Significance
Mutations in the UBQLN2 gene, which encodes the ubiquitin-like protein ubiquilin2 (UBQLN2) have been shown to cause ALS and ALS/dementia. Ubiquilin2 links familial and sporadic forms of the disease through pathology observed in the spinal cords of all ALS cases and in the brains of ALS/dementia cases with or without UBQLN2 mutations. In this communication, we develop and characterize a mouse model of mutant UBQLN2-linked dementia. We demonstrate that mutant mice develop impairment in the protein degradation pathway, abnormal protein aggregation, synaptic dysfunction, and cognitive deficits. This model provides a useful tool to further study dementia and develop rational therapies.
Abstract
Mutations in the gene encoding ubiquilin2 (UBQLN2) cause amyotrophic lateral sclerosis (ALS), frontotemporal type of dementia, or both. However, the molecular mechanisms are unknown. Here, we show that ALS/dementia-linked UBQLN2P497H transgenic mice develop neuronal pathology with ubiquilin2/ubiquitin/p62-positive inclusions in the brain, especially in the hippocampus, recapitulating several key pathological features of dementia observed in human patients with UBQLN2 mutations. A major feature of the ubiquilin2-related pathology in these mice, and reminiscent of human disease, is a dendritic spinopathy with protein aggregation in the dendritic spines and an associated decrease in dendritic spine density and synaptic dysfunction. Finally, we show that the protein inclusions in the dendritic spines are composed of several components of the proteasome machinery, including UbG76V–GFP, a representative ubiquitinated protein substrate that is accumulated in the transgenic mice. Our data, therefore, directly link impaired protein degradation to inclusion formation that is associated with synaptic dysfunction and cognitive deficits. These data imply a convergent molecular pathway involving synaptic protein recycling that may also be involved in other neurodegenerative disorders, with implications for development of widely applicable rational therapeutics.
Protein aggregates or inclusions with immunoreactivity to ubiquitin represent a common pathological hallmark in a broad range of late-onset neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson disease (PD), frontotemporal dementia (FTD), and amyotrophic lateral sclerosis (ALS) (1). However, the molecular mechanisms underlying the formation of these inclusions and their relationship to neuronal dysfunction and degeneration are poorly understood. Mutations in UBQLN2, which encodes the ubiquitin-like protein ubiquilin2 (UBQLN2), have been recently shown to cause a subset of ALS, FTD-type of dementia, or both (2, 3). Notably, mutations within and in close proximity to the PXX domain of ubiquilin2 appear to have high penetrance as shown in familial cases (2). The distribution of ubiquilin2-positive inclusions in the brain and spinal cord is well correlated with cognitive and motor symptoms of patients with diverse etiology, including chromosome 9 open reading frame 72 (C9ORF72)-linked cases (2, 4). The ubiquilin2-positive inclusions appear to cover a wide range of protein inclusions, including those without TAR DNA binding protein (TDP43) immunoreactivity (2, 4), suggesting a primary role for ubiquilin2 in inclusion formation and neurodegeneration. Therefore, understanding the pathophysiological basis of UBQLN2-linked dementia may provide mechanistic insight into the pathogenesis of neurodegenerative disorders and allow for the design of rational therapies. To this end, we developed and characterized mutant UBQLN2 transgenic mice.
Results
Development of UBQLN2P497H Transgenic Mice.
Human UBQLN2 is an intronless gene. We analyzed the UBQLN2 promoter activity using chloramphenicol acetyltransferase (CAT) reporter system and constructed a 10.8-kb EcoRI/BamHI human UBQLN2 genomic DNA transgene with a P497H mutation, which was identified in a large family with ALS and dementia (2) (Fig. S1 A and B). Transgenic mice were developed by microinjection of the transgene into fertilized eggs.
We identified 13 transgenic founders and performed initial genetic analysis to select a founder line with a relatively higher copy number of the transgene (Fig. S1C). The human UBQLN2P497H transgene appeared to be integrated into the mouse Y chromosome in this line, because the transgene cotransmitted with the Y chromosome and all of the transgenic mice were males (n > 200). The expression profiles of the human UBQLN2P497H transgene and mouse endogenous ubqln2 showed similar expression levels with relatively higher expression in the brain and spinal cord, but much lower in the liver (Fig. S1D).
Cognitive Deficits in UBQLN2P497H Transgenic Mice.
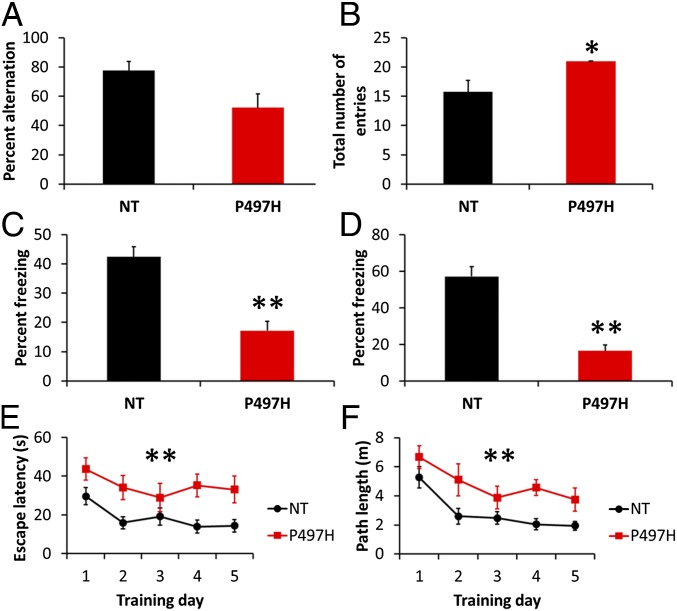
Two major categories of symptoms have been observed in patients with UBQLN2 mutations, which involve motor and cognitive functions. We did not observe gross motor abnormalities through the lifetime (>600 d, n > 40) of the UBQLN2P497H transgenic mice. Y-maze test for spontaneous alternation revealed good spatial memory in the nontransgenic controls (78%), but near random arm selection (52%) for transgenic mice, although this did not reach statistical significance (Fig. 1A). The number of arm entries was significantly higher for transgenic mice compared with controls (Fig. 1B). Similar performance in the Y maze has been observed in the transgenic mice of AD (5).
Fig. 1.
Cognitive deficits in UBQLN2P497H mice. (A) Spatial learning and memory of UBQLN2P497H and nontransgenic (NT) control mice (11–13 mo) was assessed by spontaneous alternation in the Y maze (n = 4–7 per group). UBQLN2P497H mice exhibited random arm selection (52%) instead of the spontaneous alternation of control mice (P = 0.09). (B) The total number of arm entries was higher for UBQLN2P497H mice, indicating that they do not have a motor deficit. (C and D) Delay fear conditioning was tested by assessing freezing behavior to the same context (C) and to the conditioned stimulus (CS) in a novel context (D) 24 h after training. The average percent freezing is shown as an index of delay fear memory (n = 5–7 per group). Note that the UBQLN2P497H mice showed significantly lower levels of freezing than control mice in delay fear conditioning. (E and F) Mice were trained with six trials (three blocks of two trials) per day for 5 d in the Morris water maze. The average latency and path length to reach the hidden platform is plotted across training days (n = 7–11 per group). ANOVA indicated a significantly shorter escape latency for the NT mice (F1,16 = 7.2, P = 0.02), and a decrease in escape latency across sessions (F4,64 = 9.2, P < 0.0001). ANOVA also indicated a significantly shorter path length for the NT mice (F1,16 = 7.4, P = 0.01), and a decrease in path length across sessions (F4,64 = 15.6, P < 0.0001). Each data point represents the mean ± SEM. The longer escape latency and longer path length of the UBQLN2P497H mice is indicative of cognitive impairment. *P < 0.01, **P < 0.0001 versus NT compared by ANOVA and post hoc Fisher's protected least significant difference (PLSD) test.
Temporal memory function was tested by a delay fear conditioning task that assesses freezing behavior to the same context and to a conditioned stimulus (CS) in a novel context 24 h after training. The transgenic mice showed significantly decreased freezing in response to the context and CS compared with control mice (Fig. 1 C and D).
Spatial memory was tested using the Morris water maze. The time and distance to reach a visible platform did not show significant differences between genotypes, suggesting equal visual acuity, swimming ability, and motivation to reach the platform. In the hidden platform version, mice showed an improvement in the time to find the platform, but significant differences were found between the transgenic mice and controls in acquisition rates across all training sessions (Fig. 1 E and F), with UBQLN2P497H mice taking more time and longer path lengths to reach the platform.
Ubiquilin2-Immunoreactive Inclusions in the Hippocampus and Other Brain Regions of Transgenic Mice.
Ubiquilin2-immunoreactive protein aggregates have been shown to correlate with motor and cognitive symptoms in patients with UBQLN2 mutations. In the spinal cord, we observed some ubiquilin2-immunoreactive aggregates predominantly located around the small interneurons in the Rexed lamina II of the dorsal horns (Fig. S2 A–D). The distribution pattern of these aggregates was similar to that in the patients with UBQLN2 mutations (Fig. S2 E and F), and they were also immunoreactive for ubiquitin and p62 (Fig. S2 G–I). We did not observe skeinlike inclusions, the characteristic pathology of ALS, in anterior horn motor neurons, which might explain the lack of motor symptoms.
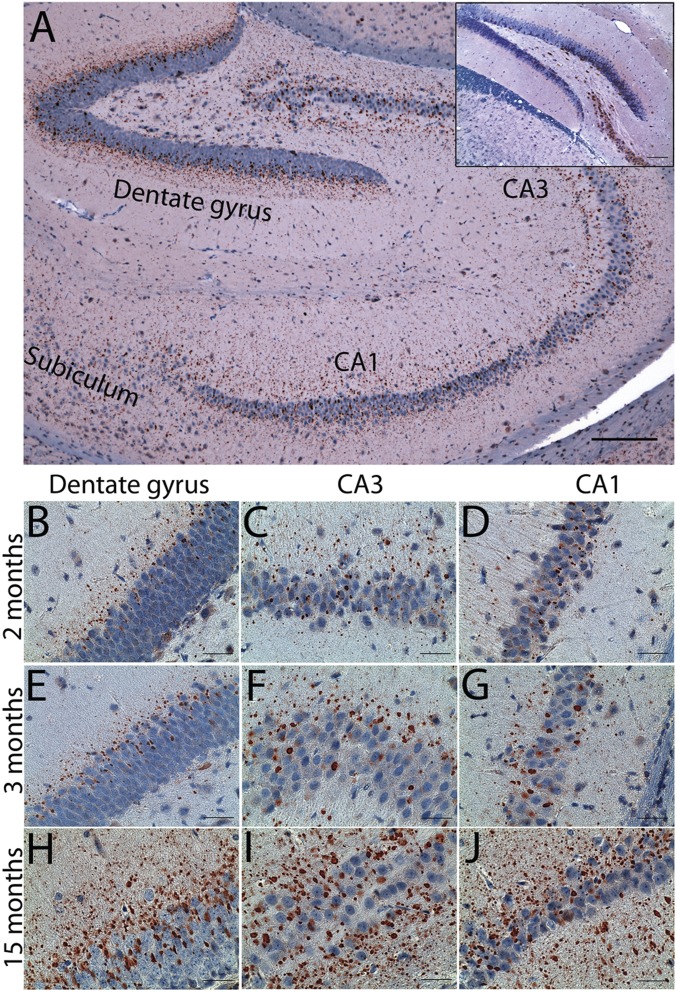
The peculiar pathology in ALS/dementia patients with UBQLN2 mutations is the presence of ubiquilin2 aggregates, which is most prominent in the hippocampus, especially in the molecular layer of the dentate gyrus (2). This pathology is also present in some ALS/dementia patients without UBQLN2 mutations (2). Recently, this pathology has been shown to be a common feature in the C9ORF72-linked ALS/dementia and frontotemporal lobar degeneration with TDP43 positive inclusions (FTLD-TDP) cases (4), which account for over 10% of familial FTD and over 60% of familial ALS/dementia (3). We performed initial pathological analysis using 4-mo-old transgenic mice. We observed striking ubiquilin2 pathology in the hippocampus (Fig. 2A), which is similar to that in human patients (2).
Fig. 2.
Ubiquilin2 pathology in transgenic mice. Immunohistochemistry with the ubiquilin2-N antibody (2) was performed using brain sections from UBQLN2P497H transgenic mice of different ages. (A) An overall image shows ubiquilin2-positive aggregates in the hippocampus of a 4-mo-old mouse. (B–J) Representative ubiquilin2-positive aggregates in the dentate gyrus (B, E, and H), CA3 (C, F, and I), and CA1 (D, G, and J) from mice of 2 mo (B–D), 3 mo (E–G), and 15 mo (H–J) are shown. Ubiquilin2 pathology is shown in transgenic, but not nontransgenic mice (Inset), even though diffused cytosolic ubiquilin2 reactivity is well shown in some neuronal cells, especially in the pyramidal neurons. (Scale bars: A, 200 µm; B–J, 50 µm.)
The ubiquilin2-positive aggregates in the dentate gyrus were prominent in the lower one-third of the molecular layer and upper one-third of the granule cell layer, with size ranging from 1 µm to 10 µm (Fig. 2A). Their size progressively decreased from the granule cell layer to the molecular layer. The ubiquilin2 aggregates were prominent in cornu ammonis (CA) subfields CA1 to CA4 and subiculum, with their distribution localized mostly to regions proximal to neurons (Fig. 2A).
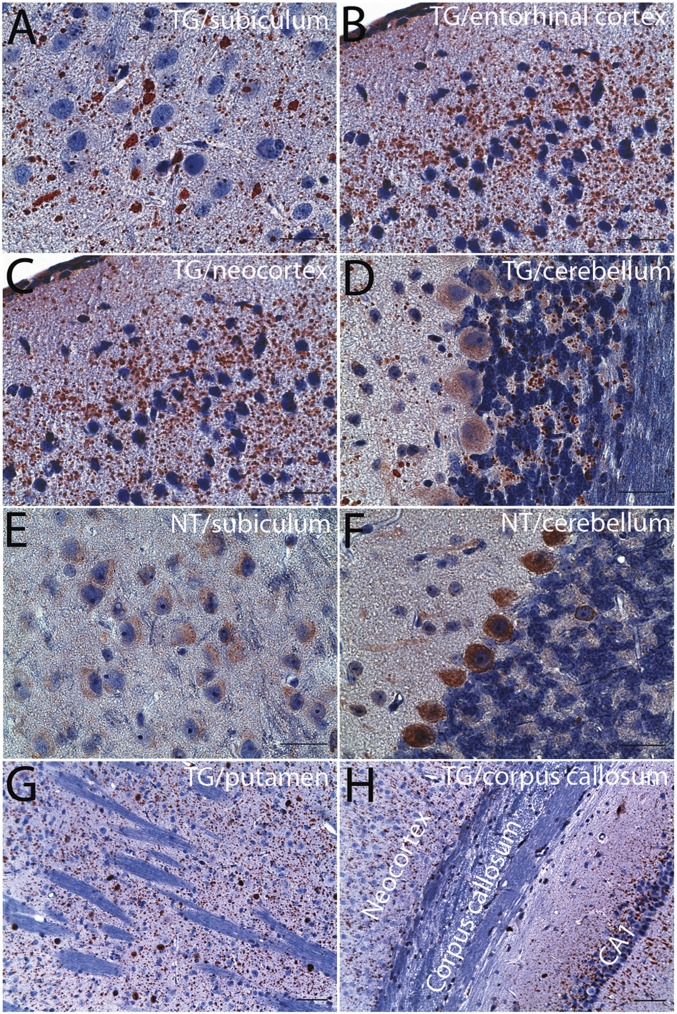
We carried out a chronological examination of the development of ubiquilin2 pathology in transgenic mice at different time points, ranging from 1 mo to 15 mo. The number and size of the aggregates increased with age (Fig. 2 B–J). In the 1-mo-old mice, a limited number of small ubiquilin2 aggregates could be found in the dentate gyrus and CA1–CA4 (Fig. S3). In addition to the most prominent ubiquilin2 pathology in the hippocampus, we also observed widespread ubiquilin2 pathology, which was progressively more severe with age in other parts of the brain, especially in frontal and temporal lobes (Fig. 3 and Fig. S4). This pathology was not observed in control mice, even at advanced time points (Fig. 2). Notably, the ubiquilin2 aggregates were predominant in the neuropil of the gray matter, but not in the white matter representing axonal tracts (Fig. 3 and Fig. S4), suggesting that these aggregates are likely to be dendritic, rather than axonal. The density of the aggregates was proportionally associated with the density of the neurons. Similar to the pathology of the FTD cases, the ubiquilin2 pathology was most prominent in layers 2 and 3 of frontal and temporal cortex in the neocortex region (Fig. S4).
Fig. 3.
Ubiquilin2 pathology in other regions of the brain. Immunohistochemistry with a ubiquilin2 antibody (ubiquilin2-N) was performed on the brain sections from 15-mo-old UBQLN2P497H transgenic (TG), and age- and sex-matched nontransgenic mice (NT). Representative images from subiculum (A), entorhinal cortex (B), neocortex (C), cerebellum (D), putamen (G), and corpus callosum (H) of TG mice are indicated. Representative images from subiculum (E) and cerebellum (F) of NT mice are shown for comparison. Ubiquilin2 aggregates are predominantly present in the gray matter (A–D), but not white matter (D, G, and H). (Scale bars: A–F, 50 µm; G and H, 100 µm.)
In addition to ubiquilin2, several other ALS-linked proteins, such as TDP43, FUS RNA binding protein (FUS), and valosin containing protein (VCP), have also been associated with ALS and FTD (3). Optineurin (OPTN), another ALS- and dementia-linked protein, is related to ubiquitin-conjugated protein clearance (3). We found VCP and OPTN, but not FUS and TDP43, in the ubiquilin2 aggregates (Fig. S5). Consistent with the ubiquilin2 pathology in the patients with UBQLN2 mutations, ubiquilin2-containing aggregates in the transgenic mice were also immunoreactive for ubiquitin and p62 (Fig. S6).
Dendritic Spinopathy in UBQLN2P497H Transgenic Mice.
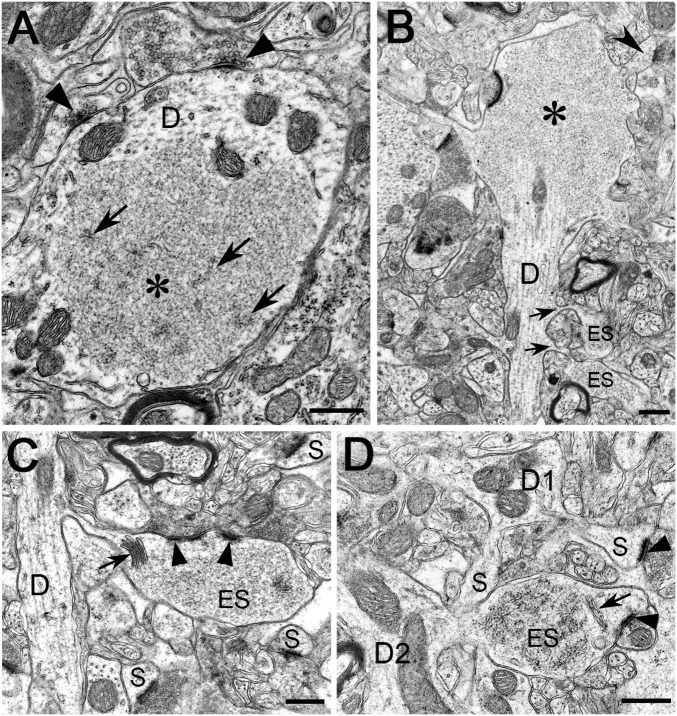
To further characterize the ubiquilin2 aggregates, we analyzed brain sections by electron microscopy (EM). Control mice were completely free of aggregates. By contrast, abnormal structures with the appearance of protein aggregates were prominent in mutant mice. By standard EM, the aggregates were characterized by a fine granulofibrillar structure and lacked a surrounding membrane. They were situated in the somatodendritic compartment of neurons, mostly lying free in the cytoplasm of dendritic spines and sometimes dendritic stems, but occasionally also in the nucleus (Fig. 4 and Fig. S7). Out of several hundred aggregates obtained from different regions of the brain, none were located in nerve fibers, axon terminals, or glial cells. Aggregates were often seen adjacent to microtubules, endoplasmic reticulum cisterns, and other cell organelles, but occasionally they contained cytoplasmic organelles, primarily multivescicular bodies, lysosomes, and mitochondria, within their interior (Fig. S7). However, the most striking finding was the presence of aggregates in the dendritic spines of the hippocampal formation and cerebral cortex. A sizeable proportion of these easily identifiable postsynaptic profiles appeared conspicuously enlarged, measuring four to five times their normal diameter. Moreover, the frequency of occurrence of the enlarged spines was roughly commensurate with the density of the neuropil aggregates observed by immunohistochemistry (Fig. S8). Strikingly, the interior of many dendritic spines and stems in the hippocampal formation and the cerebral cortex were occupied by an individual granulofibrillar aggregate. Usually, whereas the postsynaptic density was well preserved, the spine apparatus might be pushed to one side of the profile. Thus, pathologically, the neuronal fine structure can be described as a “dendritic spinopathy,” which is characterized by protein inclusions primarily present in the dendritic spines. This specific type of pathology has apparently not been previously reported in neurodegenerative disorders.
Fig. 4.
Protein aggregates in dendritic spines and stems. (A–D) Electron microscopy was performed using the hippocampus and the frontal cortex of a 15-mo-old UBQLN2P497H transgenic mouse. Representative protein aggregates containing membrane-free and flocculent-to-granulofibrillar material are illustrated in the dendritic spines and stems (A–D). (A) The inclusion (asterisk) occupies a large area of an obliquely sectioned, midsized dendritic trunk (indicated by D). Some filamentous elements within the inclusion are indicated by arrows; axodendritic synapses are indicated by arrowheads. The fine structure of mitochondria (unlabeled) appears unaltered. (B) A large granulofibrillar inclusion (asterisk) fills the ballooning portion of a small stem dendrite (indicated by D). Two enlarged spines (ES) containing flocculent material emanate from the dendritic trunk (arrows) and a normal spine (arrowhead) from the ballooned portion. (C) A small dendritic trunk (indicated by D) contains organelles with unchanged fine structure and emits a swollen, thin-necked spine (ES) filled by granulofibrillar material. Three spines with normal fine structure are labeled (S). Split postsynaptic density is indicated by arrowheads. A hypertrophic spine apparatus is indicated by an arrow. (D) A small dendritic trunk (D1) emits two spines (S) and has apparently normal ultrastructure, whereas another small dendrite (D2) has an enlarged spine (ES) with granulofibrillar contents. A spine apparatus is indicated by an arrow and synaptic junctions by arrowheads. (Scale bars: 0.5 µm.)
Defect of Synaptic Function in UBQLN2P497H Transgenic Mice.
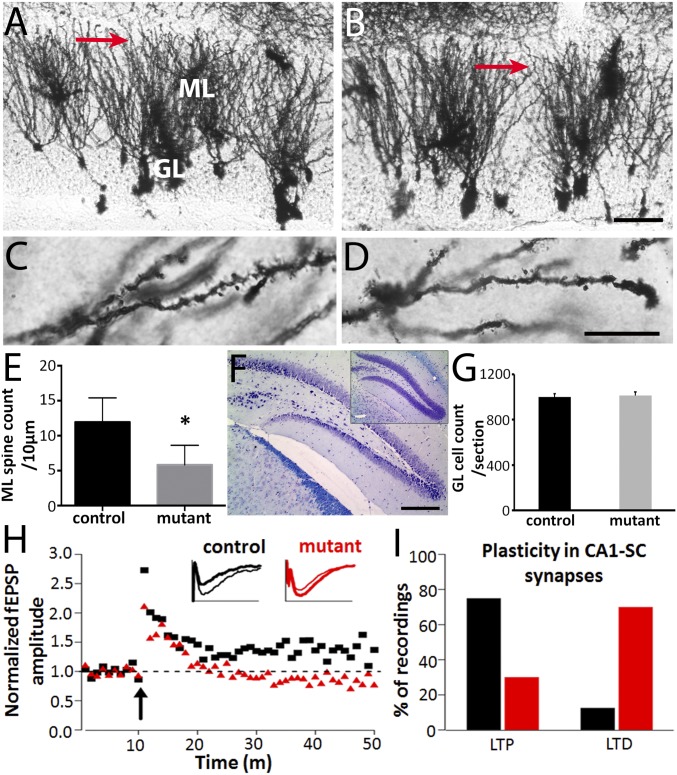
Synaptic failure has been implicated in other neurodegenerative disorders, such as AD (6). We performed Golgi staining to assess dendritic spine density. We observed a marked decrease in the density of dendritic spines in the molecular layer of the dentate gyrus in mutant mice, compared with controls (Fig. 5 A–E). However, Bielschowsky silver stain did not reveal the presence of degenerating neurons. Moreover, we did not observe a difference in the granule cell count in the dentate gyrus (Fig. 5 F and G). This is in keeping with previous reports of models of human neurodegeneration, such as, spinal and bulbar muscular atrophy, AD, and Huntington disease, where extensive neuronal pathology and neuronal/synaptic dysfunction were noted in the absence of any obvious neuronal loss (7–10).
Fig. 5.
Altered synaptic plasticity in the UBQLN2P497H mice. Representative images of the molecular layer of dentate gyrus of 15-mo-old control (A and C) and UBQLN2 transgenic mice (B and D). The red arrows in A and B indicate the areas magnified to display the spines presented in C and D. GL, granule cell layer of dentate gyrus; ML, molecular layer of dentate gyrus. (Scale bar in B, 50 µm, also applies to A; and in D, 10 µm, also applies to C.) Statistical analysis indicates that there was a significant decrease in spine density in the molecular layer of dentate gyrus of UBQLN2 transgenic mice compared with control mice (E, *P < 0.01). (F and G) Neuronal cell count in the granule cell layer of the dentate gyrus after cresyl violet and Luxol fast blue staining did not reveal any differences between transgenic and control mice (n = 3–4; P = 0.75). (Scale bar: 500 μm in F.) Extracellular recordings were performed from acute hippocampal slices in 3-mo-old transgenic mice and age-matched controls. (H) Average time course of the field potential (normalized to the pretrain response) recorded in CA1 stratum radiatum in response to stimulation (arrow) of the Schaffer collateral pathway. Inset traces: responses from individual slices from control (black traces) and transgenic (red traces) animals recorded before (thick traces) and 40 min after (thin traces) tetanic stimulation (four times 1-s long 100-Hz trains, 30-s intervals). (I) Summary of the type of synaptic plasticity observed 40 min after the tetanic stimulation in 8 control (black bars) and 10 transgenic (red bars) slices, measured from the excitatory postsynaptic potential (EPSP) slopes. The majority (75%) of the recordings from control mice produced LTP; this was not the case in transgenic animals, in which the same protocol induced LTP in only 13% of the cases. In slices from transgenic animals, the polarity of the synaptic modulation was actually reversed and led to LTD in the large majority (70%) of the recordings.
AD-related amyloid-β oligomers isolated from cells (11) and the brains of AD cases (12) have been shown to inhibit long-term potentiation (LTP) and enhance long-term depression (LTD) when injected into the rodent hippocampus. To explore the potential link between the ubiquilin2-related spinopathy and cognitive deficits, and to identify the pathophysiological basis of dementia, we analyzed hippocampal slices from UBQLN2P497H transgenic mice using an electrophysiological approach. We found that the mutant mice showed remarkably decreased LTP and increased LTD compared with age- and sex-matched controls (Fig. 5 H and I), suggesting that the ubiquilin2-containing inclusions in dendritic spines impair the effective handling of signal transmission in postsynaptic dendrites.
Impairment of Ubiquitin–Proteasome System in UBQLN2P497H Transgenic Mice.
Postsynaptic functions are regulated by finely tuned turnover of the synaptic proteins via the ubiquitin–proteasome system (UPS) (13). We have previously shown that mutant UBQLN2 impairs the UPS in vitro (2). To test this in vivo, we generated double transgenic mice expressing both UBQLN2P497H and UbG76V–GFP, a representative ubiquitinated protein substrate (UBQLN2P497H/UbG76V–GFP) (14). Similar to the observations in vitro, we found accumulation of UbG76V–GFP in the brain of UBQLN2P497H/UbG76V–GFP mice compared with single UbG76V–GFP transgenic mice (Fig. S9).
Aggregation of Proteasome Subunits, Ubiquilin2 and UbG76V–GFP in Transgenic Mice.
Ubiquilin2 is a member of one of the ubiquitin-like protein families (ubiquilins). Ubiquilins are characterized by the presence of an N-terminal ubiquitin-like (UBL) domain and a C-terminal ubiquitin-associated (UBA) domain. This structural organization is characteristic of proteins that deliver ubiquitinated proteins to the proteasome for degradation. In accordance with this function, the UBL domain of the ubiquilins binds to 19S subunits of the proteasome, such as PSMD4 and ADRM1, and the UBA domain binds to ubiquitin chains that are typically conjugated to proteins marked for degradation by the proteasome (15). The disease-causing mutations identified to date are located in the middle part of ubiquilin2, leaving the UBL and UBA domains intact. Therefore, it is likely that the UBL and UBA domains of mutant ubiquilin2 may bind to the proteasome and ubiquitinated substrates, respectively, but mutations may affect efficient clearance of ubiquitinated proteins. Moreover, the binding of the UBL domain of mutant ubiquilin2 to proteasomes may also prevent other UBL-containing proteins and ubiquitinated substrates from binding to the proteasomes, therefore further impairing the UPS.
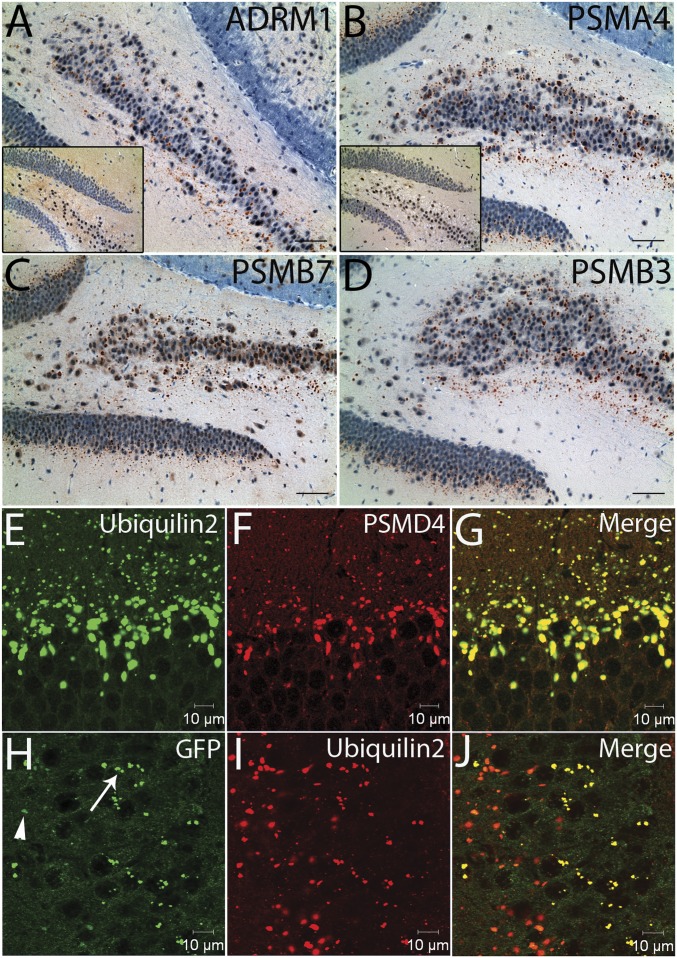
We performed immunohistochemistry on the transgenic mouse brain sections using antibodies against several proteasome subunits, including PSMD4 (19S-S5A, RPN10), ADRM1 (19S-ADRM1, RPN13), PSMA4 (20S-α3), PSMB7 (20S-β2), and PSMB3 (20S-β3). We noted immunoreactive aggregates, which shared similar morphology and distribution pattern to ubiquilin2-positive aggregates (Fig. 6 A–D). As expected, proteasome subunits, ubiquilin2 and UbG76V–GFP were colocalized in the protein aggregates in the UBQLN2P497H/UbG76V–GFP double transgenic mice (Fig. 6 E–J and Fig. S9 B–J). These data support the notion that mutant ubiquilin2 is able to bridge UbG76V–GFP and the proteasomes, but UbG76V–GFP is not efficiently degraded, resulting in dysfunctional proteasome complexes in dendritic spines, thereby leading to synaptic dysfunction.
Fig. 6.
Proteasome subunits in protein aggregates of the UBQLN2P497H transgenic mice. (A–D) Immunohistochemistry was performed on hippocampal sections using antibodies to 26S proteasome subunits. Representative images show the protein aggregates containing subunits of 19S regulatory particle (ADRM1) and 20S enzymatic core particle (PSMA4, PSMB7, and PSMB3) in transgenic mice, but not in nontransgenic controls (Inset). (E–G) Colocalization of proteasome subunits, ubiquilin2 and ubiquitinated protein substrate in protein aggregates of the UBQLN2P497H/UbG76V–GFP double transgenic mice. Confocal microscopy was performed on the hippocampal sections of the UBQLN2P497H/UbG76V–GFP double transgenic mice using antibodies to ubiquilin2, proteasome subunits (PSMD4), and GFP as indicated. Protein aggregates containing proteasome subunits, ubiquilin2 and GFP are shown in the dentate gyrus (E–G) and CA3 (H–J). The majority of the protein aggregates showed weak to moderate (arrowhead) GFP immunoreactivity; some aggregates showed strong (arrow) GFP immunoreactivity (H–J).
Discussion
Here, we report the development and comprehensive characterization of transgenic mice expressing an ALS/dementia-linked UBQLN2 mutation. These mice recapitulate several clinical and pathological features of dementia observed in the human patients. We note that the motor neuron phenotype and pathology are not recapitulated in these mice. However, this does not appear to be unusual, because mutations in several genes, including C9ORF72, VCP, FUS, TDP43, or UBQLN2 can cause either ALS or FTD, or both, in human patients (16). It is also worth noting that mouse motor neurons may be more resistant to the same pathogenic assaults affecting human motor neurons. For example, mice completely deficient of alsin did not develop an apparent motor neuron phenotype (17); in transgenic mice overexpressing SOD1G93A, it appears that the mutant protein needs to reach a high enough threshold, which is at least three times that of mouse endogenous sod1 for the disease phenotype to develop (18). The expression level of our mutant ubiquilin2 in transgenic mice was similar to that of mouse endogenous ubqln2. It remains to be tested if the transgenic mice with higher levels of mutant UBQLN2 expression develop motor phenotype.
The correlation between protein aggregates and neurodegenerative diseases has long been recognized, but a causal relationship has been debated (1). In the present study, we confirm previous in vitro findings in our new mouse model by showing evidence that ubiquitinated substrates are not effectively degraded by the proteasomes, resulting in the formation of ubiquilin2-immunoreactive protein complexes (aggregates) and functional blockage of the UPS in dendritic spines. These aggregates also contain elements of the proteasome machinery, and their sequestration could impair synaptic protein recycling. In neuronal cells, dynamic control of protein stability is crucial for the development, function, maintenance, and strength of synapses, and the UPS plays an important role in the regulation of synaptic proteins (13). It has been shown that depletion of the 26S proteasomes in mice leads to neurodegeneration, and therefore they are essential for normal neuronal homeostasis and survival (19). Remarkably, proteasomal impairment in individual neurons is sufficient to block activity-induced spine outgrowth (20). Moreover, the UPS is required for mammalian long-term memory formation (21). Furthermore, activity-dependent protein degradation is critical for the formation and stability of fear memory (22). Consistent with these data, we observed a marked reduction in spine density, synaptic dysfunction, and cognitive impairment in our mouse model.
Our data suggest a unifying hypothesis of interplay between aggregate formation and UPS failure in close proximity to synapses. Normally, ubiquitinated substrates and related proteins, such as, ubiquilin2 form functional protein complexes with the proteasomes. However, if ubiquitinated substrates cannot be effectively cleared by the proteasomes due to genetic and/or posttranslational defects, this will lead to functional blockage of the UPS (Fig. S10). In this situation, the UBL domain of the mutant ubiquilin2 could still bind to the 19S proteasome lid, but not deliver the cargo, thereby preventing other UBL-containing proteins or ubiquitinated substrates from binding to the proteasomes, further impairing the UPS function. This could elicit a self-perpetuating pathogenic cascade of events that both accelerates the accumulation of the toxic protein and impairs essential regulatory functions of the UPS. Thus, it is conceivable that if this positive-feedback cycle continues, the aggregates would increase in number and size over time and be observed as a pathological hallmark in diseased tissues. Consistent with this hypothesis, proteasome subunits and ubiquitin have been previously observed in protein aggregates associated with various neurodegenerative diseases (23); and inhibition of the UPS by the disease-associated oligomers of amyloid-β, α-synuclein, prion, or polyQ repeats have also been suggested (24–28). These data support the notion that the failure of clearance of ubiquitinated proteins in specific cellular compartments, such as dendritic spines, may represent a common defect shared by a wide spectrum of neurodegenerative disorders, although the causes and upstream pathways are different.
Dementia is the major disabling symptom in patients with several neurodegenerative disorders, including AD. ALS-associated dementia is typically FTD type, which represents the third most common neurodegenerative disease after AD and PD, and it is as common as AD in presenile dementia (29). Currently, there are no effective therapies, largely because the precise pathogenic mechanisms remain poorly defined. Hippocampal ubiquilin2 pathology, primarily characterized by dendritic spinopathy, is a common feature in C9ORF72-linked ALS/dementia and FTLD-TDP cases, indicating a shared pathogenic mechanism in these subtypes of FTD with divergent causes (2, 4). Development and characterization of appropriate animal models have been a research priority to understand pathogenesis of neurodegenerative disorders and to test therapeutic strategies. Our transgenic mice display several clinical aspects of dementia (deficits in cognitive tasks) and pathologic features (prominent ubiquilin2 aggregates in relevant brain regions). Moreover, reduction in dendritic spine density and impairment of synaptic plasticity, thought to underlie defects in learning and memory, are also present in these mice. Therefore, this mouse model may provide a valuable tool for further studies of FTD and other dementia-related neurodegenerative disorders and for the testing of rational therapeutic approaches as well.
Materials and Methods
Development of UBQLN2 Transgenic Mice.
The 11.8-kb transgene contained a 7.0-kb promoter and a short 5′ untranslated region, UBQLN2 coding sequence, 1.2-kb 3′ untranslated region, and a 0.6-kb fragment following the UBQLN2 polyA signal. The transgene was released from plasmid by restriction digestion, agarose-gel purified, and microinjected into fertilized eggs derived from a zygote of a C57BL/6 (B6) × SJL cross.
Detailed methods for generation and characterization of transgenic mice are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Neurological Disorders and Stroke (NS050641, NS078504, NS070142, NS081474, NS064091, NS009904, NS078287, and AG13854), the National Institute on Aging (AG20506), the Les Turner ALS Foundation, the Vena E. Schaff ALS Research Fund, a Harold Post Research Professorship, the Herbert and Florence C. Wenske Foundation, the David C. Asselin MD Memorial Fund, the Help America Foundation, the George Link, Jr. Foundation, and a Les Turner ALS Foundation/Herbert C. Wenske Foundation Professorship. Part of this work was performed at Northwestern University Transgenic and Targeted Mutagenesis Laboratory supported by the National Cancer Institute (NCI) (CA060553) and at the Cell Imaging Facility supported by NCI (CA060553).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405741111/-/DCSupplemental.
References
- 1.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Deng H-X, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brettschneider J, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012;123(6):825–839. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King DL, Arendash GW. Behavioral characterization of the Tg2576 transgenic model of Alzheimer’s disease through 19 months. Physiol Behav. 2002;75(5):627–642. doi: 10.1016/s0031-9384(02)00639-x. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 7.Adachi H, et al. Transgenic mice with an expanded CAG repeat controlled by the human AR promoter show polyglutamine nuclear inclusions and neuronal dysfunction without neuronal cell death. Hum Mol Genet. 2001;10(10):1039–1048. doi: 10.1093/hmg/10.10.1039. [DOI] [PubMed] [Google Scholar]
- 8.Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT. APPSw transgenic mice develop age-related A beta deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol. 1997;56(9):965–973. doi: 10.1097/00005072-199709000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Irizarry MC, et al. Abeta deposition is associated with neuropil changes, but not with overt neuronal loss in the human amyloid precursor protein V717F (PDAPP) transgenic mouse. J Neurosci. 1997;17(18):7053–7059. doi: 10.1523/JNEUROSCI.17-18-07053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martín-Aparicio E, et al. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J Neurosci. 2001;21(22):8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 12.Shankar GM, et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9(11):826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 14.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18(5):538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 15.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453(7194):481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fecto F, Siddique T. Making connections: Pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia. J Mol Neurosci. 2011;45(3):663–675. doi: 10.1007/s12031-011-9637-9. [DOI] [PubMed] [Google Scholar]
- 17.Deng HX, et al. Distal axonopathy in an alsin-deficient mouse model. Hum Mol Genet. 2007;16(23):2911–2920. doi: 10.1093/hmg/ddm251. [DOI] [PubMed] [Google Scholar]
- 18.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 19.Bedford L, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28(33):8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton AM, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74(6):1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Salon M, et al. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14(11):1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- 22.Jarome TJ, Werner CT, Kwapis JL, Helmstetter FJ. Activity dependent protein degradation is critical for the formation and stability of fear memory in the amygdala. PLoS ONE. 2011;6(9):e24349. doi: 10.1371/journal.pone.0024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ii K, Ito H, Tanaka K, Hirano A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J Neuropathol Exp Neurol. 1997;56(2):125–131. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 25.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448(7154):704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 26.Deriziotis P, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011;30(15):3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmanouilidou E, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein oligomers are targeted to, and impair, the 26S proteasome. Neurobiol Aging. 2010;31(6):953–968. doi: 10.1016/j.neurobiolaging.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2008;29(11):1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratnavalli E, Brayne C, Dawson K, Hodges JR. The prevalence of frontotemporal dementia. Neurology. 2002;58(11):1615–1621. doi: 10.1212/wnl.58.11.1615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.