Significance
Bioethanol produced from waste biomass from crops has the potential to provide a sustainable alternative to petroleum-based transportation fuel that does not compete with human food supply. The main obstacle to this approach is the resistance of this biomass to digestion. Thus, expensive energetic pretreatment and high enzyme inputs are needed to increase digestion. In this study, we screened a population of randomly mutated plants for digestibility with the aim of identifying novel factors that impact on this trait. We found a number of mutants with high digestibility and no impairments in growth or fitness. These mutants show a range of alterations in cell-wall composition, and we have mapped and characterized the mutant with the highest increase in digestibility.
Keywords: lignocellulosic biofuel, matrix polysaccharides, lignin, feruloylation
Abstract
Lignocellulosic plant biomass is an attractive feedstock for the production of sustainable biofuels, but the commercialization of such products is hampered by the high costs of processing this material into fermentable sugars (saccharification). One approach to lowering these costs is to produce crops with cell walls that are more susceptible to hydrolysis to reduce preprocessing and enzyme inputs. To deepen our understanding of the molecular genetic basis of lignocellulose recalcitrance, we have screened a mutagenized population of the model grass Brachypodium distachyon for improved saccharification with an industrial polysaccharide-degrading enzyme mixture. From an initial screen of 2,400 M2 plants, we selected 12 lines that showed heritable improvements in saccharification, mostly with no significant reduction in plant size or stem strength. Characterization of these putative mutants revealed a variety of alterations in cell-wall components. We have mapped the underlying genetic lesions responsible for increased saccharification using a deep sequencing approach, and here we report the mapping of one of the causal mutations to a narrow region in chromosome 2. The most likely candidate gene in this region encodes a GT61 glycosyltransferase, which has been implicated in arabinoxylan substitution. Our work shows that forward genetic screening provides a powerful route to identify factors that impact on lignocellulose digestibility, with implications for improving feedstock for cellulosic biofuel production.
Concerns over greenhouse-gas emissions and the sustainability of liquid transportation fuel supplies have led to a rapid expansion of global biofuel production in recent years. Current biofuel production is dominated by bioethanol produced by fermentation of starch or sucrose from food crops and by biodiesel produced by transesterification of plant or animal oils. Although the production of such “first-generation” biofuels can be efficient, there is widespread concern that further expansion will exacerbate anticipated problems with global food security through direct competition for resources. In addition, these crops often require high inputs and consequently have a relatively high carbon footprint (1). A promising alternative to first-generation biofuel is the use of nonfood lignocellulosic plant biomass, available as agricultural waste from food crops or produced from low input, nonfood plants such as perennial grasses (2).
Lignocellulosic biomass is principally composed of plant secondary cell walls and comprises ∼70% polysaccharides, which can be converted into simple sugars for fermentation (3). The main challenge in producing bioethanol from lignocellulosic biomass is that these polysaccharides occur as part of a complex and indigestible macromolecular material that contains high amounts of lignin. The saccharification (conversion into simple sugars) of lignocellulose therefore requires energy demanding pretreatments and high enzyme inputs, making bioethanol production a costly process (4). Improving the ease and yield of cell-wall saccharification could provide a way of reducing these costs.
One approach to increasing saccharification is to produce crops with cell walls that are more susceptible to hydrolysis, and a range of studies have been carried out to investigate the impact of altering cell-wall components on saccharification. Most of these studies have focused on lignin, which is generally considered to be a major determinant of cell-wall digestibility due to the coating of cell polysaccharides with this complex and insoluble polymer (5). It has been shown that altering both lignin content (6–11) and lignin structure (12, 13) in the cell wall can affect saccharification. However, it has also been shown that alterations in cell-wall components other than lignin can affect biomass digestibility. For example, Penning et al. have recently shown that quantitative trait loci (QTLs) for lignin abundance are independent of those for saccharification in a maize recombinant inbred population (14). In particular, altering cellulose production, deposition, or crystallinity can affect saccharification (15–17) because cellulose typically constitutes around one third of the total mass of plants and the insoluble crystalline cellulose fibers are hard to digest (18). Alterations in matrix polysaccharide content and composition can also affect saccharification (19, 20) by changing the extractability and/or architecture of the cell wall. Furthermore, in grasses, matrix polysaccharides are able to cross-link to lignin and each other via ferulic acid esters (21), and a reduction in these linkages can increase saccharification (22, 23).
The majority of studies investigating the determinants of lignocellulose recalcitrance have taken a reverse genetics approach, disrupting genes that play key roles in cell-wall biosynthesis or expressing genes that encode wall-modifying enzymes. However, the complexity of plant cell-wall biosynthesis and the large number of genes involved, many of which are unknown, mean it is important that we explore saccharification potential in an empirical manner to gain a better understanding of factors that can impact this trait. Indeed, it has been estimated that 10–15% of Arabidopsis thaliana genes (∼2,500–4,000) are related to cell-wall biology (24) whereas it has been estimated that only 121 genes have been experimentally validated as cell wall-related (25). Furthermore, studies on the digestibility of maize stover for animal feed show that an extremely large number of QTLs impact on this trait (26). In this context, classical (or forward) genetic screens benefit from a more empirical approach to gene discovery and have great potential to identify new or unexpected factors that impact on a phenotype. Here, we present results from screening a chemically mutagenized population of the model grass species Brachypodium distachyon (hereafter, Brachypodium) for mutant lines showing increased saccharification in the presence of a mixture of polysaccharide-degrading enzymes.
Results
Identification of Putative Mutants with Increased Saccharification.
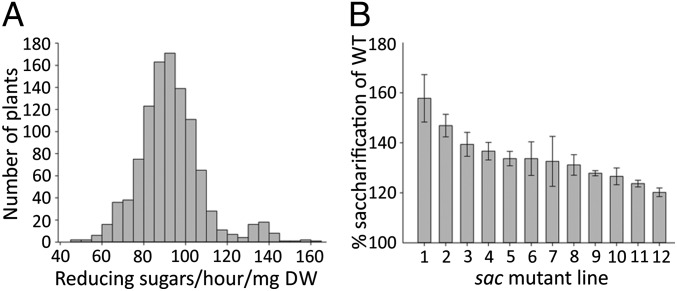
A population of 2,400 chemically mutagenized M2 generation Brachypodium plants were grown to maturity, and the straw was screened for enzymatic saccharification (release of sugar) using a high-throughput robotic platform (27). The conditions for saccharification used an alkaline pretreatment, followed by digestion with an industrial enzyme mixture that contained a variety of carbohydrate-degrading enzymes and thus would release a range of saccharides (28). Saccharification was measured by colorimetric detection of reducing sugar equivalents. The screen revealed a normal distribution for variation in saccharification across the population (Fig. 1A) (P = 0.496, Kolmogorov–Smirnov test), with a maximum saccharification 67.03% higher than wild-type (WT) and a minimum saccharification 30.31% lower than WT. From this population, 37 putative mutant plants were selected that had saccharification values greater than 20% above WT and where this difference was significant (P ≤ 0.01). To examine the heritability of this high-saccharification phenotype, the offspring of these 37 plants were subject to saccharification analysis, yielding 12 independent lines with heritable increases in saccharification (Fig. 1B), which we have named saccharification1 (sac1) to sac12.
Fig. 1.
Sugar release from Brachypodium stems after 0.5 M NaOH pretreatment at 90 °C and enzymatic hydrolysis. (A) Histogram showing distribution in saccharification of the whole mutant population in the first screen. (B) Saccharification of putative mutants with a heritable increase in saccharification seen in the second screen. Data represent mean ± SD and n = 10. DW, dry weight.
Increased Saccharification Is Associated with a Range of Cell-Wall Chemotypes.
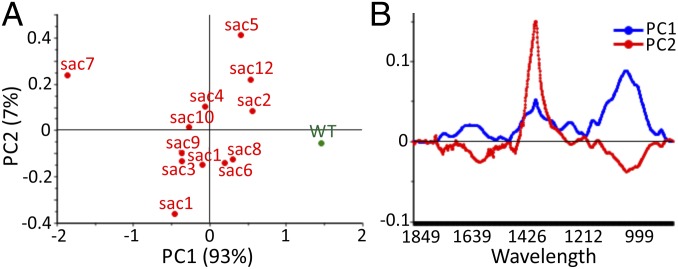
To investigate the molecular changes underlying the increases in saccharification seen in the sac lines, we analyzed their cell-wall composition in a number of ways. Fourier transform infrared (FTIR) spectroscopy has been applied extensively to study plant cell walls and is a useful tool for revealing general alterations in wall composition and for grouping plants based on similarity in wall composition (29). Powdered cell-wall samples from the 12 sac lines and WT plants were subject to FTIR, and the data were analyzed by principal component analysis (PCA). The PCA clearly separates all of the sac lines from WT based on principal component (PC) 1, with sac7 showing the most extreme separation (Fig. 2A). Loading plots show how much each wavelength contributes to the separation of the samples on each PC. Fig. 2B shows the loading plot for PC1; it has a large peak at wavelengths corresponding to cell-wall polysaccharides (950–1,100 cm−1) (30, 31). Interestingly, the sac lines themselves showed a greater separation from one another on PC2 (Fig. 2A). The loading plot for PC2 is shown in Fig. 2B and exhibits a large peak at wavelengths shown to correspond to lignin, polysaccharides, and p-hydroxycinnamic acids (1,320–1,432 cm−1) (30–32).
Fig. 2.
FTIR analysis of WT and sac stems. (A) Principal component analysis (PCA) of baseline corrected and peak normalized FTIR spectra in the range 1,850–850 cm−1, n = 10. (B) Loading plots of PC1 and PC2 from the PCA in A.
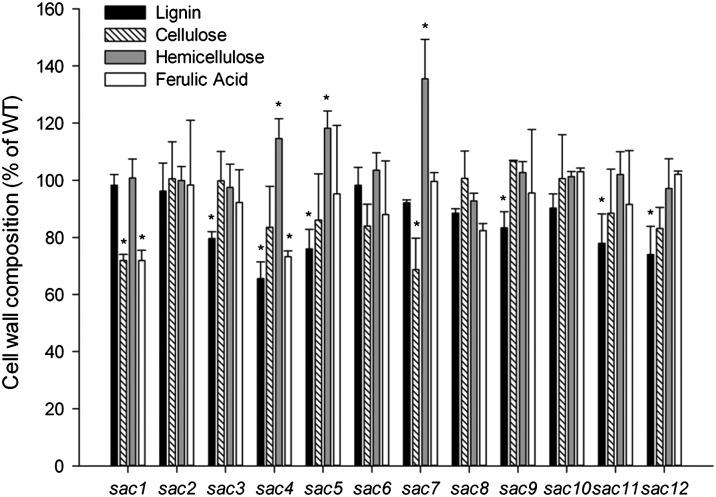
To further investigate the cell walls of the sac lines, a general analysis of lignin, cellulose, matrix polysaccharides, and ferulic acid was undertaken. Examination of lignin content using the acetyl bromide method (33) revealed that none of the lines had an increase in lignin content over WT whereas six lines (sac3, -4, -5, -9, -11, and -12) had significantly lower lignin content than WT (Fig. 3). Lignin structure of the sac lines was investigated by thioacidolysis. We found that six of the lines had alterations in the ratio of lignin thioacidolysis monomers compared with WT; sac1, -4, -6, and -7 all showed a significant decrease in S:G ratio and an increase in the percentage of H monomers (% H units) whereas sac2 and sac9 exhibited a significant increase only in % H units (P ≤ 0.01) (Table S1). Measurement of crystalline cellulose content by the anthrone-sulfuric acid method (34) revealed that most of the lines had similar crystalline cellulose content to WT whereas sac1 and sac7 both showed significantly reduced levels (Fig. 3). Matrix polysaccharide content and composition were measured by hydrolysis into component monosaccharides, followed by analysis by high-performance anion exchange chromatography (HPAEC). Three lines (sac4, sac5, and sac7) exhibited significantly higher total matrix polysaccharide content than WT whereas none showed a significant reduction (Fig. 3). Table S2 shows the composition of matrix polysaccharides in the sac lines. sac7 plants showed the largest increase in matrix polysaccharide content, most of which was accounted for by increases in xylose, with glucose, arabinose, and mannose also significantly higher than in WT. Similarly, the sac4 and sac5 lines exhibited significant increases in xylose, arabinose, and mannose compared with WT, with fucose also being significantly increased in sac4.
Fig. 3.
Lignin, cellulose, matrix polysaccharide, and ferulic acid content of stem AIR of the 12 sac lines, expressed as percentage of WT. Data represent mean ± SD and n = 3. Asterisk indicates a significant difference (P ≤ 0.05) compared with WT.
Analysis of the sac lines by immunocarbohydrate microarray analysis was carried out using antibodies that recognize a range of known grass cell-wall epitopes in polymers extracted from the cell wall (Fig. S1). This analysis suggested that there was a large amount of variation in the binding of the LM12 antibody, which recognizes esterified feruloyl groups (35), among the sac lines and WT. We therefore decided to extract and quantify cell-wall ester-bound ferulic acid, and found that sac1 and sac4 had significantly lower ester-bound ferulic acid content than WT (Fig. 3).
Development, Fitness, and Morphology.
We deliberately excluded severely dwarfed plants from the initial screen to avoid complications arising from plants exhibiting large developmental changes and because such mutants may not be so useful from a biotechnological perspective. Phenotyping of the sac lines revealed that the majority were similar to WT in their development and fitness (Fig. S2). Most lines showed similar percentage germination to WT (91%), the exceptions being sac8 and sac12, which showed lower germination (75% and 71%, respectively). Some lines showed differences in height and biomass compared with WT; sac3, -5, and -11 had significant increases in shoot biomass, with sac5 and sac11 also having significant increases in plant height. Conversely, sac1 and sac7 showed significant reductions in plant height which, in sac7, was accompanied by reduced biomass. Four lines (sac2, -3, -5, and -11) showed a significant increase in the number of seeds produced compared with WT, but none showed a significant reduction in seed number. We performed three-point bending tests to test the stem strength (maximum force the stem can withstand) and stiffness (initial force required to bend the stem) and found that none of the sac lines showed reductions in stem strength or stiffness compared with WT (Fig. S3).
Mapping the Causal Mutations.
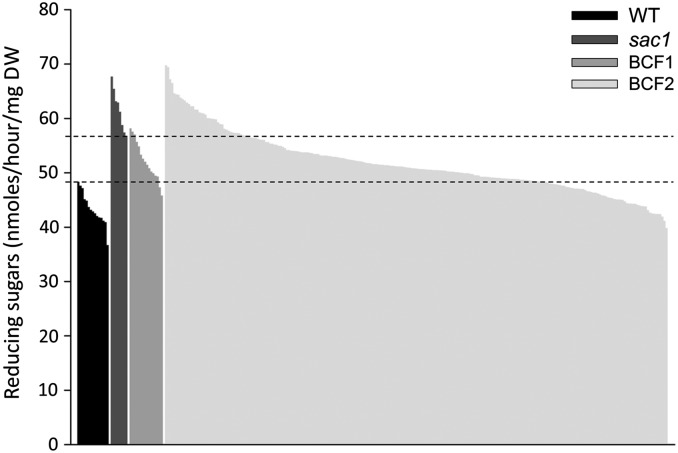
Because such screening for saccharification mutants has not previously been reported, it is important to assess whether the approach is sufficiently robust to allow the responsible gene loci to be identified. We used bulked segregant analysis approach to examine the cosegregation of phenotype and genetic lesions in segregating F2 backcrossed populations (BCF2) of the putative mutants (36, 37). For the purpose of this study, we report mapping of the sac1 mutant because it showed the largest increase in saccharification. Saccharification analysis of the BCF2 population, along with WT, sac1, and BCF1 plants, revealed that the BCF1 plants had an intermediate level of saccharification between WT and sac1 (Fig. 4), suggesting incomplete dominance of the WT allele at the position of the causal mutation. Using the lowest saccharification value of the sac1 mutant plants and the highest saccharification value of the WT plants as threshold values, we calculated the observed percentages of BCF2 plants with saccharification values resembling plants homozygous WT, heterozygous and homozygous mutant for the causal mutation. This analysis showed that 25.31% of the BCF2 plants showed a saccharification value comparable with homozygous WT plants whereas 54.77% showed values comparable with heterozygous plants and 19.92% showed values comparable with homozygous sac1 plants (Fig. 4). These values were not significantly different from the 1:2:1 ratio expected for a single segregating locus (χ2, P = 0.47).
Fig. 4.
Saccharification analysis to determine homozygous mutants of a BCF2 population from a backcross between WT and sac1. Level of saccharification of WT and sac1 plants, BCF1 plants, and the BCF2 population resulting from a self of one of the BCF1 plants is presented. Dotted lines show thresholds for the lowest sac1 saccharification and highest WT saccharification.
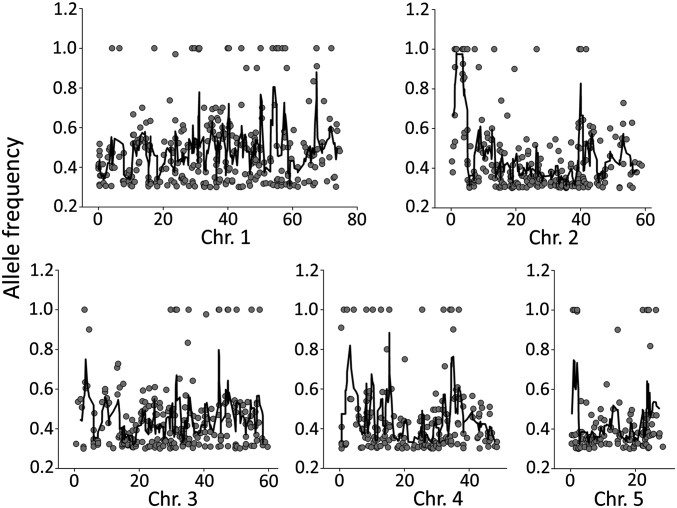
DNA from the top 40 high-saccharification BCF2 plants was extracted and pooled for whole-genome sequencing. After alignment of the sequence data to the reference Brachypodium genome, SNPs were called and filtered to retain only mutations that did not occur in the WT, mutations with high quality scores, and mutations resembling those induced by sodium azide (38). The allele frequency of each SNP (i.e., the percentage of reads for the alternative allele) was plotted for each chromosome (Fig. 5), revealing a single peak of high allele frequencies on chromosome 2, with a cluster of 14 SNPs, each with an allele frequency of ≥0.9. The peak spanned 2.6 Mbp and covered 362 annotated genes. Within the 14-SNP cluster, 10 SNPs were in noncoding regions and, of the 4 remaining, 3 were in exons and only 2 would cause a change in the encoded amino acid sequence (Table S3). One SNP caused a glycine-to-serine substitution in Bradi2g01480, predicted to encode a glycosyltransferase family 61 enzyme. The other SNP caused a change from proline to leucine in Bradi2g02520, which encodes a protein of unknown function. Sorts intolerant from tolerant (SIFT) analysis, which provides a score for how likely a mutation is to affect the function of a gene based on alignment of the protein to similar proteins (39), was performed for these two mutations. This analysis suggested that the SNP in the GT61 protein would disrupt structure and function drastically whereas the SNP in Bradi2g02520 would have little impact. It is possible that the whole-genome sequencing has missed SNPs within this region; however, given the high coverage of the sequencing, this seems unlikely. The predicted effect of the GT61 mutation on amino acid sequence and structure/function of the protein compared with the other SNPs identified within the region, makes the GT61 mutation the most likely candidate to be responsible for the observed phenotype, and we decided to explore this hypothesis further.
Fig. 5.
Allele frequency plots for the five chromosomes of the Brachypodium sac1 mutant. A sac1 plant was backcrossed to WT, and DNA from the homozygous mutants of the resulting BCF2 population was pooled and whole genome-sequenced. SNPs with an allele frequency above 0.3 were called and plotted against position in the chromosome. Gray circles indicate SNP positions on the chromosome and their allele frequencies. The black line shows a five point moving average of the SNP allele frequencies.
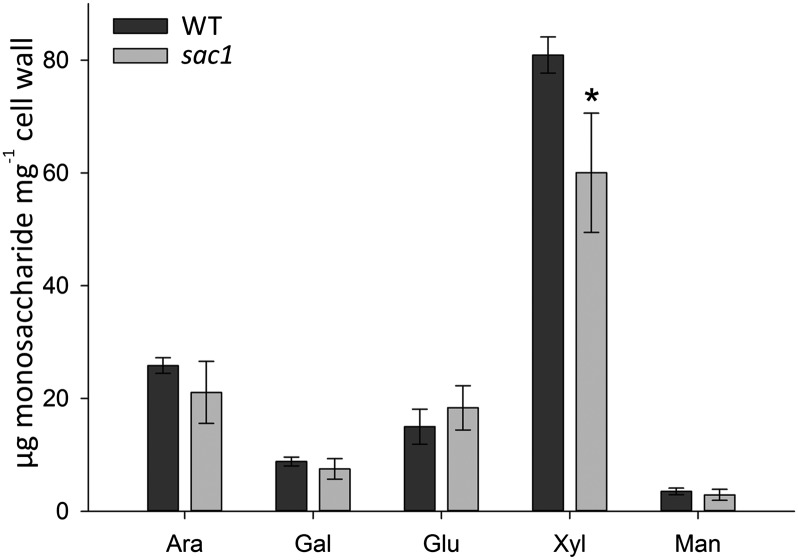
Phylogenetic analysis of the GT61 family in various monocot and dicot plants has shown the Bradi2g01480 protein to be part of grass-specific clade C.III (40). Clade C is highly expanded in grasses and contains rice and wheat xylan arabinosyltransferases (XATs) and XYLOSYL ARABINOSYL SUBSTITUTION OF XYLAN (XAX) in rice (40, 41), both involved in arabinoxylan substitution. Chiniquy et al. (40) recently showed that a xax1 mutant in rice exhibits a large reduction in ferulic acid content and an increase in saccharification. Interestingly, the sac1 mutant shows a very similar phenotype to that of the rice xax1 mutant, making the SNP in Bradi2g01480 a likely candidate for the causal mutation. The rice xax1 mutant is reported to show a minor 2.6% reduction in matrix polysaccharide xylose content when assessed by 2 M TFA hydrolysis of alcohol insoluble residue (AIR). In our analysis using this method, there was no significant difference in xylose content of sac1 AIR (Table S2). However, serial extractions of sac1 AIR using 1,2-cyclohexanedinitrilotetraacetic acid (CDTA) and Na2CO4 to remove pectinaceous fractions, followed by extraction with 4 M KOH, were used to produce a xylan-enriched fraction. Compositional analysis of this fraction revealed a significantly lower level of xylose in sac1 compared with WT (P = 0.03, t test) (Fig. 6), lending further support to the hypothesis that the sac1 phenotype results from the mutation in a XAX1 ortholog in Brachypodium.
Fig. 6.
Monosaccharide composition of a xylan-enriched fraction from WT and sac1 plants. A xylan-enriched fraction was extracted from AIR using 4 M KOH after pectin removal. Monosaccharides were quantified by hydrolyzing with 2 M TFA, followed by separation and quantification by HPAEC. Data represent mean ± SD and n = 3. Asterisk indicates a significant difference (P ≤ 0.05) compared with WT. Ara, arabinose; Gal, galactose; Glu, glucose; Man, mannose; Xyl, xylose.
Discussion
Brachypodium was chosen for this study because its small genome size, short life cycle, and simple diploid genetics make it practical for carrying out mutant screens (42). In addition, Brachypodium’s fully sequenced genome has significant colinearity with those of major cereal crops, and the species is also seen as a good model for biomass grasses (43). The notable differences in wall composition between monocots and dicots (3) make A. thaliana a less suitable model for the cereal straw and grasses that will form much of the biomass for biofuel applications. In this study, we screened a population of chemically mutagenized Brachypodium plants and identified 12 putative mutant lines that exhibited heritable improvements in saccharification with industrial polysaccharide-degrading enzyme mixtures. We analyzed a range of cell-wall and growth characteristics of these lines to get a better understanding of the mutant phenotypes. It is apparent from the cell-wall composition analysis (Fig. 3) that the increased saccharification phenotype is accompanied by a range of different cell-wall modifications among the sac lines.
Lines with Altered Lignin Content.
It is widely accepted that lignin plays a major role in the recalcitrance of lignocellulose to digestion, and many studies have shown that a reduction in lignin content results in increased saccharification [reviewed in Li et al. (44)]. In line with these studies, six of the sac lines (sac3, -4, -5, -9, -11, and -12) showed significantly lower levels of lignin than WT. In three lines (sac3, sac11, and sac12), lignin content was the only cell-wall component to be altered significantly from WT. Although the work presented here confirms a major role for lignin in determining saccharification, it is notable that a number of the sac lines, including the two with highest saccharification (sac1 and sac2), showed no significant differences in lignin content compared with WT, suggesting that additional factors can be important for saccharification efficiency.
Lines with Reduced Lignin and Alterations in Other Cell-Wall Components.
In the sac4, -5, and -9 lines, the reduced lignin content was accompanied by alterations in other cell-wall components. Both sac4 and sac5 showed significant increases in matrix polysaccharides. Such increases in these polysaccharides have been reported in other plants with altered lignin content (7, 45) and could be a compensatory effect to make up for any loss in cell-wall strength caused by the reduction in lignin. The increase in matrix polysaccharides might also contribute to the increased saccharification of these lines because our saccharification assay does not distinguish between the breakdown of cellulose and matrix polysaccharides.
Lines with Alterations in Cell-Wall Components Other than Lignin.
A number of the sac lines showed little or no change in lignin content but exhibited alterations in other wall components. Among these lines, sac1 showed the largest increase in saccharification. One of the features of sac1 cell walls was a significant decrease in crystalline cellulose content compared with WT, which was also observed in sac7. Highly crystalline cellulose is recalcitrant to digestion whereas the amorphous regions and matrix polysaccharides released by TFA treatment as “matrix polysaccharides” are easier to digest. The majority of glucose released by the TFA treatment in Brachypodium will be derived from amorphous cellulose because grass secondary cell walls contain only minor amounts of xyloglucan and mixed linkage glucan (MLG) (3). The sac7 plants showed a significant increase in glucose released by TFA treatment (Table S2), which may indicate an increase in the ratio of amorphous to crystalline cellulose in this line and could contribute to the high saccharification. The reduction in crystalline cellulose in sac7 was accompanied by a significant increase in matrix polysaccharides, characterized by significant increases in arabinose, mannose, and especially xylose, which suggests an increase in arabinoxylan and mannan content. Interestingly, the differences in matrix polysaccharide composition revealed by TFA extraction followed by HPAEC were not apparent in the immunocarbohydrate microarray analysis (Fig. S1). The antibodies used for this analysis recognize relatively specific epitopes, and the binding efficiency of the LM11 antibody, which recognizes arabinoxylans, is affected by the amount of arabinose substitution and does not bind well to sorghum or maize arabinoxylan (46). The differences in matrix polysaccharide composition detected by HPAEC were not severe and so may have been too subtle for detection by antibodies.
No difference was seen in glucose content of the TFA extract between sac1 and WT plants, which suggests a reduction in total cellulose content in this line due to the reduction in crystalline cellulose content. The sac1 cell walls also showed a significant reduction in the amount of ester-bound ferulic acid, a feature also seen in sac4. In cell walls of grasses, ferulic acid esters are abundant on arabinosyl side chains of arabinoxylans (21). These feruloyl esters may be oxidatively dimerized to form cross-links between or within arabinoxylan chains and also between arabinoxylan and lignin (21). It is therefore thought that ferulic esters could play a role in the recalcitrance of plant cell walls to digestion, and a reduction in these residues has been shown to result in increased saccharification in a number of plant species (22, 23, 47). The alkaline pretreatment used in this study is likely to result in partial hydrolysis of the ferulate ester linkages within the cell wall and may therefore be more effective for the sac1 and sac4 lines, which have lower ferulic acid content.
Lines with Altered Lignin Structure.
A number of the sac lines had alterations in lignin structure. In most cases, this alteration consisted of a reduction in thioacidolysis S:G ratio accompanied by an increase in % H units (sac1, -4, -6, and -7) whereas sac2 and sac9 showed just an increase in % H units. A number of studies have shown transgenic and natural accessions with reduced S:G ratio, no reduction in lignin content, and an increase in digestibility [switchgrass (13), ryegrass (48), maize (49), tall fescue (50), and A. thaliana (7)]. The most straightforward hypothesis for the improved saccharification of alkaline-pretreated sac1, -2, -4, -6, -7, and -9 lines is their higher frequency in G and/or H units. Studies have shown that about 50% of native grass lignins can be solubilized in alkali due to their high frequency in terminal units (51, 52). These terminal units are prominently H and G units whereas S units are essentially internal. Accordingly, increasing the frequency of H and G units in grass lignin makes the lignin polymers more susceptible to alkaline pretreatment.
Mapping One of the Responsible Mutations.
To verify that our screening approach can provide information on genes involved in the digestibility of plant biomass, we have mapped the responsible mutation of the sac1 line to a narrow genomic region. The most likely responsible mutation within this region is an SNP in a gene encoding a glycosyltransferase, family 61 (Bradi2g01480). GT61 proteins have been implicated previously in arabinoxylan substitution due to genes encoding this family of proteins being more highly expressed and numerous in grasses than dicots (53). Furthermore, members of GT61 clade C have been shown to introduce the arabinosyl side chains onto arabinoxylans (AX) (41). The sac1 mutant shows a reduction in ferulic acid content and reduced xylose in its xylan-enriched cell-wall fraction, characteristics also reported for the xax1 mutant in rice (40). XAX1 is a GT61 family clade C member that has been proposed to catalyze the transfer of xylosyl residues on to arabinose side chains of AX (40). The authors hypothesized that the observed reduction in ferulic acid content in the xax1 mutant is due to loss of xylose substitutions that are required for the feruloylation of neighboring arabinosyl residues, resulting in decreased activity of the feruloyltransferase enzyme(s) (40). This hypothesis remains to be experimentally validated, and the authors (40) alternatively suggested that the xylosyl residues might render the arabinose-bound ferulic acid less susceptible to feruloyl esterases; reductions in xylosyl substitution may therefore lead to increased rates of loss of ferulic acid esters in the xax1 plants. We propose that the Brachypodium Bradi2g01480 gene encodes an enzyme with a similar function to XAX1 in rice, due to the similar reductions in xylose and ferulic acid content in the mutants, and the grouping of these enzymes together in clade C of the GT61 family by phylogenetic analysis. We believe that the increase in saccharification in this mutant is primarily due to the reduction in ferulic acid content.
The work described in this study has implications for breeding strategies of biofuel crops because it indicates that a number of different routes can be adopted to increase saccharification and potentially thereby lower processing costs. This finding raises the possibility of combining more than one such modification in a plant to obtain even higher saccharification. Importantly, our screen has enabled the identification of plants with increased digestibility but with no major impairment in growth, development, or stem strength. Indeed, a number of the sac lines showed increased height, biomass, and/or seed production under glasshouse conditions, raising the possibility of eventually producing crops with both improved yield and processability. Although future work will target the identification and confirmation of responsible genes and translational studies in crops grown under field conditions, this study gives a positive indication of what could be achieved in this area to improve the sustainability and efficiency of lignocellulosic biofuel production.
Materials and Methods
Plant Growth and Material.
The mutant B. distachyon population consisted of 2,400 M2 plants in the Bd21 background that had been chemically mutagenized with sodium azide according to the protocol described in ref. 54. Details of plant growth are given in SI Materials and Methods. Internodes from the primary and secondary stems of mature and dried plants were used for all experiments, except where stated otherwise. All internodes excluding the first were cut into 4-mm lengths and ground using a custom-made robotic platform (Labman Automation) [see Gomez et al. (27)].
Saccharification Analysis.
Ground stem material from above was weighed into four replicate 4-mg samples per plant in a 96-deep-well plate using the robotic platform mentioned above. Saccharification analysis of this plant material was performed using a liquid handling robotic platform (Tecan Evo 200; Tecan Group Ltd.) (27), as described in SI Materials and Methods.
Selection of High-Saccharification Putative Mutants.
Plants were selected from the first-round screen if saccharification was 20% higher than WT and significant at P ≤ 0.01 using a t test. To examine heritability, 12 seeds were sown from each putative mutant selected in the first screen and the plant material tested for saccharification as described above. The saccharification trait was considered to be heritable if all of the offspring showed a similar saccharification trait as observed in the first screen.
Fourier Transform Infrared Spectroscopy.
Fourier transform infrared (FTIR) spectra were obtained for WT and the 12 sac lines for the wavelength range 850–1,850 cm−1 using a Spectrum One (Perkin-Elmer). Three spectra were collected for each sample, and the triplicate-averaged spectrum was used for principal component analysis (PCA). Further details are described in SI Materials and Methods.
Cell-Wall Composition Analysis.
All cell-wall composition analyses were carried out on alcohol insoluble residue (AIR) and in biological triplicates. For details on how AIR was obtained, see SI Materials and Methods. Lignin content was quantified using Foster et al.’s (33) acetyl bromide method. Crystalline cellulose content was analyzed using Foster et al.’s (34) anthrone-sulfuric acid method. Matrix polysaccharide compositional analysis of AIR and a xylan-enriched fraction extracted from AIR was performed by hydrolyzing matrix polysaccharides into component monosaccharides and analyzing using HPAEC. The xylan-enriched fraction was extracted from AIR as described in ref. 40. Ferulic acid was extracted using 1 M NaOH and detected and quantified using HPLC. Details of all four analyses are described in SI Materials and Methods. Lignin composition was measured by thioacidolysis followed by analysis of the H, G, and S monomer products by GC-MS of their trimethylsilyated derivatives, as described by Lapierre et al. (55). Immunocarbohydrate microarray analysis was performed as described in Alonso-Simón et al. (56).
Stem Mechanical Properties.
The strength and stiffness of sac and WT stems was assessed using three-point bending tests. Stem lengths of 5 cm were cut from the third internode of dried plant material, and bending tests were performed on this material using a universal testing machine. For further details, see SI Materials and Methods.
Mapping.
The sac1 line was backcrossed to WT, the resulting BCF1 progeny allowed to self and 250 of the BCF2 seeds sown. The BCF2 plants were subject to saccharification analysis as described in SI Materials and Methods. DNA from the top 40 BCF2 plants with the highest saccharification was extracted, pooled, and sent for whole-genome sequencing by The Genome Analysis Centre (TGAC). Homo- and heterozygous SNPs were called and filtered to exclude low-quality SNPs. For further details on the mapping process, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Sébastien Antelme (Institut National de la Recherche Agronomique, Versailles) for cultivating the Brachypodium plants for the mutagenized population. We thank Dr. Avtar Matharu and Chris Mortimer (Green Chemistry, University of York) for use of the universal testing device for stem strength and stiffness analysis. P.E.M. is funded by a studentship from the Biotechnology and Biological Sciences Research Council (BBSRC). Development of the automated saccharification platform was funded by The European Commission's Seventh Framework Programme (FP7) project Renewall (211982) and by BBSRC projects BB/G016178 and BB/G016194. Development of the Brachypodium mutant collection was financed in part by FP7 RENEWALL and Knowledge-Based Bio Economy project CELLWALL (to H.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414020111/-/DCSupplemental.
References
- 1.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103(30):11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somerville C, Youngs H, Taylor C, Davis SC, Long SP. Feedstocks for lignocellulosic biofuels. Science. 2010;329(5993):790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11(3):301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Harris D, DeBolt S. Synthesis, regulation and utilization of lignocellulosic biomass. Plant Biotechnol J. 2010;8(3):244–262. doi: 10.1111/j.1467-7652.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- 5.Vanholme R, Van Acker R, Boerjan W. Potential of Arabidopsis systems biology to advance the biofuel field. Trends Biotechnol. 2010;28(11):543–547. doi: 10.1016/j.tibtech.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25(7):759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 7.Van Acker R, et al. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels. 2013;6(46) doi: 10.1186/1754-6834-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansfield SD, Kang K-Y, Chapple C. Designed for deconstruction: Poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012;194(1):91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K, et al. An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell. 2012;24(7):3135–3152. doi: 10.1105/tpc.112.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouvier d’Yvoire M, et al. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 2013;73(3):496–508. doi: 10.1111/tpj.12053. [DOI] [PubMed] [Google Scholar]
- 11.Shadle G, et al. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry. 2007;68(11):1521–1529. doi: 10.1016/j.phytochem.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Studer MH, et al. Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci USA. 2011;108(15):6300–6305. doi: 10.1073/pnas.1009252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu C, et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA. 2011;108(9):3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penning BW, et al. Genetic determinants for enzymatic digestion of lignocellulosic biomass are independent of those for lignin abundance in a maize recombinant inbred population. Plant Physiol. 2014;165(4):1475–1487. doi: 10.1104/pp.114.242446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo DK, Stork J, DeBolt S, Maiti IB. Manipulating cellulose biosynthesis by expression of mutant Arabidopsis proM24:CESA3(ixr1-2) gene in transgenic tobacco. Plant Biotechnol J. 2013;11(3):362–372. doi: 10.1111/pbi.12024. [DOI] [PubMed] [Google Scholar]
- 16.Harris D, Stork J, Debolt S. Genetic modification in cellulose-synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. Glob Change Biol Bioenergy. 2009;1(1):51–61. [Google Scholar]
- 17.Abramson M, Shoseyov O, Shani Z. Plant cell wall reconstruction toward improved lignocellulosic production and processability. Plant Sci. 2010;178(2):61–72. [Google Scholar]
- 18.Gomez LD, Steele-King CG, McQueen-Mason SJ. Sustainable liquid biofuels from biomass: The writing’s on the walls. New Phytol. 2008;178(3):473–485. doi: 10.1111/j.1469-8137.2008.02422.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee C, Teng Q, Huang W, Zhong R, Ye ZH. Down-regulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol. 2009;50(6):1075–1089. doi: 10.1093/pcp/pcp060. [DOI] [PubMed] [Google Scholar]
- 20.DeMartini JD, et al. Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energ Environ Sci. 2013;6(3):898–909. [Google Scholar]
- 21.de O Buanafina MM. Feruloylation in grasses: Current and future perspectives. Mol Plant. 2009;2(5):861–872. doi: 10.1093/mp/ssp067. [DOI] [PubMed] [Google Scholar]
- 22.Buanafina MM, Langdon T, Hauck B, Dalton S, Morris P. Expression of a fungal ferulic acid esterase increases cell wall digestibility of tall fescue (Festuca arundinacea) Plant Biotechnol J. 2008;6(3):264–280. doi: 10.1111/j.1467-7652.2007.00317.x. [DOI] [PubMed] [Google Scholar]
- 23.Bartley LE, et al. Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol. 2013;161(4):1615–1633. doi: 10.1104/pp.112.208694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mewalal R, Mizrachi E, Mansfield SD, Myburg AA. Cell wall-related proteins of unknown function: Missing links in plant cell wall development. Plant Cell Physiol. 2014;55(6):1031–1043. doi: 10.1093/pcp/pcu050. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Ye C-Y, Bisaria A, Tuskan GA, Kalluri UC. Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci. 2011;181(6):675–687. doi: 10.1016/j.plantsci.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Truntzler M, et al. Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet. 2010;121(8):1465–1482. doi: 10.1007/s00122-010-1402-x. [DOI] [PubMed] [Google Scholar]
- 27.Gomez LD, Whitehead C, Barakate A, Halpin C, McQueen-Mason SJ. Automated saccharification assay for determination of digestibility in plant materials. Biotechnol Biofuels. 2010;3:23. doi: 10.1186/1754-6834-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez LD, Bristow JK, Statham ER, McQueen-Mason SJ. Analysis of saccharification in Brachypodium distachyon stems under mild conditions of hydrolysis. Biotechnol Biofuels. 2008;1:15. doi: 10.1186/1754-6834-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fagard M, Hofte H, Vernhettes S. Cell wall mutants. Plant Physiol Biochem. 2000;38(1-2):15–25. [Google Scholar]
- 30.Kacurakova M, Capek P, Sasinkova V, Wellner N, Ebringerova A. FT-IR study of plant cell wall model compounds: Pectic polysaccharides and hemicelluloses. Carbohydr Polym. 2000;43(2):195–203. [Google Scholar]
- 31.Xiao B, Sun XF, Sun RC. Chemical, structural, and thermal characterizations of alkali-soluble lignins and hemicelluloses, and cellulose from maize stems, rye straw, and rice straw. Polym Degrad Stabil. 2001;74(2):307–319. [Google Scholar]
- 32.Guo G-L, Hsu D-C, Chen W-H, Chen W-H, Hwang W-S. Characterization of enzymatic saccharification for acid-pretreated lignocellulosic materials with different lignin composition. Enzyme Microb Technol. 2009;45(2):80–87. [Google Scholar]
- 33.Foster CE, Martin TM, Pauly M. Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass). Part I: Lignin. J Vis Exp. 2010;(37):e1745. doi: 10.3791/1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster CE, Martin TM, Pauly M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part II: Carbohydrates. J Vis Exp. 2010;(37):e1745. doi: 10.3791/1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen HL, et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem. 2012;287(47):39429–39438. doi: 10.1074/jbc.M112.396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abe A, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30(2):174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 37.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drake JW, Baltz RH. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- 39.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiniquy D, et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA. 2012;109(42):17117–17122. doi: 10.1073/pnas.1202079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders N, et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA. 2012;109(3):989–993. doi: 10.1073/pnas.1115858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brkljacic J, et al. Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 2011;157(1):3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garvin DF, et al. An SSR-based genetic linkage map of the model grass Brachypodium distachyon. Genome. 2010;53(1):1–13. doi: 10.1139/g09-079. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Weng J-K, Chapple C. Improvement of biomass through lignin modification. Plant J. 2008;54(4):569–581. doi: 10.1111/j.1365-313X.2008.03457.x. [DOI] [PubMed] [Google Scholar]
- 45.Allison GG, Morris C, Clifton-Brown J, Lister SJ, Donnison IS. Genotypic variation in cell wall composition in a diverse set of 244 accessions of Miscanthus. Biomass Bioenergy. 2011;35(11):4740–4747. [Google Scholar]
- 46.McCartney L, Marcus SE, Knox JP. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem. 2005;53(4):543–546. doi: 10.1369/jhc.4B6578.2005. [DOI] [PubMed] [Google Scholar]
- 47.Grabber JH, Ralph J, Hatfield RD. Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J Agric Food Chem. 1998;46(7):2609–2614. [Google Scholar]
- 48.Tu Y, et al. Functional analyses of caffeic acid O-Methyltransferase and Cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne) Plant Cell. 2010;22(10):3357–3373. doi: 10.1105/tpc.109.072827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piquemal J, et al. Down-regulation of caffeic acid o-methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 2002;130(4):1675–1685. doi: 10.1104/pp.012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, et al. Transgenic down-regulation of caffeic acid O-methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea) Funct Plant Biol. 2004;31(3):235–245. doi: 10.1071/FP03254. [DOI] [PubMed] [Google Scholar]
- 51.Lapierre C. 2010. Determining lignin structure by chemical degradations. Lignin and Lignans, Advances in Chemistry, eds Heitner C, Dimmel D, Schmidt JA (CRC, Boca Raton, FL), pp 11–48.
- 52.Lapierre C, Monties B, Rolando C. Thioacidolyses of diazomethane-methylated pine compression wood and wheat straw in situ lignins. Holzforschung. 1988;42(6):409–411. [Google Scholar]
- 53.Mitchell RAC, Dupree P, Shewry PR. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 2007;144(1):43–53. doi: 10.1104/pp.106.094995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalmais M, et al. A TILLING platform for functional genomics in Brachypodium distachyon. PLoS ONE. 2013;8(6):e65503. doi: 10.1371/journal.pone.0065503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lapierre C, Pollet B, Rolando C. New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res Chem Intermed. 1995;21(3-5):397–412. [Google Scholar]
- 56.Alonso-Simón A, et al. High-throughput microarray profiling of cell wall polymers during hydrothermal pre-treatment of wheat straw. Biotechnol Bioeng. 2010;105(3):509–514. doi: 10.1002/bit.22546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.