of Ac-TYProxN-NH2 peptides. The order of stereoelectronic effects F ≥ benzoates > OH ∼ N3 ≥ sulfonates was observed in both series. Benzoates exhibited lower overall effects on cis-trans isomerism in Ac-TAProxN-NH2 peptides than they did in Ac-TYProxN-NH2 peptides, although in 4S peptides the effects of benzoates were still greater than those of fluorine in Ac-TAProxN-NH2 peptides. Interestingly, even in Ac-TAProxN-NH2 peptides, which lack the possibility of an aromatic-proline interaction, the 3JαN of Alacis less than 5 Hz in 4S-substituted prolines indicated significant conformational restriction in the main chain (φ torsion angle) at alanine, consistent with type VIa1 β-turn formation. These data suggest that, within the context of a cis amide bond, an endo ring pucker more effectively promotes type VIa1 β-turns than an exo ring pucker does. Similarly, Asntrans also exhibited a chemical shift that was dependent on the stereochemistry and electronics of substitution (more downfield for electron-withdrawing 4R substitutents, more upfield for electron-withdrawing 4S substitutents), as it did in Ac-TYProxN-NH2 peptides, suggesting that these backbone effects were inherent to proline conformational restriction via 4-substitution.
The effects of sterics on switching proline conformational preferences were readily observed when the atom directly attached to the proline ring was substituted with larger atoms or was multiply substituted (Figure 7c). The 4S-N-phthalimide-substituted proline exhibited a strong preference for trans amide bond, despite the electronic effect of two carbonyls bound to the nitrogen. The trisubstituted nitrogen is apparently highly sterically demanding, leading to a conformational preference for trans amide bond despite the electron-withdrawing nature of the phthalimide. Similarly, replacement of an oxygen in an aryl ether with sulfur neutralized the conformational preferences of both 4R and 4S substitution, and substitution with selenium reversed the conformational preferences, with the 4S-selenophenyl ether exhibiting a higher population of trans amide bond than the 4R-selenophenyl ether. Likewise, for mercaptoproline, the 4S configuration had a greater preference for trans amide bond than the 4R configuration, both in thiol and thiolate protonation states (Figure 7d, Table 3), as has been seen previously.39 In addition, the 4S-iodoproline had a higher Ktrans/cis than the bromo, chloro, or fluoro derivatives, consistent with reduced stereoelectronic and increased steric effects for the iodide compared to the smaller halides.
Oxidation of the thiophenyl ether also resulted in an increase in sterically directed conformational effects (Figure 7d). For the thiophenyl ether, the 4S and 4R configurations exhibited similar Ktrans/cis. Oxidation of the thioether to the sulfoxide led to similar conformational preferences, despite the presence of an additional electron-withdrawing oxygen and increased positive charge on sulfur that would be expected to enhance a stereoelectronic effect. Further oxidation to the sulfone led to a clear sterically dependent preference of 4S for trans conformation and 4R for cis conformation relative to proline, despite the even greater electron-withdrawing effects of the sulfone.28b
Two ionizable proline derivatives exhibited particularly noteworthy pH-dependent conformational preferences. First, 4-aminoproline exhibited non-standard conformational preferences when protonated, with the 4S substitution having a greater trans preference than the 4R substitution, despite an ammonium having a stronger stereoelectronic effect than an amine.33d, 33f These effects of anomalously high trans amide bond stabilization from an endo-favoring residue have been previously described for protonated 4S-aminoproline in both acetylated amino acid methyl esters and in polyproline helices.33i, 70 This effect has been described as due to hydrogen bonding between the ammonium and the proline backbone carbonyl (here, the carbonyl conjugated to the Asn amide) for the protonated 4S-aminoproline, an interaction not possible for the 4R-aminoproline. Indeed, the chemical shift of Asntrans is highly aberrant for protonated 4S-aminoproline, consistent with a hydrogen bond between the ammonium and the carbonyl, with δ = 8.93 ppm for protonated 4S-aminoproline compared to δ = 8.08 ppm for hyp, δ = 8.18 for flp, and δ = 7.91 for 4S-azidoproline, typical values for electron-withdrawing 4S-substituents (Table 1, Table 3 Figure 6c). Interestingly, although the acetamide of 4-aminoproline also showed evidence of this intramolecular hydrogen bonding in the acetylated amino acid methyl esters, in our work herein the 4S-guanidinium-substituted proline did not exhibit this same effect, despite similarly favorable geometry and electrostatics.
Protein phosphorylation is an important post-translational modification whose structural effects are not well understood. Bielska and Zondlo observed that serine/threonine phosphorylation of peptides from the tau proline-rich domain induced a conformational change to polyproline helix, but the structural basis for this conformational change was not described.45g, h Phosphorylation could introduce conformational restriction on proline through steric effects, stereoelectronic effects, or through attractive or repulsive interactions with the peptide backbone. The TYProxN and TAProxN peptide contexts could be used to better understand the nature of the effects of phosphorylation on protein structure. Comparison of the phosphorylated hydroxyproline peptides with the non-phosphorylated hydroxyproline peptides suggested that phosphorylation indeed may induce a stereoelectronic effect, with a greater effect in the monoanionic form than the dianionic form. These data are consistent with the idea that phosphorylation leads to conformational restriction through a combination of stereoelectronic effects and either repulsive or attractive backbone interactions, as the largest structural effects of phosphorylation are in the dianionic form. Interestingly, the neutral diethylphosphate exhibited a modestly reduced apparent stereoelectronic effect compared to the monoanionic or dianionic phosphates or to hydroxyproline, indicating that the diethyl phosphate is a more sterically demanding substituent.
Beyond these initial parameters on cis-trans isomerism, coupling constants, and amide proton chemical shift, the conformational preferences for a subset of peptides were evaluated more thoroughly with additional information provided by TOCSY data. Analysis of proline side chain chemical shifts revealed apparent signatures for proline ring pucker that are consistent with the effects on Ktrans/cis described above. For example, substitutions with larger Ktrans/cis than Pro also had a larger Δδ of the diastereotopic proline beta protons, whether the effect was due to an electron-withdrawing 4R substitutent or to a sterically demanding 4S substituent. In contrast, the opposing modifications that led to smaller Ktrans/cis than Pro (4S-electron-withdrawing, 4R-sterically demanding) had smaller Δδ of the diastereotopic beta protons. These trends were observed in both TYProxN and TAProxN peptides. A similar dichotomy was observed in the Δδ of the diastereotopic proline gamma protons, with 4S-electron-withdrawing and 4R-sterically demanding substituents (lower Ktrans/cis) exhibiting a larger Δδ of the diastereotopic gamma protons, while the opposite substitutions (higher Ktrans/cis) displayed a smaller Δδ. Additional trends consistent with conformational preferences were observed in the N-terminal acetyl protons and the Thr methyl protons. Changes in ring pucker are also supported by changes in coupling patterns of Hβ and Hγ protons (see the Supporting Information for details). In total, the NMR data indicate that the effects of proline substitution are translated over the structure of the entire peptide as a function of the stereochemistry, stereoelectronic effects, and sterics of the 4-substituent.
To further examine the effects of proline substitution on peptide structure, a series of peptides containing 4R-, 4S- and 4,4-disubstituted prolines was examined by circular dichroism (Figure 9). The circular dichroism signatures of the peptides will predominantly represent the conformation of peptides with the major trans amide bond, particularly for peptides with residues with a strong preference for an exo ring pucker (4R stereochemistry). The CD data were consistent with NMR data indicating that proline substitution modulates peptide main chain conformation as a function of both stereochemistry and electronics of substitution. In particular, proline 4R and 4S substitution led to conformational effects that were divergent in opposite directions from proline and with the stereoelectronic effects of azidoproline more modest in a peptide context than those of hydroxyproline, fluoroproline, or acetylated hydroxyproline.
Figure 9.
CD spectra of TYXN peptides indicating the effect of proline substitutions on modulating peptide conformation. (a) Ac-TYPN-NH2; (b) Ac-TYHypN-NH2(1) (red circles) and Ac-TYhypN-NH2(4) (blue squares); (c) Ac-TYFlpN-NH2(27) (red circles) and Ac-TYflpN-NH2(26) (blue squares); (d) Ac-TYP(4R-azido)N-NH2(18) (red circles) and Ac-TYP(4S-azido)N-NH2(21) (blue squares); (e) Ac-TYP(4R-OAc)N-NH2(58) (red circles) and Ac-TYP(4S-OAc)N-NH2(59) (blue squares); (f) Ac-TYP(4-oxo)N-NH2(91); (g) Ac-TYP(4,4-F2)N-NH2(101) and (h) Ac-TYP(4,4-dithiolane)N-NH2(102). CD data were collected in water with 25 mM KF and 20 mM phosphate pH 7.0. CD data were background corrected but were not smoothed. CD spectra with error bars are shown in the Supporting Information.
Interestingly, 4,4-disubstitutued prolines, whose Ktrans/cis values were similar to proline herein and previously in acetylated amino acid methyl esters, exhibited CD spectra in peptides that were substantially different from each other and/or proline.17b, c 4,4-Difluoroproline (101) in a peptide exhibited a CD spectrum similar to proline (Figures 9a and 9g), suggestive of its potential application at the site of proline with minimal thermodynamic effect on structure. In contrast, peptides containing 4-oxoproline (91) (Figure 9f) and proline-4,4-dithiolane (102) (Figure 9h) exhibited CD spectra that were substantially different from those of proline, as was also seen in their NMR spectra (Figure 7b). The 4,4-dithiolane in particular exhibited both unique NMR and CD spectra that were distinct from those of any other peptide examined in this study, and that in particular were very different from that of 4-oxoproline, presumably due to the greater steric demand of the 4,4-dithiolane. These data suggest that potentially distinct conformational effects may be induced with disubstituted prolines in peptides.
Proline derivatives as handles for IR, fluorescence, and 19F and 77Se NMR spectroscopy
Proline editing is capable of incorporating a range of functional groups into peptides, including derivatives that can serve as spectroscopic probes. The cyanophenyl ether derivative synthesized above includes an aryl nitrile, which could potentially be employed as a probe for either infrared or fluorescence spectroscopy, as has been described for 4-cyanophenylalanine.71 Analysis of the IR spectrum (Figure 10) of the peptide Ac-TAP(4S-OPhCN)N-NH2(145) in H2O revealed a strong IR absorbance at 2233 cm-1, a spectroscopically quiet region in the IR of proteins, with resultant high signal to noise. These data suggest that cyanophenyl hydroxyprolines may be useful infrared probes via site-specific incorporation at a proline residue.
Figure 10.
FTIR spectrum of Ac-TAP(4S-OPhCN)N-NH2(145) in H2O (ν = 2233 cm-1). The full FTIR spectrum is in the Supporting Information.
Due to extended conjugation of the nitrile, cyanophenyl ethers also exhibit strong fluorescence. In order to understand the relationship of peptide structure to fluorescence, the fluorescence spectra of the peptides Ac-TYP(4R-OPhCN)N-NH2(35), Ac-TYP(4S-OPhCN)N-NH2(36), and Ac-TAP(4S-OPhCN)N-NH2(145) were examined (Figure 11 and Supporting Information) and compared to the control peptide Ac-TYPN-NH2. The fluorescence excitation and emission spectra of cyanophenylhydroxyproline-containing peptides reveal a strong absorbance with a broad excitation maximum λmax = 262 nm and fluorescence emission at λmax = 295 nm. Comparison of the data of tyrosine- versus alanine-containing peptides indicates evidence of fluorescence resonance energy transfer or quenching between cyanophenylhydroxyproline and tyrosine: the tyrosine-containing peptides exhibited both substantially reduced fluorescence compared to the alanine-containing peptide and stereochemistry-dependent fluorescence intensities. Comparing Ac-TAP(4S-OPhCN)N-NH2 and Ac-TYPN-NH2, cyanophenyl hydroxyproline had 7.5-fold greater fluorescence intensity than tyrosine with excitation at 265 nm. Based on its fluorescence emission wavelength, cyanophenyl hydroxyprolines are expected to be appropriate FRET donors for tryptophan, as has been described for cyanophenylalanine.72
Figure 11.
Fluorescence of peptides in 5 mM HEPES buffer pH 7.5 with 100 mM NaCl and 2 mM MgCl2. Data were collected with band widths of 3 nm. (a) Fluorescence excitation spectrum of Ac-TAP(4S-OPhCN)N-NH2(145) with detection of emission at 300 nm. (b) Fluorescence emission spectra of Ac-TAP(4S-OPhCN)N-NH2 (blue circles) and Ac-TYPN-NH2 (green squares) (50 μM peptide) with excitation at 265 nm. (c) Fluorescence emission spectra of Ac-TAP(4S-OPhCN)N-NH2 (blue circles) and Ac-TYP(4S-OPhCN)N-NH2(36) (magenta triangles) with excitation at 265 nm, showing the effect of tyrosine in quenching fluorescence. (d) Fluorescence emission spectra of Ac-TYP(4R-OPhCN)N-NH2(35) (red circles), Ac-TYP(4S-OPhCN)N-NH2(36) (magenta triangles), and Ac-TYPN-NH2 (green squares). The fluorescence emission spectra of these peptides, with excitation at 265 nm and at 280 nm, with error bars, are in the Supporting Information.
Proline editing was employed to incorporate a range of substitutions into peptides. Among these modifications were derivatives that incorporate spin-1/2 nuclei that could be used as NMR probes, specifically probes that might be exploited site-specifically without background from natural sources.73 19F NMR spectra were collected for fluorinated proline peptides (Figure 12), including those containing flp (26), Flp (27), 4,4-difluoroproline (101, 134), pentafluorophenyl ethers (33, 34), and perfluoro-tert-butyl ethers (55, 56). 19F NMR spectra are characterized in general by excellent chemical shift dispersion, a feature which was observed in fluorinated peptides, with an overall chemical shift range of over 100 ppm in the fluorinated peptides: δ from −70 ppm (perfluoro-tert-butyl ethers) to −178 ppm (fluoroprolines), with difluoroprolines (∼ −100 ppm) and aryl fluorines (∼ −160 ppm) intermediate (Table 6). The 19F NMR spectra corroborated the Ktrans/cis values observed by 1H NMR. The perfluoro-tert-butyl hydroxyproline peptides were noteworthy for the fluorine signal existing as a sharp singlet, with 9 equivalent fluorines, suggesting potential applications as a magnetic resonance probe with high signal to noise.44b, 46
Figure 12.
19F NMR spectra of peptides in 5 mM phosphate buffer pH 4 with 25 mM NaCl in 90% H2O/10% D2O at 23 °C. The data were collected with 1H decoupling. (a) Ac-TYFlpN-NH2(27); (b) Ac-TYflpN-NH2(26); (c) Ac-TYP(4,4-F2)N-NH2(101); (d) Ac-TAP(4,4-F2)N-NH2(134); (e) Ac-TYP(4R-O-C6F5)N-NH2(33); (f) Ac-TYP(4S-O-C6F5)N-NH2(34); (g) Ac-TYP(4R-OC(CF3)3)N-NH2(55); (h) Ac-TYP(4S-OC(CF3)3)N-NH2(56).
Table 6.
19F NMR chemical shifts. Chemical shifts are listed in order from upfield to downfield based on δ in the trans conformation. n.d. = not determined due to spectral overlap.
| Peptide | δtrans, ppm | Multiplicity (J, Hz) | δcis, ppm | Multiplicity (J, Hz) |
|---|---|---|---|---|
|
|
−177.1 | s | −177.9 | s |
|
|
−173.2 | s | −173.5 | s |
|
|
−163.2 | dd (24, 24) | −163.6 | dd (20, 20) |
|
|
−161.9 | t (24) | −162.1 | t (20) |
|
|
−156.5 | d (24) | −156.1 | d (20) |
|
|
−163.4 | dd (24, 24) | −163.6 | dd (24, 24) |
|
|
−162.4 | t (24) | −162.9 | t (24) |
|
|
−156.3 | d (24) | −156.3 | n.d. |
|
|
−100.6 | s | −103.2, −103.9 | d (252) |
| −94.8, −95.4 | d (252) | |||
|
|
−100.8, −101.4 | d (248) | −104.7, −105.3 | d (252) |
| −99.7, −100.3 | d (248) | −94.3, −94.9 | d (252) | |
|
|
−70.7 | s | −70.8 | s |
|
|
−70.0 | s | −70.7 | s |
Interestingly, in the 4,4-difluoroproline-containing peptides (Figure 12cd), the diastereotopic fluorines exhibited almost identical chemical shifts when in the trans conformation, appearing as a pseudo-singlet for Ac-TYP(4,4-F2)N-NH2 (101, Figure 12c) due to Δδ < 2JFF and exhibiting strong second order effects due to similar chemical shifts (Δδ ∼ 2JFF) in Ac-TAP(4,4-F2)N-NH2 (134, Figure 12d). In contrast, the diastereotopic fluorines exhibited substantial chemical shift dispersion (Δδ = 8-10 ppm) and 2JFF (= 252 Hz) coupling of the geminal fluorines in the cis conformation. This amide conformation-dependent chemical shift dispersion was also observed in Ac-TAP(4,4-F2)N-NH2, indicating that the dispersion was not due to interaction with the tyrosine aromatic ring but was an inherent effect dependent on amide trans versus cis conformation. These results suggest that 4,4-difluoroproline could potentially be a useful magnetic probe of prolyl amide cis-trans isomerism, which is difficult to easily identify within proteins.74 Notably, 4,4-difluoroproline may be incorporated in expressed proteins in proline auxotrophic bacteria, although curiously no 19F NMR spectrum has been reported for these proteins.17c, 18b
Selenium is a redox-sensitive trace element present in proteins as selenocysteine and selenomethionine.75 77Se NMR has emerged as a useful spectroscopic probe due to the rare nature of selenium in proteins, its ready substitution for sulfur, and its large chemical shift dispersion that is highly sensitive to oxidation state. These properties result in information-rich NMR spectra with good signal to noise due to the absence of background 77Se signals, despite only 7.6% natural abundance of 77Se and a relatively low gyromagnetic ratio.76 To examine the possible application of selenoproline analogues as NMR probes, the 77Se NMR spectrum of the selenophenyl proline peptide Ac-TYP(4R-SePh)N-NH2(41) was collected (Figure 13). The proton-decoupled data exhibit a sharp singlet at 299.9 ppm, compared to a chemical shift of 462.8 ppm for the reference compound PhSeSePh, consistent with large chemical shift dispersion in 77Se NMR that makes it useful for probing oxidation and biological interactions.
Figure 13.
77Se NMR spectrum of Ac-TYP(4R-SePh)N-NH2(41) (2 mM) (δ = 299.9 ppm) at 23 °C in D2O. The data were collected with 1H decoupling. The full spectrum is in the Supporting Information.
Discussion
We have described a practical approach to the synthesis of peptides with proline residues stereospecifically modified at the 4-position with diverse functionalities and structural preferences. Proline editing involves the incorporation of the inexpensive amino acid Fmoc-4R-Hydroxyproline by standard solid phase synthesis, protection in situ in an automated manner on solid phase, and subsequent synthesis of the rest of the peptide. The fully synthesized peptide is then selectively deprotected at the hydroxyproline, and the hydroxyproline selectively and stereospecifically modified via reactions including Mitsunobu, sulfonylation, SN2, acylation, and oxidation. The critical difference between solid phase modification via proline editing and solution phase reactions is the elimination of the cumbersome need for protecting group chemistry and manipulation on the amine and carboxylic acid functional groups, with the peptide main chain serving as the “protecting group.” In addition, because the modification reactions are performed on solid phase, the reactions are conducted with excess reagents that are washed away after each synthetic step, without the usual need in solution synthesis for the separation, isolation, and purification after each reaction step. In proline editing, the only purification step is standard HPLC purification required after any peptide synthesis. This practical aspect allows the rapid conduct of multistep synthetic sequences compared to similar sequences by solution phase chemistry. Moreover, typically solution syntheses of modified prolines are conducted on the more stable Boc- or Cbz-protected amines, then converted to Fmoc protection for solid phase peptide synthesis. Combining the practical advantages of solid phase synthesis with the lack of protection/deprotection/reprotection steps involving the amine and carboxylic acid, proline editing synthetic sequences are both fewer steps and more easily conducted than the sequences to prepare the equivalent Fmoc amino acids.
Proline editing provides access to a wide range of 4-substituted prolines with diverse substitution and control of stereochemistry, with both 4R and 4S substitution demonstrated for most proline derivatives. In total, peptides with 123 different 4-substituted proline amino acids were synthesized, of which 56 were novel amino acids never previously described, and many more were amino acids synthesized and used only once or just a small number of times. In contrast to proline editing, synthesis of each proline amino acid as an Fmoc amino acid for application in peptide synthesis would involve 5-9 steps of solution phase synthesis per amino acid, and would also involve the repetitive solid phase synthesis of each peptide with a different proline amino acid separately. In contrast, proline editing is both practical, with no solution phase synthesis, and diversity oriented, with a single fully synthesized peptide capable of being modified to generate a series of peptides with divergent proline substituents. Notably, the approach is general for several classes of reactions, and thus could be used to incorporate a broad range of additional functionalities not described in this work.
Functionalities incorporated include electrophiles (sulfonates, halides, ketone, maleimide, NHS ester, bromoacetate), nucleophiles (amine, thiol, aminoxy, 1,2-aminothiol), bioorthogonal functionalities (azide, alkyne, alkene, aryl halides, tetrazine), entities for polymerization in biomaterials (acrylate), affinity groups (biotin, RGD), fluorescence and IR spectroscopic probes (aryl nitrile, azide), NMR probes (77Se, 19F), hydrophobic groups, and native amino acid functional groups. Moreover, the reactive functional groups could be modified on solid phase and in neutral aqueous solution to further diversify the peptides, using reactions including azidealkyne coupling, Sonogashira and Suzuki-Miyaura cross-couplings, Diels-Alder reaction, Grubbs olefin cross-metathesis, native chemical ligation, reductive amination, tetrazine-transcyclooctene ligation, oxime formation, and reductive amination. Combining these reactions with a wide array of available reactive partners provides substantial opportunities for even broader functionalization of peptides containing these amino acids (e.g. rapid solution incorporation of furan, alkene, and fluorine functional groups demonstrated in oxime reactions in solution). Most notably, the range of functional groups incorporated in 4-substituted prolines included several different derivatives whose reactivities were orthogonal to one another, and thus which were capable of selective reaction in the presence of other functional groups capable of alternative selective reaction. This work suggests the possibility of modifying a peptide with multiple functional groups capable of independent modification, and thus the possibility of multiple functionalizations of a single peptide in aqueous solution. As proof of principle of this concept, three parallel bioorthogonal reactions were conducted in aqueous solution using three peptides containing different functional groups in the presence of three different reactive small molecules, with rapid (30 minutes) and selective modification of all three peptides observed with excellent conversion.
4-Substitution of proline residues induces side chain conformational preferences that bias peptide and protein main chain conformation, in a manner dependent on the stereochemistry of the substitution and on the sterics versus electronics of the substitution. Specifically, proline 4R-substitution with electron-withdrawing groups or 4S-substitution with sterically demanding groups leads to a preference for the proline exo ring pucker, which leads to a stronger preference for trans amide bond and for more compact φ, ψ torsion angles (αR, PPII). In contrast, proline 4S-substitution with electron-withdrawing groups or 4R-substitution with sterically demanding groups leads to a preference for the proline endo ring pucker, which leads to a greater relative preference for a cis amide bond or for more extended φ, ψ torsion angles. Due to these strong conformational biases, a variety of 4-substituted prolines have been broadly used to control peptide and protein structure, particularly within collagen mimetics but also in medicinal chemistry, biomaterials, and globular proteins. Proline editing was employed to incorporate 4-substituted prolines with divergent steric and electronic properties of substitution, and thus with a wide range of conformational properties. The effects of 4-susbstitution were quantified for 109 proline derivatives within a peptide context, which differs in context from simple acetylated amino acid methyl esters, which lack adjacent amino acids whose conformations could affect or be affected by the proline derivatives, and from the tertiary structure of collagen triple helices. NMR analysis of these proline derivatives revealed a continuum of conformational effects for the 4-substituted prolines described herein, and most notably indicated that the effects of proline modification were propagated beyond the proline amino acid to the entire tetrapeptide, with structural differences observed in all amino acids in the Ac-TYProxN-NH2 peptides that were a function of stereochemistry, sterics, and electronics of substitution. This work specifically allowed quantification of the structural effects of a wide range of proline 4-substitutents, including a majority that have not been previously examined, with 1.3 kcal/mol tunability in the free energy of cis-trans isomerism observed (9-fold change in Ktrans/cis) in the proline derivatives studied and in general distinct structures observed for the 4R- versus 4S-substituted diastereomers of the peptides in all cases.
Among the derivatives synthesized were modified proline residues that contain amino acid functional groups and could function as structurally constrained mimics of the encoded amino acids Cys, Asp, Glu, Phe, Lys, Arg, Ser, and Thr, as well as the post-translationally modified amino acids phosphoserine and phosphothreonine (Figure 14).21b, 21e, 21l, 31d, 33b, c, 77 Conformationally constrained amino acids and amino acid mimetics have a long history in medicinal chemistry to identify ligand binding conformations and to optimize ligands for high selectivity for targets, and have been also applied in protein and materials design to tune protein stability and function. The canonical amino acids have multiple available conformations, considering both φ, ψ space and χ space, which is important to their plasticity to adopt different structures. In contrast, specific target binding usually involves the conformational entropy cost of adopting defined conformations. Limiting the available main chain and side chain conformations via conformationally restricted amino acid mimetics provides the possibility of tailored, structurally defined functions. This is particularly true given the greater flexibility of non-proline amino acids and the ability of proline-substituted amino acids to adopt most of the major protein secondary structure conformations, including α-helix and PPII via exo-favoring residues or more extended conformations via endo-favoring residues. Conformations where the main chain amide is a hydrogen bond donor are obviously not accessible via these proline-based amino acid mimetics, although this limitation is less significant in view of the critical role of loops and turns in the conformations of bound ligands, particularly those bound to GPCRs and other receptors.68d, 78 Indeed, much of the original work to synthesize novel 4-substituted proline amino acids was conducted to increase ligand binding to GPCRs, integrin receptors, enzymes, and other proteins. Optimization of χ space in particular has proven fruitful in controlling ligand conformation and binding, due to the coupling of side chain conformation to main chain conformation.3a-c Therefore, numerous examples have been developed of conformationally restricted versions of the canonical amino acids, including a substantial number of proline-derived amino acid mimetics, and these amino acid mimetics have proven highly valuable in diverse applications, particularly in medicinal chemistry.5d, 21b, 21d-g, 21i, j, 21l, 31c, d, 33b-k, 48b, c, 77, 79 The 4R- and 4S-substitituted proline amino acid derivatives described herein provide the ability to broadly functionalize proline beyond the minimal natural proline modifications, and to easily incorporate proteinogenic functional groups within structured contexts.
Figure 14.
Structured amino acid mimetics. Peptides with all functionalities herein were synthesized with both 4R (shown) and 4S stereochemistries. NMR data indicating the structural differences between 4R and 4S substitution are in Table 1 and the Supporting Information.
The key limitations of proline editing are the dependence on 4-substitution (due to the availability of commercial amino acid) and the dependence for direct modification of the proline ring on reactivity at an unactivated secondary carbon. Thus, while numerous SN2 and Mitsunobu reactions were achieved successfully, several nucleophiles exhibited no significant product formation, including most significantly carbon nucleophiles (cyanide, alkyne, malonate) that were unreactive with sulfonates on solid phase. These functionalities potentially could be incorporated via the more activated bromoacetate electrophile, as was done using phenyl acetylene as a nucleophile in an SN2 reaction, and was done under milder conditions for sodium azide, thiophenol, and isobutylamine as SN2 nucleophiles. However, the ester linkage thus employed, while highly general, could be limiting for some applications, most notably situations in which the peptide would be exposed to esterases. The amide linkage with 4-aminoproline, demonstrated with the biotin derivative, could potentially be an alternative in these cases. Furthermore, while all of these reactions were conducted on functionally rich and relatively sterically hindered peptides (Ac-TYProxN-NH2, containing protected alcohol, phenol, and side chain (trityl-)amide functional groups and an aromatic-proline interaction that is persistent in organic solvents; and, in some cases, on the trp cage), obviously some reactions (most notably oxidation reactions) will be incompatible with certain functional groups (Met, Cys, and potentially Trp) in peptides.
Conclusion
The work herein describes proline editing, a general and highly practical approach to the synthesis of peptides modified at the 4-position of proline to incorporate a broad range of functional groups to control peptide structure, reactivity, and function. Potential applications of these functional groups were demonstrated in rapid, high yielding, and orthogonal modifications of peptides on solid phase and in neutral aqueous solution. Proline editing was also applied to broadly interrogate the effects of 4R- and 4S-substitutions on peptide structure within a peptide context. We expect this work to have broad applications in new approaches to synthesize structured functional peptides, to understand steric and stereoelectronic effects on peptide and protein structure, and to incorporate novel functionalities into peptides and proteins.
Supplementary Material
Scheme 21.
Three parallel bioorthogonal reactions in one solution.a, b
a 50 μM 50, 50 μM 78, and 10 μM 82 in 5 mM phophate pH 7.5 with 25 mM NaCl at 37 °C in the presence of 50 mM each 3-methylbutyraldehyde and trans-cyclooctene and 40 mM cyclopentadiene. HPLC, top: starting material peptides; bottom: crude HPLC chromatogram after 30 minutes. All products were confirmed by isolation, mass spectrometry, and comparison to authentic material synthesized independently.
b RN = Ac-Thr-Tyr-, RC = -Asn-NH2.
Acknowledgments
We thank NSF (CAREER, CHE-0547973), NIH (RR17716), and the University of Delaware for support of this work. We thank Professor Joseph Fox and Ramajeyam Selvaraj for help with tetrazine synthesis and for the synthesis of trans-cyclooctene. We thank Gasirat Tririya for initial work in the development of Mitsunobu reactions on peptides on solid phase using 4-nitrobenzoic acid.
Abbreviations
- ACE
angiotensin converting enzyme
- ADDP
azodicarboxylic acid dipiperidide
- Alloc
allyloxycarbonyl
- AllocCl
allylchloroformate
- αR
right-handed α-helix conformation
- Azp
4R-azidoproline
- azp
4S-azidoproline
- Boc
tert-butoxycarbonyl
- 15-C-5
15-crown-5
- DAST
diethylaminosulfur trifluoride
- DIAD
diisopropylazodicarboxylate
- DIC
N,N-diisopropylcarbodiimide
- DIPEA
diisopropylethylamine
- DMAP
4-dimethylamino pyridine
- EWG
electron-withdrawing group
- Flp
4R–fluoroproline
- flp
4S–fluoroproline
- Fmoc
9-fluorenylmethoxycarbonyl
- HBTU
O-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- Hyp
4R–hydroxyproline
- hyp
4S–hydroxyproline
- Ms
methylsulfonyl
- Mst
2,4,6,-trimethylbenzenesulfonyl
- NHS
N-hydroxysuccinimide
- Ns
4-nitrobenzenesulfonyl
- Nu
nucleophile
- OBzCF3
4-trifluoromethyl benzoate ester
- OBzNO2
4-nitrobenzoate ester
- OMst
2,4,6–trimethyl phenylsulfonate
- OPhBr
4-bromophenyl ether
- OPhCN
4-cyanophenyl ether
- OPhI
4-iodophenyl ether
- OMs
methanesulfonate
- ONs
nitrobenzenesulfonate
- OTs
toluenesulfonate
- PDC
pyridinium dichromate
- Ph
phenyl
- pivaloyl
trimethyl acetyl
- PPII
type II polyproline helix
- Prox
4-substituted proline
- SAc
thioacetyl
- SPhNO2
4-nitrophenyl thioether
- TBAF
tetra-n-butylammonium fluoride
- TBS
tert-butyldimethylsilyl
- TBSCl
tert-butyldimethylsilyl chloride
- TES
triethylsilane
- TFA
trifluoroacetic acid
- TIS
triisopropylsilane
- Trt
triphenylmethyl (trityl)
- Ts
4-methylbenzenesulfonyl
Footnotes
Supporting Information Available: Experimental procedures, NMR spectra (1H, 19F, 77Se), HPLC chromatograms, CD, fluorescence and IR spectroscopies, and characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).(a) Young TS, Schultz PG. J Biol Chem. 2010;285:11039. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu CC, Schultz PG. Ann Rev Biochem. 2010;79:413. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]; (c) Wu X, Schultz PG. J Am Chem Soc. 2009;131:12497. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Link AJ, Mock ML, Tirrell DA. Curr Opin Biotech. 2003;14:603. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]; (e) Yoder NC, Kumar K. Chem Soc Rev. 2002;31:335. doi: 10.1039/b201097f. [DOI] [PubMed] [Google Scholar]; (f) DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Ann Rev Biochem. 1999;68:779. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]; (g) Neumann H, Hazen JL, Weinstein J, Mehl RA, Chin JW. J Am Chem Soc. 2008;130:4028. doi: 10.1021/ja710100d. [DOI] [PubMed] [Google Scholar]; (h) Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. J Am Chem Soc. 2011;133:11418. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]; (i) Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Curr Opin Chem Biol. 2010;14:774. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Chiu HP, Kokona B, Fairman R, Cheng RP. J Am Chem Soc. 2009;131:13192. doi: 10.1021/ja903631h. [DOI] [PubMed] [Google Scholar]; (k) Struthers MD, Cheng RP, Imperiali B. Science. 1996;271:342. doi: 10.1126/science.271.5247.342. [DOI] [PubMed] [Google Scholar]; (l) Merkel L, Budisa N. Org Biomol Chem. 2012;10:7241. doi: 10.1039/c2ob06922a. [DOI] [PubMed] [Google Scholar]; (m) Salwiczek M, Nyakatura EK, Gerling UIM, Ye SJ, Koksch B. Chem Soc Rev. 2012;41:2135. doi: 10.1039/c1cs15241f. [DOI] [PubMed] [Google Scholar]
- (2).(a) Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]; (c) Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]; (d) Pochan DJ, Schneider JP, Kretsinger J, Ozbas B, Rajagopal K, Haines L. J Am Chem Soc. 2003;125:11802. doi: 10.1021/ja0353154. [DOI] [PubMed] [Google Scholar]; (e) Dalal S, Balasubramanian S, Regan L. Nat Struct Biol. 1997;4:548. doi: 10.1038/nsb0797-548. [DOI] [PubMed] [Google Scholar]; (f) Zondlo NJ, Schepartz A. J Am Chem Soc. 1999;121:6938. [Google Scholar]; (g) Neidigh JW, Fesinmeyer RM, Andersen NH. Nature Struct Biol. 2002;9:425. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]; (h) am Ende CW, Meng HY, Ye M, Pandey AK, Zondlo NJ. ChemBioChem. 2010;11:1738. doi: 10.1002/cbic.201000056. [DOI] [PubMed] [Google Scholar]; (i) Zondlo SC, Gao F, Zondlo NJ. J Am Chem Soc. 2010;132:5619. doi: 10.1021/ja100862u. [DOI] [PubMed] [Google Scholar]
- (3).(a) Hruby VJ, Li G, Haskell-Luevano C, Shenderovich M. Biopolymers. 1997;43:219. doi: 10.1002/(SICI)1097-0282(1997)43:3<219::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]; (b) Cowell SM, Lee YS, Cain JP, Hruby VJ. Curr Med Chem. 2004;11:2785. doi: 10.2174/0929867043364270. [DOI] [PubMed] [Google Scholar]; (c) Hruby VJ. Acc Chem Res. 2001;34:389. doi: 10.1021/ar990063q. [DOI] [PubMed] [Google Scholar]; (d) Anil B, Song B, Tang Y, Raleigh DP. J Am Chem Soc. 2004;126:13194. doi: 10.1021/ja047119i. [DOI] [PubMed] [Google Scholar]; (e) Chapman RN, Dimartino G, Arora PS. J Am Chem Soc. 2004;126:12252. doi: 10.1021/ja0466659. [DOI] [PubMed] [Google Scholar]; (f) Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891. [Google Scholar]; (g) Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. J Am Chem Soc. 2005;127:2974. doi: 10.1021/ja0456003. [DOI] [PubMed] [Google Scholar]
- (4).Milner-White EJ, Bell LH, Maccallum PH. J Mol Biol. 1992;228:725. doi: 10.1016/0022-2836(92)90859-i. [DOI] [PubMed] [Google Scholar]
- (5).(a) Grathwohl C, Wüthrich K. Biopolymers. 1976;15:2025. doi: 10.1002/bip.1976.360151012. [DOI] [PubMed] [Google Scholar]; (b) Wu WJ, Raleigh DP. Biopolymers. 1998;45:381. doi: 10.1002/(SICI)1097-0282(19980415)45:5<381::AID-BIP6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]; (c) Pal D, Chakrabarti P. J Mol Biol. 1999;294:271. doi: 10.1006/jmbi.1999.3217. [DOI] [PubMed] [Google Scholar]; (d) Halab L, Lubell WD. J Am Chem Soc. 2002;124:2474. doi: 10.1021/ja012442w. [DOI] [PubMed] [Google Scholar]; (e) Taylor CM, Hardré R, Edwards PJB, Park JH. Org Lett. 2003;5:4413. doi: 10.1021/ol035711r. [DOI] [PubMed] [Google Scholar]; (f) Zarrinpar A, Bhattacharyya RP, Lim WA. Sci STKE. 2003;2003:re8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]; (g) Bhattacharyya R, Chakrabarti P. J Mol Biol. 2003;331:925. doi: 10.1016/s0022-2836(03)00759-9. [DOI] [PubMed] [Google Scholar]; (h) Zondlo NJ. Acc Chem Res. 2012 doi: 10.1021/ar300087y. A.S.A.P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Dunbrack RL, Jr, Karplus M. J Mol Biol. 1993;230:543. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]; (b) Lovell SC, Word JM, Richardson JS, Richardson DC. Proteins. 2000;40:389. [PubMed] [Google Scholar]
- (7).(a) Shoulders MD, Satyshur KA, Forest KT, Raines RT. Proc Natl Acad Sci USA. 2010;107:559. doi: 10.1073/pnas.0909592107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vitagliano L, Berisio R, Mastrangelo A, Mazzarella L, Zagari A. Protein Sci. 2001;10:2627. doi: 10.1110/ps.ps.26601a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jenkins CL, Raines RT. Nat Prod Rep. 2002;19:49. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- (9).(a) Eberhardt ES, Panasik N, Jr, Raines RT. J Am Chem Soc. 1996;118:12261. doi: 10.1021/ja9623119. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J Am Chem Soc. 2001;123:777. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]; (c) DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J Am Chem Soc. 2002;124:2497. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]; (d) Hodges JA, Raines RT. J Am Chem Soc. 2003;125:9262. doi: 10.1021/ja035881z. [DOI] [PubMed] [Google Scholar]
- (10).Alabugin IV, Zeidan TA. J Am Chem Soc. 2002;124:3175. doi: 10.1021/ja012633z. [DOI] [PubMed] [Google Scholar]
- (11).Shoulders MD, Hodges JA, Raines RT. J Am Chem Soc. 2006;128:8112. doi: 10.1021/ja061793d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lorenzen S, Peters B, Goede A, Preissner R, Frommel C. Proteins: Struct, Funct, Bioinf. 2005;58:589. doi: 10.1002/prot.20342. [DOI] [PubMed] [Google Scholar]
- (13).(a) Schmid FX. Adv Protein Chem. 2002;59:243. doi: 10.1016/s0065-3233(01)59008-7. [DOI] [PubMed] [Google Scholar]; (b) Dugave C, Demange L. Chem Rev. 2003;103:2475. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- (14).(a) Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Proc Natl Acad Sci USA. 2002;99:1899. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Andreotti AH. Biochemistry. 2003;42:9515. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]; (c) Eckert B, Martin A, Balbach J, Schmid FX. Nature Struct Mol Biol. 2005;12:619. doi: 10.1038/nsmb946. [DOI] [PubMed] [Google Scholar]; (d) Lummis SCR, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Nature. 2005;438:248. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- (15).(a) Yaffe MB, Schutkowski M, Shen MH, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Science. 1997;278:1957. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]; (b) Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. Nature. 1999;399:784. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- (16).(a) Hinderaker MP, Raines RT. Protein Sci. 2003;12:1188. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hodges JA, Raines RT. Org Lett. 2006;8:4695. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. Nat Chem Biol. 2010;6:615. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Zondlo NJ. Nat Chem Biol. 2010;6:567. doi: 10.1038/nchembio.413. [DOI] [PubMed] [Google Scholar]; (e) Krow GR, Shoulders MD, Edupuganti R, Gandla D, Yu F, Sonnet PE, Sender M, Choudhary A, DeBrosse C, Ross CW, Carroll P, Raines RT. J Org Chem. 2012;77:5331. doi: 10.1021/jo300700a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).(a) Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]; (b) Shoulders MD, Kamer KJ, Raines RT. Bioorg Med Chem Lett. 2009;19:3859. doi: 10.1016/j.bmcl.2009.03.168. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Renner C, Alefelder S, Bae JH, Budisa N, Huber R, Moroder L. Angew Chem Int Ed. 2001;40:923. [PubMed] [Google Scholar]; (d) Barth D, Milbrandt AG, Renner C, Moroder L. ChemBioChem. 2004;5:79. doi: 10.1002/cbic.200300702. [DOI] [PubMed] [Google Scholar]; (e) Malkar ND, Lauer-Fields JL, Borgia JA, Fields GB. Biochemistry. 2002;41:6054. doi: 10.1021/bi012071w. [DOI] [PubMed] [Google Scholar]; (f) Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. J Am Chem Soc. 2003;125:11500. doi: 10.1021/ja036673+. [DOI] [PubMed] [Google Scholar]; (g) Levins CG, Schafmeister CE. J Am Chem Soc. 2003;125:4702. doi: 10.1021/ja0293958. [DOI] [PubMed] [Google Scholar]; (h) Doi M, Nishi Y, Uchiyama S, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. J Am Chem Soc. 2003;125:9922. doi: 10.1021/ja035997v. [DOI] [PubMed] [Google Scholar]; (i) Kim W, McMillan RA, Snyder JP, Conticello VP. J Am Chem Soc. 2005;127:18121. doi: 10.1021/ja054105j. [DOI] [PubMed] [Google Scholar]; (j) Kim W, Hardcastle KI, Conticello VP. Angew Chem, Int Ed. 2006;45:8141. doi: 10.1002/anie.200603227. [DOI] [PubMed] [Google Scholar]; (k) Crespo MD, Rubini M. PLoS One. 2011;6:e19425. doi: 10.1371/journal.pone.0019425. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Holzberger B, Marx A. J Am Chem Soc. 2010;132:15708. doi: 10.1021/ja106525y. [DOI] [PubMed] [Google Scholar]; (m) Zheng TY, Lin YJ, Horng JC. Biochemistry. 2010;49:4255. doi: 10.1021/bi100323v. [DOI] [PubMed] [Google Scholar]; (n) Limapichat W, Lester HA, Dougherty DA. J Biol Chem. 2010;285:8976. doi: 10.1074/jbc.M109.060939. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Kumin M, Sonntag LS, Wennemers H. J Am Chem Soc. 2007;129:466. doi: 10.1021/ja067148o. [DOI] [PubMed] [Google Scholar]; (p) Erdmann RS, Wennemers H. J Am Chem Soc. 2010;132:13957. doi: 10.1021/ja103392t. [DOI] [PubMed] [Google Scholar]; (q) Arnold U, Hinderaker MP, Köditz J, Golbik R, Ulbrich-Hofmann R, Raines RT. J Am Chem Soc. 2003;125:7500. doi: 10.1021/ja0351239. [DOI] [PubMed] [Google Scholar]; (r) Naduthambi D, Zondlo NJ. J Am Chem Soc. 2006;128:12430. doi: 10.1021/ja0648458. [DOI] [PubMed] [Google Scholar]
- (18).(a) Buechter DD, Paolella DN, Leslie BS, Brown MS, Mehos KA, Gruskin EA. J Biol Chem. 2003;278:645. doi: 10.1074/jbc.M209364200. [DOI] [PubMed] [Google Scholar]; (b) Kim WY, George A, Evans M, Conticello VP. ChemBioChem. 2004;5:928. doi: 10.1002/cbic.200400052. [DOI] [PubMed] [Google Scholar]; (c) Merkel L, Schauer M, Antranikian G, Budisa N. ChemBioChem. 2010;11:1505. doi: 10.1002/cbic.201000295. [DOI] [PubMed] [Google Scholar]
- (19).(a) Mauger AB, Witkop B. Chem Rev. 1966;66:47. [Google Scholar]; (b) Remuzon P. Tetrahedron. 1996;52:13803. [Google Scholar]; (c) Mauger AB. J Nat Prod. 1996;59:1205. doi: 10.1021/np9603479. [DOI] [PubMed] [Google Scholar]
- (20).(a) Eder U, Sauer G, Weichert R. Angew Chem Int Ed. 1971;10:496. [Google Scholar]; (b) Hajos ZG, Parrish DR. J Org Chem. 1974;39:1615. [Google Scholar]; (c) List B, Lerner RA, Barbas CF. J Am Chem Soc. 2000;122:2395. [Google Scholar]; (d) List B. Tetrahedron. 2002;58:5573. [Google Scholar]; (e) Bock DA, Lehmann CW, List B. Proc Natl Acad Sci USA. 2010;107:20636. doi: 10.1073/pnas.1006509107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Quintard A, Langlois JB, Emery D, Mareda J, Guenee L, Alexakis A. Chem Eur J. 2011;17:13433. doi: 10.1002/chem.201102757. [DOI] [PubMed] [Google Scholar]; (g) Schmid MB, Zeitler K, Gschwind RM. Chem Sci. 2011;2:1793. [Google Scholar]; (h) Tanzer EM, Zimmer LE, Schweizer WB, Gilmour R. Chem Eur J. 2012;18:11334. doi: 10.1002/chem.201201316. [DOI] [PubMed] [Google Scholar]; (i) Sculimbrene BR, Morgan AJ, Miller SJ. J Am Chem Soc. 2002;124:11653. doi: 10.1021/ja027402m. [DOI] [PubMed] [Google Scholar]; (j) Fiori KW, Puchlopek ALA, Miller SJ. Nat Chem. 2009;1:630. doi: 10.1038/nchem.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).(a) Flippenanderson JL, Gilardi R, Karle IL, Frey MH, Opella SJ, Gierasch LM, Goodman M, Madison V, Delaney NG. J Am Chem Soc. 1983;105:6609. [Google Scholar]; (b) Krapcho J, Turk C, Cushman DW, Powell JR, Deforrest JM, Spitzmiller ER, Karanewsky DS, Duggan M, Rovnyak G, Schwartz J, Natarajan S, Godfrey JD, Ryono DE, Neubeck R, Atwal KS, Petrillo EW. J Med Chem. 1988;31:1148. doi: 10.1021/jm00401a014. [DOI] [PubMed] [Google Scholar]; (c) Smith EM, Swiss GF, Neustadt BR, McNamara P, Gold EH, Sybertz EJ, Baum T. J Med Chem. 1989;32:1600. doi: 10.1021/jm00127a033. [DOI] [PubMed] [Google Scholar]; (d) Karanewsky DS, Badia MC, Cushman DW, Deforrest JM, Dejneka T, Lee VG, Loots MJ, Petrillo EW. J Med Chem. 1990;33:1459. doi: 10.1021/jm00167a028. [DOI] [PubMed] [Google Scholar]; (e) Bridges RJ, Stanley MS, Anderson MW, Cotman CW, Chamberlin AR. J Med Chem. 1991;34:717. doi: 10.1021/jm00106a037. [DOI] [PubMed] [Google Scholar]; (f) Bhagwat SS, Fink CA, Gude C, Chan K, Qiao Y, Sakane Y, Berry C, Ghai RD. Bioorg Med Chem Lett. 1994;4:2673. [Google Scholar]; (g) Kolodziej SA, Nikiforovich GV, Skeean R, Lignon MF, Martinez J, Marshall GR. J Med Chem. 1995;38:137. doi: 10.1021/jm00001a019. [DOI] [PubMed] [Google Scholar]; (h) Bisang C, Weber C, Robinson JA. Helv Chim Acta. 1996;79:1825. [Google Scholar]; (i) Bellier B, McCortTranchepain I, Ducos B, Danascimento S, Meudal H, Noble F, Garbay C, Roques BP. J Med Chem. 1997;40:3947. doi: 10.1021/jm970439a. [DOI] [PubMed] [Google Scholar]; (j) Lerner C, Siegrist R, Schweizer E, Diederich F, Gramlich V, Jakob-Roetne R, Zurcher G, Borroni E. Helv Chim Acta. 2003;86:1045. [Google Scholar]; (k) Lafrance D, Caron S. Org Process Res Dev. 2012;16:409. [Google Scholar]; (l) Qu HC, Cai MY, Mayorov AV, Grieco P, Zingsheim M, Trivedi D, Hruby VJ. J Med Chem. 2009;52:3627. doi: 10.1021/jm801300c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).(a) Poulsen N, Sumper M, Kröger N. Proc Natl Acad Sci USA. 2003;100:12075. doi: 10.1073/pnas.2035131100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kühlberg A, Haid M, Metzger S. J Biol Chem. 2010;285:31484. doi: 10.1074/jbc.M109.035428. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, Verli H, Pauly M, Bacic A, Olsen CE, Ulvskov P, Petersen BL, Somerville C, Iusem ND, Estevez JM. Science. 2011;332:1401. doi: 10.1126/science.1206657. [DOI] [PubMed] [Google Scholar]; (d) Taylor CM, Karunaratne CV, Xie N. Glycobiology. 2012;22:757. doi: 10.1093/glycob/cwr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fmoc-Hyp-OH is $135/25 g (Chem Impex). In contrast, other 4-substituted proline analogs that are commercially available are substantialy more expensive e.g. Fmoc-(2S,4R)-Flp (1 g, $ 337, Bachem), Fmoc-(2S,4S)-flp (1 g, $ 337, Bachem), Fmoc-L-(3,4-dehydroproline) (1 g, $ 470, Chem Impex), Fmoc-L-4,4-F2-proline (1 g, $ 524, Polypeptide Group), and Fmoc-(2S,4R)-proline(4-OPh) (1 g, $ 458, Polypeptide Group)
- (24).Thomas KM, Naduthambi D, Tririya G, Zondlo NJ. Org Lett. 2005;7:2397. doi: 10.1021/ol0506720. [DOI] [PubMed] [Google Scholar]
- (25).Meng HY, Thomas KM, Lee AE, Zondlo NJ. Biopolymers (Peptide Sci) 2006;84:192. doi: 10.1002/bip.20382. [DOI] [PubMed] [Google Scholar]
- (26).(a) Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. J Mol Biol. 1988;201:161. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]; (b) Yao J, Dyson HJ, Wright PE. J Mol Biol. 1994;243:754. doi: 10.1016/0022-2836(94)90045-0. [DOI] [PubMed] [Google Scholar]; (c) Yao J, Feher VA, Espejo BF, Reymond MT, Wright PE, Dyson HJ. J Mol Biol. 1994;243:736. doi: 10.1016/0022-2836(94)90044-2. [DOI] [PubMed] [Google Scholar]; (d) Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. J Mol Biol. 1998;279:449. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- (27).Hutchinson EG, Thornton JM. Protein Sci. 1994;3:2207. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).(a) Thomas KM, Naduthambi D, Zondlo NJ. J Am Chem Soc. 2006;128:2216. doi: 10.1021/ja057901y. [DOI] [PubMed] [Google Scholar]; (b) Forbes CR, Zondlo NJ. Org Lett. 2012;14:464. doi: 10.1021/ol202947f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Isidro-Llobet A, Alvarez M, Albericio F. Chem Rev. 2009;109:2455. doi: 10.1021/cr800323s. [DOI] [PubMed] [Google Scholar]
- (30).Gomez-Vidal JA, Forrester MT, Silverman RB. Org Lett. 2001;3:2477. doi: 10.1021/ol016104b. [DOI] [PubMed] [Google Scholar]
- 31.(a) Patchett AA, Witkop B. J Am Chem Soc. 1957;79:185. [Google Scholar]; (b) Rüeger H, Benn MH. Can J Chem. 1982;60:2918. [Google Scholar]; (c) Natchus MG, Bookland RG, De B, Almstead NG, Pikul S, Janusz MJ, Heitmeyer SA, Hookfin EB, Hsieh LC, Dowty ME, Dietsch CR, Patel VS, Garver SM, Gu F, Pokross ME, Mieling GE, Baker TR, Foltz DJ, Peng SX, Bornes DM, Strojnowski MJ, Taiwo YO. J Med Chem. 2000;43:4948. doi: 10.1021/jm000246e. [DOI] [PubMed] [Google Scholar]; (d) Tamaki M, Han GX, Hruby VJ. J Org Chem. 2001;66:1038. doi: 10.1021/jo005626m. [DOI] [PubMed] [Google Scholar]
- (32).(a) Andreatta RH, Nair V, Robertson AV, Simpson WR. J Aust J Chem. 1967;20:1493. [Google Scholar]; (b) Gomez-Vidal JA, Silverman RB. Org Lett. 2001;3:2481. doi: 10.1021/ol0161054. [DOI] [PubMed] [Google Scholar]; (c) Sonntag LS, Schweizer S, Ochsenfeld C, Wennemers H. J Am Chem Soc. 2006;128:14697. doi: 10.1021/ja0654938. [DOI] [PubMed] [Google Scholar]
- (33).(a) Bartra M, Romea P, Urpí F, Vilarrasa J. Tetrahedron. 1990;46:587. [Google Scholar]; (b) Webb TR, Eigenbrot C. J Org Chem. 1991;56:3009. [Google Scholar]; (c) Zhang ZY, Van Aerschot A, Hendrix C, Busson R, David F, Sandra P, Herdewijn P. Tetrahedron. 2000;56:2513. [Google Scholar]; (d) Babu IR, Ganesh KN. J Am Chem Soc. 2001;123:2079. doi: 10.1021/ja002165d. [DOI] [PubMed] [Google Scholar]; (e) Crespo L, Sanclimens G, Montaner B, Pérez-Tomás R, Royo M, Pons M, Albericio F, Giralt E. J Am Chem Soc. 2002;124:8876. doi: 10.1021/ja020364m. [DOI] [PubMed] [Google Scholar]; (f) Umashankara M, Babu IR, Ganesh KN. Chem Commun. 2003:2606. doi: 10.1039/b308581c. [DOI] [PubMed] [Google Scholar]; (g) Farrera-Sinfreu J, Giralt E, Castel S, Albericio F, Royo M. J Am Chem Soc. 2005;127:9459. doi: 10.1021/ja051648k. [DOI] [PubMed] [Google Scholar]; (h) Sanclimens G, Crespo L, Giralt E, Albericio F, Royo M. J Org Chem. 2005;70:6274. doi: 10.1021/jo050720u. [DOI] [PubMed] [Google Scholar]; (i) Sonar MV, Ganesh KN. Org Lett. 2010;12:5390. doi: 10.1021/ol1021993. [DOI] [PubMed] [Google Scholar]; (j) Nanda M, Ganesh KN. J Org Chem. 2012;77:4131. doi: 10.1021/jo300070p. [DOI] [PubMed] [Google Scholar]; (k) Fillon YA, Anderson JP, Chmielewski J. J Am Chem Soc. 2005;127:11798. doi: 10.1021/ja052377g. [DOI] [PubMed] [Google Scholar]
- (34).(a) Koskinen AMP, Helaja J, Kumpulainen ETT, Koivisto J, Mansikkamaki H, Rissanen K. J Org Chem. 2005;70:6447. doi: 10.1021/jo050838a. [DOI] [PubMed] [Google Scholar]; (b) Attempts to directly synthesize the mesylate with inversion via Mitsunobu reactions with methane sulfonic acid were unsuccessful. [Google Scholar]
- (35).(a) Gerig JT, McLeod RS. J Am Chem Soc. 1973;95:5725. doi: 10.1021/ja00798a046. [DOI] [PubMed] [Google Scholar]; (b) Muller K, Faeh C, Diederich F. Science. 2007;317:1881. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- (36).(a) Hudlicky M, Merola JS. Tet Lett. 1990;31:7403. [Google Scholar]; (b) Avent AG, Bowler AN, Doyle PM, Marchand CM, Young DW. Tet Lett. 1992;33:1509. [Google Scholar]; (c) Demange L, Ménez A, Dugave C. Tet Lett. 1998;39:1169. [Google Scholar]; (d) Chorghade MS, Mohapatra DK, Sahoo G, Gurjar MK, Mandlecha MV, Bhoite N, Moghe S, Raines RT. J Fluorine Chem. 2008;129:781. doi: 10.1016/j.jfluchem.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Doi M, Nishi Y, Kiritoshi N, Iwata T, Nago M, Nakano H, Uchiyama S, Nakazawa T, Wakamiya T, Kobayashi Y. Tetrahedron. 2002;58:8453. [Google Scholar]
- (37).(a) Mitsunobu O. Synthesis. 1981:1. [Google Scholar]; b Hughes DL. Org Prep Proced Int. 1996;28:127. [Google Scholar]; (c) Swamy KCK, Kumar NNB, Balaraman E, Kumar K. Chem Rev. 2009;109:2551. doi: 10.1021/cr800278z. [DOI] [PubMed] [Google Scholar]; (d) Wisniewski K, Koldziejczyk AS, Falkiewicz B. J Pept Sci. 1998;4:1. doi: 10.1002/(sici)1099-1387(199802)4:1<1::aid-psc132>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- (38).Vergnon AL, Pottorf RS, Player MR. J Comb Chem. 2004;6:91. doi: 10.1021/cc0300356. [DOI] [PubMed] [Google Scholar]
- (39).Cadamuro SA, Reichold R, Kusebauch U, Musiol HJ, Renner C, Tavan P, Moroder L. Angew Chem Int Ed. 2008;47:2143. doi: 10.1002/anie.200704310. [DOI] [PubMed] [Google Scholar]
- (40).(a) Shang S, Tan Z, Dong S, Danishefsky SJ. J Am Chem Soc. 2011;133:10784. doi: 10.1021/ja204277b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Townsend SD, Tan ZP, Dong SW, Shang SY, Brailsford JA, Danishefsky SJ. J Am Chem Soc. 2012;134:3912. doi: 10.1021/ja212182q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).(a) Verbiscar AJ, Witkop B. J Org Chem. 1970;35:1924. doi: 10.1021/jo00831a045. [DOI] [PubMed] [Google Scholar]; (b) Eswarakrishnan V, Field L. J Org Chem. 1981;46:4182. [Google Scholar]; (c) Kemp DS, Curran TP, Davis WM, Boyd JG, Muendel C. J Org Chem. 1991;56:6672. [Google Scholar]; (d) Reddie KG, Carroll KS. Curr Opin Chem Biol. 2008;12:746. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- (42).(a) Dirksen A, Hackeng TM, Dawson PE. Angew Chem Int Ed. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; (b) Liu F, Stephen AG, Fisher RJ, Burke TR., Jr Bioorg Med Chem Lett. 2008;18:1096. doi: 10.1016/j.bmcl.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu F, Stephen AG, Waheed AA, Aman MJ, Freed EO, Fisher RJ, Burke TR., Jr ChemBioChem. 2008;9:2000. doi: 10.1002/cbic.200800281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Shoulders MD, Guzei IA, Raines RT. Biopolymers. 2008;89:443. doi: 10.1002/bip.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).(a) Sebesta DP, Orourke SS, Pieken WA. J Org Chem. 1996;61:361. [Google Scholar]; (b) Jiang ZX, Yu YB. J Org Chem. 2007;72:1464. doi: 10.1021/jo0616308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).(a) Bisel P, Al-Momani L, Muller M. Org Biomol Chem. 2008;6:2655. doi: 10.1039/b800083b. [DOI] [PubMed] [Google Scholar]; (b) Lyu PC, Sherman JC, Chen A, Kallenbach NR. Proc Natl Acad Sci USA. 1991;88:5317. doi: 10.1073/pnas.88.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cornish VW, Kaplan MI, Veenstra DL, Kollman PA, Schultz PG. Biochemistry. 1994;33:12022. doi: 10.1021/bi00206a003. [DOI] [PubMed] [Google Scholar]; (d) Sigman MS, Jacobsen EN. J Am Chem Soc. 1998;120:4901. [Google Scholar]; (e) Guerin DJ, Miller SJ. J Am Chem Soc. 2002;124:2134. doi: 10.1021/ja0177814. [DOI] [PubMed] [Google Scholar]; (f) Formaggio F, Baldini C, Moretto V, Crisma M, Kaptein B, Broxterman QB, Toniolo C. Chem Eur J. 2005;11:2395. doi: 10.1002/chem.200400892. [DOI] [PubMed] [Google Scholar]; (g) Bielska AA, Zondlo NJ. Biochemistry. 2006;45:5527. doi: 10.1021/bi052662c. [DOI] [PubMed] [Google Scholar]; (h) Brown AM, Zondlo NJ. Biochemistry. 2012;51:5041. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- (46).(a) Dalvit C. Prog Nucl Magn Reson Spectrosc. 2007;51:243. doi: 10.1016/j.pnmrs.2023.07.001. [DOI] [PubMed] [Google Scholar]; (b) Papeo G, Giordano P, Brasca MG, Buzzo F, Caronni D, Ciprandi F, Mongelli N, Veronesi M, Vulpetti A, Dalvit C. J Am Chem Soc. 2007;129:5665. doi: 10.1021/ja069128s. [DOI] [PubMed] [Google Scholar]
- (47).Jenkins CL, McCloskey AI, Guzei IA, Eberhardt ES, Raines RT. Biopolymers. 2005;80:1. doi: 10.1002/bip.20164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).(a) Yong YF, Kowalski JA, Lipton MA. J Org Chem. 1997;62:1540. [Google Scholar]; (b) Peterlin-Masic L, Kikelj D. Tetrahedron. 2001;57:7073. [Google Scholar]; (c) Balakrishnan S, Scheuermann MJ, Zondlo NJ. ChemBioChem. 2012;13:259. doi: 10.1002/cbic.201100638. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Balakrishnan S, Zhao C, Zondlo NJ. J Org Chem. 2007;72:9834. doi: 10.1021/jo701766c. [DOI] [PubMed] [Google Scholar]
- (49).Lee J, Chmielewski J. Chem Biol Drug Des. 2010;75:161. doi: 10.1111/j.1747-0285.2009.00929.x. [DOI] [PubMed] [Google Scholar]
- (50).(a) Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]; (b) Ruoslahti E. Ann Rev Cell Devel Biol. 1996;12:697. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- (51).(a) Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Selvaraj R, Liu SL, Hassink M, Huang CW, Yap LP, Park R, Fox JM, Li ZB, Conti PS. Bioorg Med Chem Lett. 2011;21:5011. doi: 10.1016/j.bmcl.2011.04.116. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J Am Chem Soc. 2011;133:9646. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Seitchik JL, Peeler JC, Taylor MT, Blackman ML, Rhoads TW, Cooley RB, Refakis C, Fox JM, Mehl RA. J Am Chem Soc. 2012;134:2898. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]; (f) Oh S, Jung JC, Avery MAZ. Naturforsch, B: J Chem Sci. 2008;63b:210. [Google Scholar]; (g) Kristensen TE, Hansen FK, Hansen T. Eur J Org Chem. 2009:387. [Google Scholar]
- (52).Cornish VW, Hahn KM, Schultz PG. J Am Chem Soc. 1996;118:8150. [Google Scholar]
- (53).(a) Benedetti E, Diblasio B, Pavone V, Pedone C, Felix A, Goodman M. Biopolymers. 1981;20:283. doi: 10.1002/bip.1981.360200204. [DOI] [PubMed] [Google Scholar]; (b) Kang YK, Park HS. Biopolymers. 2009;92:387. doi: 10.1002/bip.21203. [DOI] [PubMed] [Google Scholar]
- (54).Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- (55).(a) Johnson LN, Barford D. Annu Rev Biophys Biomol Struct. 1993;22:199. doi: 10.1146/annurev.bb.22.060193.001215. [DOI] [PubMed] [Google Scholar]; (b) Szilak L, Moitra J, Krylov D, Vinson C. Nature Struct Biol. 1997;4:112. doi: 10.1038/nsb0297-112. [DOI] [PubMed] [Google Scholar]; (c) Andrew CD, Warwicker J, Jones GR, Doig AJ. Biochemistry. 2002;41:1897. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]; (d) Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann V, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003;112:859. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]; (e) Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. Nucleic Acids Res. 2004;32:1037. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Balakrishnan S, Zondlo NJ. J Am Chem Soc. 2006;128:5590. doi: 10.1021/ja057692h. [DOI] [PubMed] [Google Scholar]
- (56).Zhang L, Chen X, Xue P, Sun HHY, Williams ID, Sharpless KB, Fokin VV, Jia G. J Am Chem Soc. 2005;127:15998. doi: 10.1021/ja054114s. [DOI] [PubMed] [Google Scholar]
- (57).(a) Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lim RKV, Lin Q. Chem Commun. 2010;46:1589. doi: 10.1039/b925931g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).(a) Chalker JM, Wood CSC, Davis BG. J Am Chem Soc. 2009;131:16346. doi: 10.1021/ja907150m. [DOI] [PubMed] [Google Scholar]; (b) Spicer CD, Davis BG. Chem Commun. 2011;47:1698. doi: 10.1039/c0cc04970k. [DOI] [PubMed] [Google Scholar]; (c) Spicer CD, Triemer T, Davis BG. J Am Chem Soc. 2012;134:800. doi: 10.1021/ja209352s. [DOI] [PubMed] [Google Scholar]; (d) Li N, Lim RKV, Edwardraja S, Lin Q. J Am Chem Soc. 2011;133:15316. doi: 10.1021/ja2066913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).(a) Gopi HN, Tirupula KC, Baxter S, Ajith S, Chaiken IM. ChemMedChem. 2006;1:54. doi: 10.1002/cmdc.200500037. [DOI] [PubMed] [Google Scholar]; (b) Lee KK, Park KH, Joo C, Kwon HJ, Jeon J, Jung HI, Park S, Han H, Cho M. J Phys Chem B. 2012;116:5097. doi: 10.1021/jp1085119. [DOI] [PubMed] [Google Scholar]; (c) Bonger KM, Kapoerchan VV, Grotenbreg GM, van Koppen CJ, Timmers CM, van der Marel GA, Overkleeft HS. Org Biomol Chem. 2010;8:1881. doi: 10.1039/b923556f. [DOI] [PubMed] [Google Scholar]
- (60).(a) Lin YA, Chalker JM, Floyd N, Bernardes GJL, Davis BG. J Am Chem Soc. 2008;130:9642. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]; (b) Lin YYA, Chalker JM, Davis BG. ChemBioChem. 2009;10:959. doi: 10.1002/cbic.200900002. [DOI] [PubMed] [Google Scholar]; (c) Lin YA, Chalker JM, Davis BG. J Am Chem Soc. 2010;132:16805. doi: 10.1021/ja104994d. [DOI] [PubMed] [Google Scholar]
- (61).Dirksen A, Dawson PE. Curr Opin Chem Biol. 2008;12:760. doi: 10.1016/j.cbpa.2008.10.009. [DOI] [PubMed] [Google Scholar]
- (62).(a) Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J Am Chem Soc. 2012;134:10317. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. J Am Chem Soc. 2012;134:792. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lang K, Davis L, Torres-Kolbus J, Chou CJ, Deiters A, Chin JW. NatChem. 2012;4:298. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).(a) Poethko T, Schottelius M, Thumshirn G, Hersel U, Herz M, Henriksen G, Kessler H, Schwaiger M, Wester HJ. J Nucl Med. 2004;45:892. [PubMed] [Google Scholar]; (b) Flavell RR, Kothari P, Bar-Dagan M, Synan M, Vallabhajosula S, Friedman JM, Muir TW, Ceccarini G. J Am Chem Soc. 2008;130:9106. doi: 10.1021/ja801666z. [DOI] [PubMed] [Google Scholar]; (c) Åberg O, Pisaneschi F, Smith G, Nguyen QD, Stevens E, Aboagye EO. J Fluorine Chem. 2012;135:200. [Google Scholar]
- (64).(a) Ding H, Shigenaga A, Sato K, Morishita K, Otaka A. Org Lett. 2011;13:5588. doi: 10.1021/ol202316v. [DOI] [PubMed] [Google Scholar]; (b) Wang YS, Fang XQ, Wallace AL, Wu B, Liu WSR. J Am Chem Soc. 2012;134:2950. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).(a) MacArthur MW, Thornton JM. J Mol Biol. 1991;218:397. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]; (b) Kay BK, Williamson MP, Sudol P. FASEB J. 2000;14:231. [PubMed] [Google Scholar]
- (66).Hodges JA, Raines RT. J Am Chem Soc. 2005;127:15923. doi: 10.1021/ja054674r. [DOI] [PubMed] [Google Scholar]
- (67).(a) Baker GL, Fritschel SJ, Stille JR, Stille JK. J Org Chem. 1981;46:2954. [Google Scholar]; (b) See the Supporting Information for details. [Google Scholar]
- (68).(a) Ciarkowski J, Gdaniec M, Kolodziejczyk A, Liberek B, Borremans FAM, Anteunis MJO. Int J Pept Prot Res. 1990;36:285. doi: 10.1111/j.1399-3011.1990.tb00980.x. [DOI] [PubMed] [Google Scholar]; (b) Bean JW, Kopple KD, Peishoff CE. J Am Chem Soc. 1992;114:5328. [Google Scholar]; (c) Stanger HE, Gellman SH. J Am Chem Soc. 1998;120:4236. [Google Scholar]; (d) Favre M, Moehle K, Jiang LY, Pfeiffer B, Robinson JA. J Am Chem Soc. 1999;121:2679. [Google Scholar]; (e) Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J Am Chem Soc. 2002;124:15030. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- (69).Vuister GW, Bax A. J Am Chem Soc. 1993;115:7772. [Google Scholar]
- (70).Kuemin M, Nagel YA, Schweizer S, Monnard FW, Ochsenfeld C, Wennemers H. Angew Chem Int Ed. 2010;49:6324. doi: 10.1002/anie.201001851. [DOI] [PubMed] [Google Scholar]
- (71).(a) Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J Am Chem Soc. 2003;125:405. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]; (b) Aprilakis KN, Taskent H, Raleighj DP. Biochemistry. 2007;46:12308. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]; (c) Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J Am Chem Soc. 2006;128:13984. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]; (d) Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Biochemistry. 2009;48:9040. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]; (e) Zimmermann J, Thielges MC, Seo YJ, Dawson PE, Romesberg FE. Angew Chem Int Ed. 2011;50:8333. doi: 10.1002/anie.201101016. [DOI] [PubMed] [Google Scholar]
- (72).(a) Serrano AL, Troxler T, Tucker MJ, Gai F. Chem Phys Lett. 2010;487:303. doi: 10.1016/j.cplett.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Taskent-Sezgin H, Marek P, Thomas R, Goldberg D, Chung J, Carrico I, Raleigh DP. Biochemistry. 2010;49:6290. doi: 10.1021/bi100932p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).(a) Ruiz-Cabello J, Barnett BP, Bottomley PA, Bulte JWM. NMR in Biomedicine. 2011;24:114. doi: 10.1002/nbm.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mykhailiuk PK, Afonin S, Palamarchuk GV, Shishkin OV, Ulrich AS, Komarov IV. Angew Chem Int Ed. 2008;47:5765. doi: 10.1002/anie.200801022. [DOI] [PubMed] [Google Scholar]; (c) Wadhwani P, Burck J, Strandberg E, Mink C, Afonin S, Ulrich AS. J Am Chem Soc. 2008;130:16515. doi: 10.1021/ja804928q. [DOI] [PubMed] [Google Scholar]; (d) Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RAS, Mehl RA, Pielak GJ. J Am Chem Soc. 2010;132:321. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Thomas CA, Talatya ER, Bann JG. Chem Comm. 2009:3366. doi: 10.1039/b821952d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).(a) Hawkes WC, Alkan Z. Biol Trace Elem Res. 2010;134:235. doi: 10.1007/s12011-010-8656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lobanov AV, Hatfield DL, Gladyshev VN. Biochim Biophys Acta, Gen Subj. 2009;1790:1424. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Papp LV, Lu J, Holmgren A, Khanna KK. Antioxid Redox Signaling. 2007;9:775. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- (76).(a) Duddeck H. Se-77 Nmr Spectroscopy and Its Applications in Chemistry. Advances in Solid State Nmr Studies of Materials and Polymers: A Special Volume Dedicated to Isao Ando. 2004;52:105. [Google Scholar]; (b) Mobli M, de Araujo AD, Lambert LK, Pierens GK, Windley MJ, Nicholson GM, Alewood PF, King GE. Angew Chem, Int Ed. 2009;48:9312. doi: 10.1002/anie.200905206. [DOI] [PubMed] [Google Scholar]; (c) Duddeck H. Prog Nucl Magn Reson Spectrosc. 1995;27:1. [Google Scholar]
- (77).(a) Wang Q, Sasaki NA, Potier P. Tetrahedron. 1998;54:15759. [Google Scholar]; (b) Ganorkar R, Natarajan A, Mamai A, Madalengoitia JS. J Org Chem. 2006;71:5004. doi: 10.1021/jo060210f. [DOI] [PubMed] [Google Scholar]
- (78).(a) Kee S, Jois SDS. Current Pharm Design. 2003;9:1209. doi: 10.2174/1381612033454900. [DOI] [PubMed] [Google Scholar]; (b) Tyndall JDA, Pfeiffer B, Abbenante G, Fairlie DP. Chem Rev. 2005;105:793. doi: 10.1021/cr040689g. [DOI] [PubMed] [Google Scholar]
- (79).(a) Jeannotte G, Lubell WD. J Am Chem Soc. 2004;126:14334. doi: 10.1021/ja0471222. [DOI] [PubMed] [Google Scholar]; (b) Geisler I, Chmielewski J. Bioorg Med Chem Lett. 2007;17:2765. doi: 10.1016/j.bmcl.2007.02.077. [DOI] [PubMed] [Google Scholar]; (c) Farrera-Sinfreu J, Zaccaro L, Vidal D, Salvatella X, Giralt E, Pons M, Albericio F, Royo M. J Am Chem Soc. 2004;126:6048. doi: 10.1021/ja0398621. [DOI] [PubMed] [Google Scholar]; (d) Hanessian S, McNaughtonSmith G, Lombart HG, Lubell WD. Tetrahedron. 1997;53:12789. [Google Scholar]; (e) Melendez RE, Lubell WD. J Am Chem Soc. 2004;126:6759. doi: 10.1021/ja039643f. [DOI] [PubMed] [Google Scholar]; (f) Del Valle JR, Goodman M. J Org Chem. 2003;68:3923. doi: 10.1021/jo034214l. [DOI] [PubMed] [Google Scholar]; (g) Wittelsberger A, Keller M, Scarpellino L, Patiny L, Acha-Orbea H, Mutter M. Angew Chem Int Ed. 2000;39:1111. doi: 10.1002/(sici)1521-3773(20000317)39:6<1111::aid-anie1111>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]; (h) Guitot K, Larregola M, Pradhan TK, Vasse JL, Lavielle S, Bertus P, Szymoniak J, Lequin O, Karoyan P. ChemBioChem. 2011;12:1039. doi: 10.1002/cbic.201000707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.