Significance
High levels of brown/beige fat activity protects animals against metabolic disease, but there has been little known about the precursor cells that mediate the expansion of brown or beige fat. We discovered that early B-cell factor 2 (Ebf2), a transcription factor, is selectively expressed in brown and beige fat cell precursors. Through purification of Ebf2+ cells, we identified a gene profile of brown fat precursors that can be used to distinguish these cells from other developmentally related cell types. Importantly, Ebf2 was also found to regulate the gene expression profile of brown fat precursor cells. Taken together, this study identifies Ebf2 as a highly specific marker of brown and beige preadipose cells and reveals that Ebf2 functions to control brown preadipose cell identity.
Keywords: beige adipocyte, brown adipose tissue
Abstract
Brown adipocytes and muscle and dorsal dermis descend from precursor cells in the dermomyotome, but the factors that regulate commitment to the brown adipose lineage are unknown. Here, we prospectively isolated and determined the molecular profile of embryonic brown preadipose cells. Brown adipogenic precursor activity in embryos was confined to platelet-derived growth factor α+, myogenic factor 5Cre-lineage–marked cells. RNA-sequence analysis identified early B-cell factor 2 (Ebf2) as one of the most selectively expressed genes in this cell fraction. Importantly, Ebf2-expressing cells purified from Ebf2GFP embryos or brown fat tissue did not express myoblast or dermal cell markers and uniformly differentiated into brown adipocytes. Interestingly, Ebf2-expressing cells from white fat tissue in adult animals differentiated into brown-like (or beige) adipocytes. Loss of Ebf2 in brown preadipose cells reduced the expression levels of brown preadipose-signature genes, whereas ectopic Ebf2 expression in myoblasts activated brown preadipose-specific genes. Altogether, these results indicate that Ebf2 specifically marks and regulates the molecular profile of brown preadipose cells.
Brown adipose tissue (BAT) expends energy in the form of heat in response to various stimuli, including cold exposure and certain diets (1). The thermogenic activity of brown adipocytes is mediated by uncoupling protein 1 (Ucp1), a mitochondrial membrane protein. Ucp1, when activated, dissipates the electrochemical gradient that powers ATP synthesis, which allows for low rates of ATP production despite high levels of substrate oxidation (1). Thermogenically competent Ucp1-expressing brown-like adipocytes, called beige or brite (brown-in-white) cells, also develop in certain white adipose tissue (WAT) depots in response to cold exposure (reviewed in ref. 2). In mice, BAT and/or beige fat-mediated thermogenesis suppresses high-fat-diet–associated obesity and metabolic disease (2–8). High levels of activated BAT also correlate with reduced adiposity in people (9). There is thus great hope that brown/beige fat activity can be increased to reduce obesity and metabolic disease.
BAT depots mature during the embryonic and fetal stages of mouse development. In humans, the major BAT depot is located in the interscapular region of newborns, but this tissue regresses and is absent in adults (10). However, brown and beige adipocytes are found in other depots of adult humans, particularly in the supraclavicular area, neck, and along the spinal column (10–14). Recent lineage studies showed that brown adipocytes originate from precursor cells that express myogenic factor 5 (Myf5), paired box protein 7 (Pax7), Pax3, and engrailed 1 (En1) in the somitic mesoderm (15–17). However, the factors that control brown preadipose cell fate in this “tripotent” compartment are unknown.
In the absence of molecular markers for committed brown preadipocytes, such cells can only be recognized retrospectively after their differentiation into Ucp1+ adipocytes. Here, we combined lineage and cell surface markers to prospectively isolate brown preadipose cells during embryogenesis. Global mRNA transcriptomic analyses identified a brown-preadipose–specific gene signature, which included early B-cell factor 2 (Ebf2), a critical transcriptional regulator in mature brown adipocytes (18). We found that brown/beige preadipose activity in embryos and adult fat depots was restricted to Ebf2-expressing cells. Furthermore, Ebf2 deficiency in brown preadipose cells reduced the expression of nearly all brown preadipose signature genes, whereas ectopic expression of Ebf2 in myoblasts activated brown preadipose-selective genes. Altogether, our study reveals that Ebf2 is a specific marker of brown/beige preadipose cells and that Ebf2 functions at this stage to control precursor identity.
Results
Prospective Isolation of Myf5Cre-Lineage–Marked Brown Preadipose Cells.
Specific marker genes for brown preadipose cells were previously unknown. To identify such factors, we isolated enriched populations of brown preadipose cells from mouse embryos. Brown adipocytes descend from a Myf5Cre-expressing cell lineage that also gives rise to skeletal muscle cells (17). To enable the efficient purification of Myf5Cre-lineage–marked cells by flow cytometry, we generated Myf5Cre; R26R-mTmG mice, in which Myf5Cre-expressing cells are genetically and heritably labeled by a membrane-targeted form of GFP [Myf5Cre(GFP)] (19). We noted that GFP was readily detected in somites starting from embryonic day (E) ∼E9.5 (Fig. S1A). In agreement with previous findings, the majority of peroxisome proliferator-activated receptor γ (Pparγ)-expressing adipocytes in the interscapular BAT as well as the Desmin-expressing skeletal myofibers in the trunk and limb were GFP+ at E16.5 (Fig. S1B) (17). GFP was also expressed in adipocytes within the axillary and cervical BAT (Fig. S1C) and in the majority of preadipocytes from postnatal BAT (Fig. S1D). GFP+ cells also contributed to the dorsal dermis (Fig. S1B). These data indicate that Myf5Cre cells contribute to at least three different mesodermal tissue types in late stage embryos: BAT, skeletal muscle, and dorsal dermis. Consistent with recent studies, GFP was also found in some adipocytes in the retroperitoneal and back s.c. WAT of adult mice (20) (Fig. S1E). Thus, Myf5Cre (GFP) is a selective but not specific marker of developing brown fat cells.
BAT forms prenatally in mice, but the embryonic stages associated with the morphological and molecular differentiation of brown adipocytes were unclear. To assess this differentiation, we performed hematoxylin and eosin (H&E) staining and immunohistochemistry on transverse sections of E13.5–E16.5 embryos. At E14, we detected distinctive clusters of Myf5Cre(GFP)+ presumptive brown adipogenic cells that expressed Pparγ (Fig. S2). Consistent with previous studies, BAT was easily recognized by H&E staining at E15.5 (21, 22) and contained cells that expressed the mature adipocyte marker perilipin (Fig. S2). This suggested that the dorsal interscapular region of E14 embryos is a rich source of pioneering brown adipose precursors.
We next considered whether particular cell surface markers could be combined with Myf5Cre-lineage marking to provide additional selectivity for brown preadipose cells. Platelet-derived growth factor α (Pdgfrα) is a cell surface marker of adipogenic precursors in adult WAT and muscle (23–26); this prompted us to examine Pdgfrα expression during BAT development. Lineage tracing using a PdgfrαCre allele, previously shown to mark most/all white adipocytes in adult mice (23), showed that Pdgfrα-expressing cells broadly contributed to somite-derived tissues, including BAT, skeletal muscle, and dermis (Fig. S3A). At E14.5, Pdgfrα+ cells were abundant in the dorsal anterior region of embryos and the proportion of Pdgfrα+ cells in this region declined by E15.5 as the number of perilipin-expressing adipocytes increased (Fig. S3B).
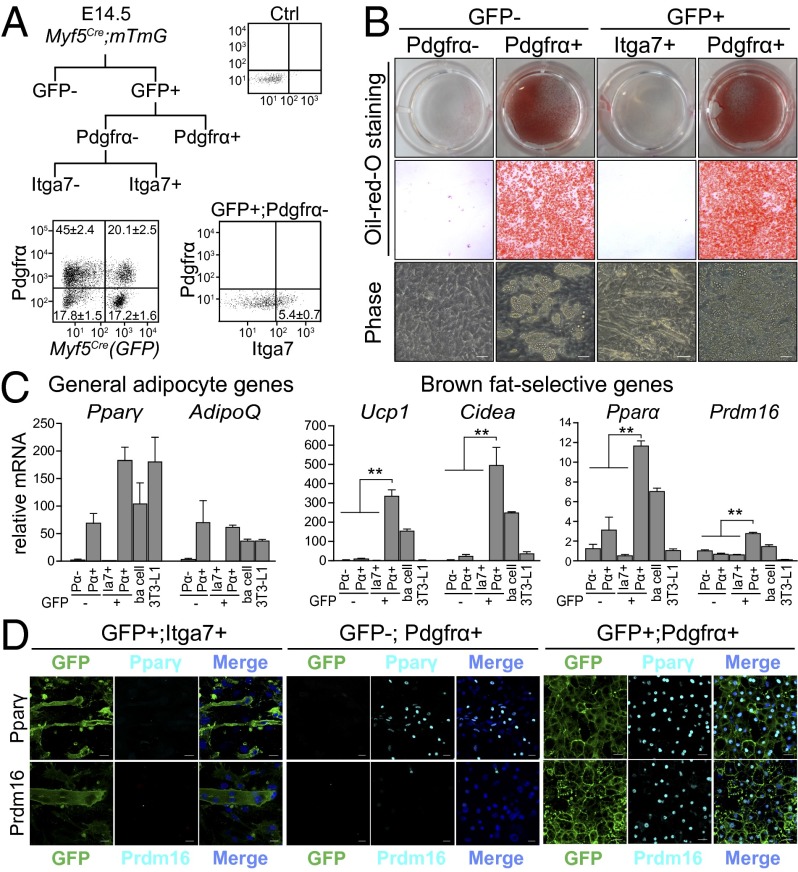
To examine the adipogenic potential of Pdgfrα+ cells in the Myf5Cre-expressing lineage, we dissociated cells from the dorsal body wall of E14.5 Myf5Cre (GFP) embryos and used fluorescence-activated cell sorting (FACS) to fractionate cells into four populations: GFP−; Pdgfrα−, GFP−; Pdgfrα+, GFP+; Pdgfrα−, and GFP+; Pdgfrα+ (Fig. 1A). Integrin alpha-7 (Itga7), a marker of skeletal myoblasts (27), was used to select against the differentiated brown adipocytes found in the GFP+; Pdgfrα− fraction. The sort-purified fractions were cultured and treated with standard adipogenic inducers to analyze their differentiation potential. Oil Red O staining for lipids revealed that a high proportion of both GFP−; Pdgfrα+ and GFP+; Pdgfrα+ cells differentiated into adipocytes, whereas GFP+; Pdgfrα−; Itga7+ cells differentiated into multinucleated muscle cells (Fig. 1B).
Fig. 1.
Myf5Cre-lineage–derived brown adipogenic precursors are Pdgfrα+. (A) Cells from E14.5 Myf5Cre; mTmG embryos were fractionated based on expression of GFP, Pdgfrα, and Itga7. The percentage of each cell fraction (of total cell number) is reported as mean ± SD, n = 3. (B) FACS-purified fractions were induced to differentiate into adipocytes and stained with Oil Red O. (C) mRNA levels of general adipogenic genes, brown-fat–selective markers in indicated cell cultures. Pα, Pdgfrα; Ia7, Itga7; ba, brown adipocyte. Values are mean ± SD, n = 3; *P < 0.05, **P < 0.01. (D) Brown adipogenic conversion of sorted precursor populations was evaluated by immunofluoresent staining of Pparγ and Prdm16.
Gene expression analysis was used to further analyze the phenotype of the differentiated cell cultures. The GFP−; Pdgfrα+ and GFP+; Pdgfrα+ cultures expressed high levels of general adipocyte genes compared with all Pdgfrα− cells. However, only GFP+; Pdgfrα+ cells expressed brown-fat–selective genes and mitochondrial genes, and at levels that were comparable to or higher than their levels in genuine brown adipocytes from an established cell line (Fig. 1C and Fig. S4A). Immunofluorescence staining showed that ∼70% of GFP+; Pdgfrα+ cells differentiated into brown adipocytes that expressed the general adipocyte marker, Pparγ, and the brown-selective factor, PR domain containing 16 (Prdm16) (Fig. 1D and Fig. S4B). GFP+; Pdgfrα+ cells did not express muscle-selective genes [myogenic differentiation 1 (MyoD) and myogenin (MyoG)] (Fig. S4C) and had lower levels of white-adipocyte–selective markers (Agt and Retn) relative to GFP−; Pdgfrα+ cells (Fig. S4D). Taken together, these results indicate that cell surface expression of Pdgfrα enriches for brown adipose precursors in a heterogenous population of embryonic Myf5Cre (GFP)+ cells.
Ebf2 Expression Identifies Brown Preadipose Cells During Development.
We then focused our analysis on freshly isolated brown adipogenic Myf5Cre(GFP)+; Pdgfrα+ cells. Using flow cytometry, we found that the Myf5Cre(GFP)+; Pdgfrα+ fraction contained only 2.6% CD31+ endothelial, 0.1% CD45+ hematopoietic, and 0.1% Itga7+ cells (Fig. S5A). Notably, 43.4% of Myf5Cre(GFP)+; Pdgfrα+ cells were CD34+, and 89.6% were CD24+ (Fig. S5A), showing that this population expresses markers that are also found on white-fat–derived precursors (23, 25). However, most embryonic Myf5Cre(GFP)+; Pdgfrα+ cells were negative for Sca1 (Fig. S5A), whereas preadipose cells in postnatal BAT and WAT were Sca1+ (Fig. S5B) (28, 29). Quantitative PCR (qPCR) analysis revealed that Pparγ and Zfp423, key adipocyte-lineage genes (30, 31), were expressed at higher levels in Myf5Cre(GFP)+; Pdgfrα+ cells relative to other sorted fractions (Fig. S5C). MyoD, the master regulator of skeletal muscle differentiation, was undetectable in Myf5Cre(GFP)+; Pdgfrα+ cells (Fig. S5C), whereas Dermo1 (also called Twist2), a dermis lineage marker (32), was highly expressed in Myf5Cre(GFP)+; Pdgfrα+ cells (Fig. S5C). These data suggest that the Myf5Cre(GFP)+; Pdgfrα+ fraction contains brown fat and dermal precursor cells.
To search for specific markers of brown adipogenic cells within the Myf5Cre(GFP)+; Pdgfrα+ fraction, we used RNA-sequencing (RNA-seq) to globally compare gene expression between Myf5Cre(GFP)+; Pdgfrα+ and Myf5Cre(GFP)+; Pdgfrα−;Itga7+ cells. This led to the identification of Ebf2 as one of the most selectively expressed transcription factors in Myf5Cre(GFP)+; Pdgfrα+ cells (Fig. S5D). Ebf2 cooperates with Pparγ in adipocytes to regulate the expression of thermogenic genes (18), but a function for Ebf2 in precursor cells was unknown. Immunofluorescence analysis showed that Ebf2 protein was present in ∼70% of sorted Myf5Cre(GFP)+; Pdgfrα+ cells both before and after adipogenic conversion and was only very rarely found in Pdgfrα− cells (<1%) (Fig. S5E).
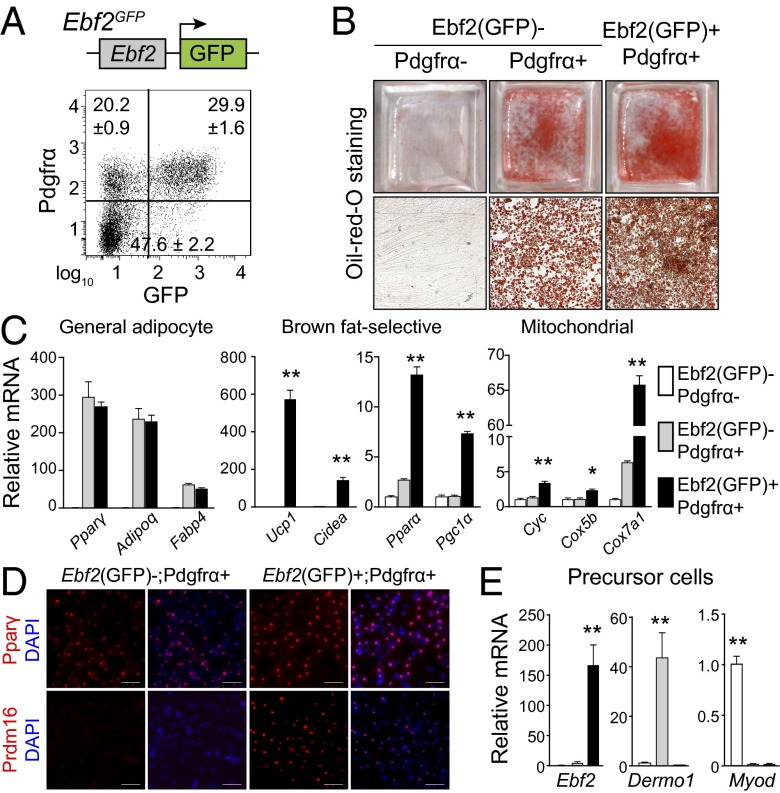
We isolated and examined the differentiation potential of Ebf2+ cells purified from the dorsal anterior region of E14.5 Ebf2-GFP (Ebf2Gfp/+) knockin mice. Virtually all Ebf2 (GFP)+ cells were also Pdgfrα+ and expressed high levels of Ebf2 mRNA (Fig. 2 A and E). Under adipogenic conditions, Ebf2(GFP)+; Pdgfrα+ and Ebf2(GFP)−; Pdgfrα+ cells differentiated into Oil Red O-stained adipocytes that expressed similar levels of general adipocyte genes (Fig. 2 B and C). However, only Ebf2(GFP)+ cells expressed brown-fat–selective genes and high levels of mitochondrial genes (Fig. 2C). Immunofluorescence staining for Pparγ and Prdm16 showed that nearly all Ebf2-expressing cells differentiated into (Pparγ+; Prdm16+) brown adipocytes (Fig. 2D). Ebf2 (GFP)+ cells isolated from adult BAT also underwent adipogenic differentiation much more efficiently than Ebf2 (GFP)− cells (Fig. S6). Importantly, Ebf2+ precursor cells did not express either MyoD or Dermo1, which mark muscle and dermal precursor cells, respectively (Fig. 2E). MyoD was uniquely expressed in Ebf2 (GFP)−; Pdgfrα− cells, whereas Dermo1 was selectively expressed in Ebf2(GFP)−; Pdgfrα+ cells (Fig. 2E). These results indicate that Ebf2 expression identifies precursor cells that are competent to undergo brown adipogenesis.
Fig. 2.
Prospective identification of Ebf2+ brown adipogenic precursors. (A) Cells from E14.5 Ebf2GFP embryos were fractionated based on GFP and Pdgfrα expression. The percentages of each population are reported as mean ± SD, n = 3. (B) Flow-sorted cell populations (from A) were induced to undergo adipocyte differentiation and stained with Oil Red O. (Scale bar, 100 µm.) (C) mRNA levels of general adipocyte markers, brown-fat–selective genes, and mitochondrial genes in indicated cultures. Values are mean ± SD, n = 3; *P < 0.05, **P < 0.01. (D) Immunofluorescence staining of Pparγ and Prdm16 in indicated cell cultures. (Scale bar, 50 µm.) (E) mRNA levels of Ebf2, Dermo1, and MyoD in freshly sorted cell fractions. Values are mean ± SD, n = 3; *P < 0.05, **P < 0.01.
Ebf2 Marks Beige Adipogenic Precursor Cells in WAT.
Beige adipocytes that develop in s.c. WAT do not originate from Myf5Cre-lineage+ precursor cells (2, 14, 17, 20, 24, 29). Our immunofluorescence experiments showed that Ebf2 was present in a subpopulation of embryonic Myf5Cre (GFP)− cells (Fig. S5E). To quantify the proportion of Ebf2-expressing cells that belong to the Myf5Cre-marked brown fat lineage, we incorporated a Myf5Cre-driven dTomato lineage reporter gene into Ebf2GFP mice (Fig. S7A). Flow cytometric analysis showed that ∼50% of the Ebf2 (GFP)+; Pdgfrα+ cells from the dorsal region of embryos were Myf5Cre (dTomato)+ (Fig. S7A). All sorted populations of Pdgfrα+ cells, whether or not they expressed Myf5Cre (dTomato) or Ebf2 (GFP), robustly differentiated into adipocytes that expressed general adipocyte genes (Fig. S7 B and C). However, only the Ebf2+ cells [both Myf5Cre (dTomato)+ and Myf5Cre (dTomato)−], activated brown-fat–selective genes (Fig. S7C). Moreover, rosiglitazone, a Pparγ activator, increased the expression of brown-fat−specific genes in both populations of Ebf2+ cells. Brown versus beige fat markers (Eva1, Pdk4, and Oplah) (14) were expressed at higher levels in Myf5Cre (dTomato)+ relative Myf5Cre (dTomato)− adipocytes (Fig. S7D). Conversely, beige- versus brown-fat cell-selective gene markers (Tmem26 and Cd137) (14) were expressed at higher levels in Myf5Cre (dTomato)− relative to Myf5Cre (dTomato)+ adipocytes (Fig. S7D). These results suggest that Ebf2 expression marks brown and beige adipogenic cells regardless of their developmental origin.
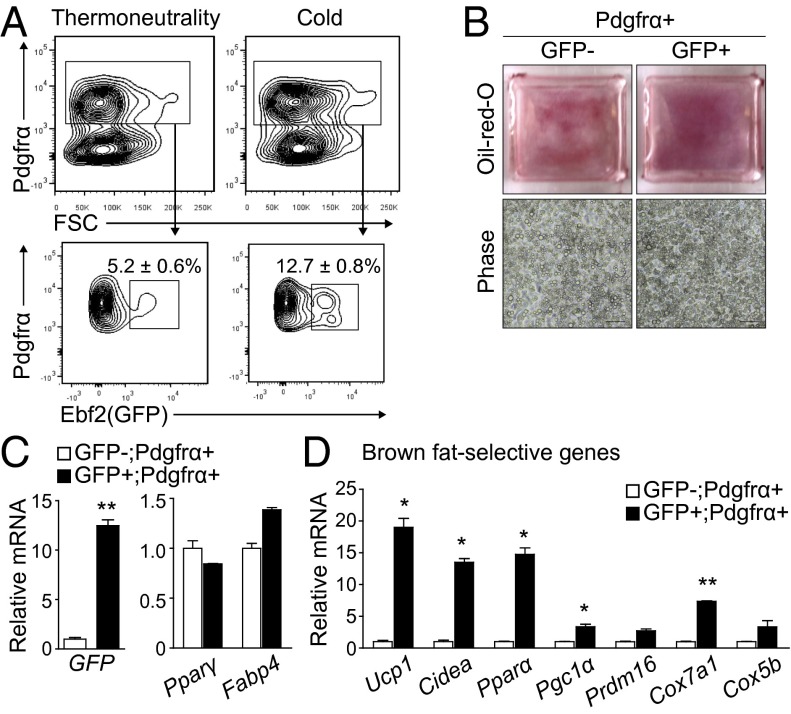
The next question was whether Ebf2 is a specific marker for beige adipogenesis in adult WAT. We used flow cytometry to analyze the SVF from the inguinal (s.c.) WAT (iWAT) of Ebf2-GFPTG mice. At thermoneutrality, 5.2 ± 0.6% of the Pdgfrα+ cells in the SVF were Ebf2 (GFP)+ (Fig. 3A). Exposure of mice to the cold for 3 d, which stimulates beige fat development, increased the proportion of Ebf2 (GFP)+ cells to 12.7 ± 0.8% (Fig. 3A). FACS-purified populations of Ebf2+; Pdgfrα+ and Ebf2−; Pdgfrα+ cells from iWAT efficiently differentiated into adipocytes that expressed very similar levels of general adipogenic genes (Fig. 3 B and C). Remarkably, however, only Ebf2 (GFP)+ cells differentiated into adipocytes that expressed brown-fat–specific genes, including Ucp1, Cidea, and Pparα (Fig. 3D). These data strongly suggest that Ebf2 is a specific marker for the beige adipogenic precursor cells in WAT that are competent to activate a brown-fat–selective gene program in response to cold.
Fig. 3.
Ebf2 marks beige adipogenic precursors in WAT. (A) FACS plots and percentage of stromal cells expressing Ebf2-GFP in iWAT from mice housed at thermoneutrality (30 °C) or cold (4 °C) for 3 d. FSC, forward scatter. The percentage is mean ± SD, n = 3 (six mice per experiment). (B) Sort-purified cell populations from iWAT were induced to undergo adipocyte differentiation and stained with Oil Red O. (C and D) mRNA levels of Gfp, general adipogenic genes, and (D) brown-fat–selective genes in cell cultures from B. Values are mean ± SD, n = 3; *P < 0.05, **P < 0.01.
Mutually Exclusive Expression of Ebf2 and MyoD in Developing Somites by E12.5.
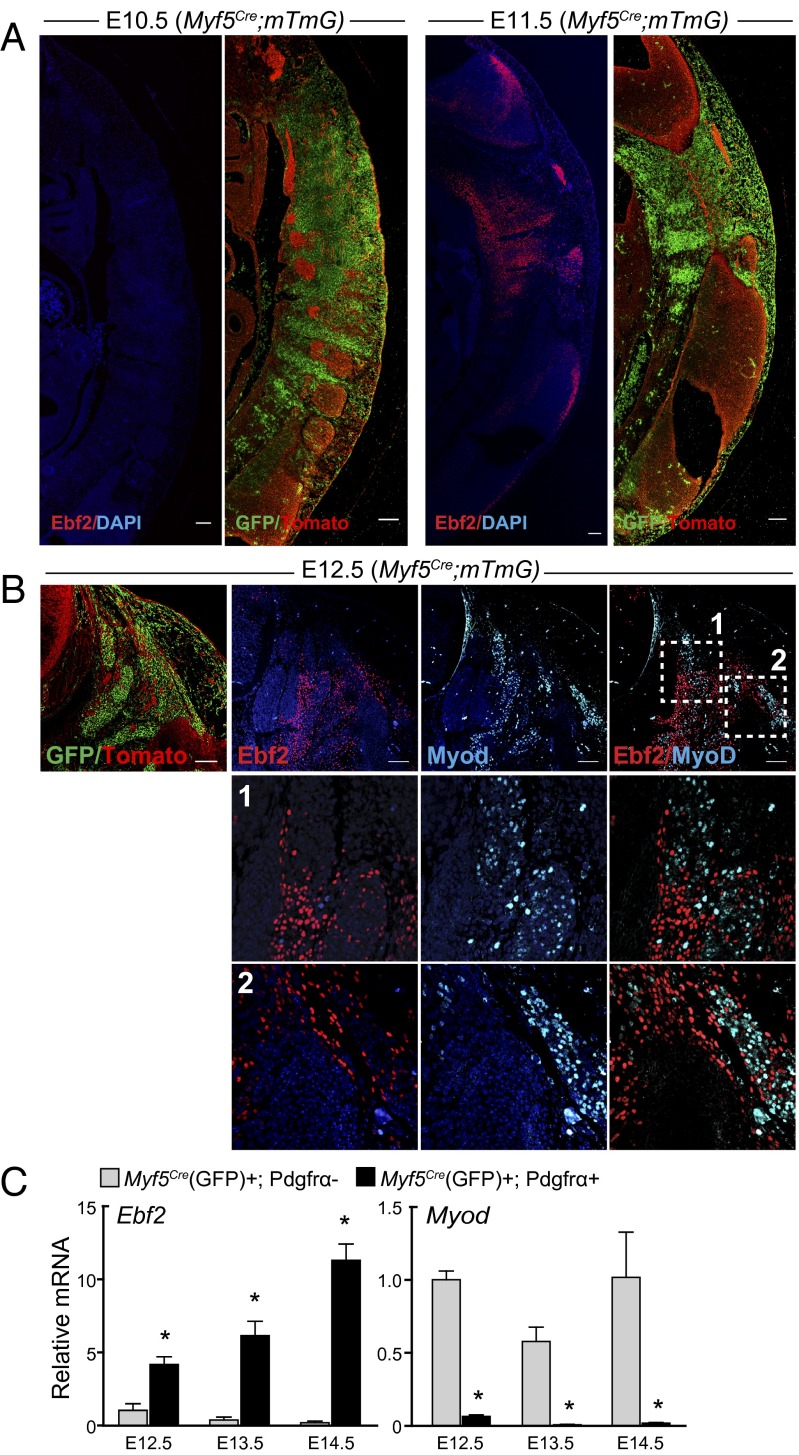
We performed immunofluorescence studies in Myf5Cre; R26R-mTmG embryos to analyze the timing and pattern of Ebf2 expression during brown fat development. At E11.5, but not at E10.5, we detected Ebf2 protein expression in a subpopulation of Myf5Cre (GFP)+ cells in anterior somites (Fig. 4A). At E12.5, there was a dramatic expansion of Ebf2-expressing Myf5Cre (GFP)+ cells (Fig. 4B). Already at this early stage, Ebf2 and MyoD were expressed in a mutually exclusive manner in the same Myf5Cre-lineage–marked compartment (Fig. 4B). qPCR analysis of FACS-purified cells confirmed that Ebf2 and MyoD expression was enriched in different embryonic cell populations at E12.5, E13.5, and E14.5. Ebf2 levels increased from E12.5 to E14.5 in Myf5Cre (GFP)+; Pdgfrα+ cells, whereas MyoD levels remained relatively constant in Pdgfrα− cells over the same interval (Fig. 4C). These results suggest that brown adipocyte and myogenic precursors have undergone lineage commitment by the time that Ebf2 protein expression is detected in Myf5Cre-lineage+ cells.
Fig. 4.
Ebf2 and MyoD are expressed in distinct cells within the somitic mesoderm. (A) Immunofluorescence staining of tdTomato, GFP (Myf5Cre), and Ebf2 (red) on adjacent sagittal sections of E10.5 and E11.5 Myf5Cre; mTmG embryos. (B) Costaining of Ebf2 (red) and MyoD (cyan) on transverse sections of anterior somites of E12.5 Myf5Cre; mTmG embryos. GFP (Myf5Cre)/tdTomato double staining is shown (far Left). White dashed boxes show magnified regions 1 and 2. (C) mRNA levels of Ebf2 and MyoD in freshly sorted cells from E12.5, E13.5, and E14.5 embryos. Values are mean ± SD, n = 3 experiments; *P < 0.05, **P < 0.01.
Ebf2 Regulates the Molecular Identity of Brown Preadipose Cells.
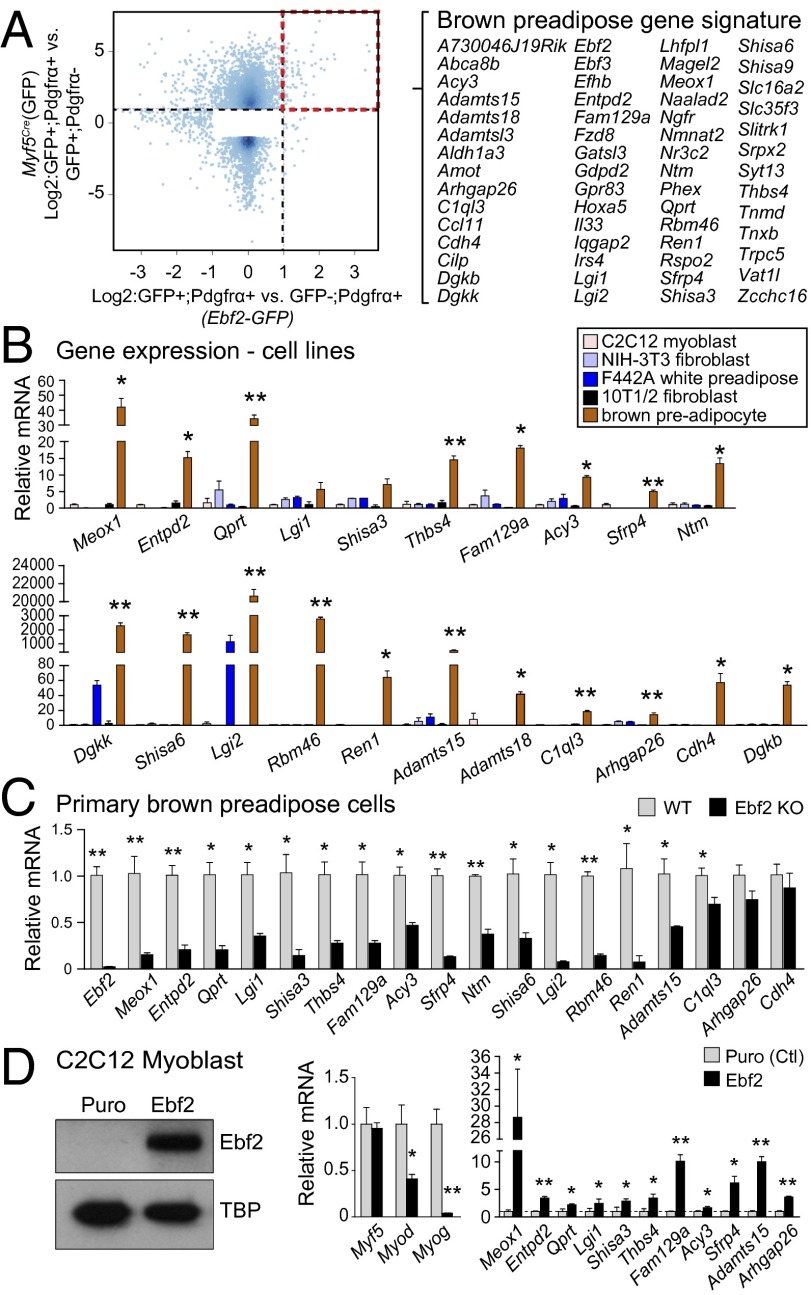
Ebf2 is required in mature (or differentiating) adipocytes to promote or maintain the expression of terminal brown-fat–specific genes (18). Our results above suggested that Ebf2 may also play a functional role in brown fat precursor cells. Because there were no molecular markers that could be used to examine the role of Ebf2 at the preadipose stage, we first sought to establish a brown-preadipose–specific gene signature that could be used to monitor cell identity. Global transcriptomic analyses identifed 58 genes whose mRNA levels were enriched by twofold or more in (i) Ebf2 (GFP)+ versus Ebf2 (GFP)− cells and in (ii) Myf5Cre (GFP)+; Pdgfrα+ versus Myf5Cre (GFP)+; Pdgfrα− cells (Fig. 5A). As a secondary filter, we profiled the expression of these 58 genes in nonadipogenic and adipogenic (brown and white) cell lines. This led us to identify 21 genes that were very selectively expressed in brown preadipose cells relative to fibroblasts (NIH 3T3, 10T1/2), white preadipose (3T3-F442A) and muscle precursor cells (C2C12) (Fig. 5B and Fig. S8A). The majority of these genes were expressed at high levels in preadipose cells and were decreased in expression during the process of adipocyte differentiation (Fig. S8B). Moreover, most of these genes had the same expression pattern as Ebf2 in that they were selectively enriched in Myf5Cre (GFP)+; Pdgfrα+ cells relative to other cell fractions isolated from the dorsal region of embryos starting from E11.5 of development (Fig. S8C).
Fig. 5.
Ebf2 regulates the brown-preadipose–specific gene signature. (A) Scatter plot showing global gene expression in freshly sorted Ebf2+ versus Ebf2− (all Pdgfrα+) precursor cells (x axis) and Pdgfrα+ versus Pdgfrα− [all Myf5Cre(GFP)+] cells (y axis). (B) mRNA levels of select signature genes in established cell lines. (C) mRNA levels of signature genes in wild-type (WT) and Ebf2-knockout (KO) brown preadipose cells. (D) C2C12 myoblasts were transduced with control (ctl) puromycin (Puro)- or Ebf2-expressing retrovirus. Western blot analysis of Ebf2 protein levels (Left). mRNA levels of muscle-specific genes and brown-preadipose–selective genes (Right). Values are mean ± SD, n = 3; *P < 0.05, **P < 0.01.
To determine whether Ebf2 was genetically required to establish brown preadipose cell identity, we analyzed the expression of the 21 brown preadipose signature genes in primary preadipose cells isolated from WT and Ebf2-knockout (KO) BAT. Strikingly, the expression of 16/21 of these genes was significantly diminished by Ebf2 deficiency, including a 70–80% reduction in the levels of Meox1, Enpd2, Qprt, Shisa3, Thbs4, Fam129a, Sfrp4, Lgi2, Rbm46, and Ren1 (Fig. 5C and Fig. S8D). By contrast, genetic deletion of Pparγ, the master regulator of differentiation in all types of adipocytes, did not affect the expression levels of any brown preadipose signature genes (Fig. S8E). These results show that Ebf2 functions independently from Pparγ in establishing and/or maintaining the gene program of brown adipose precursors.
Ectopic expression of Ebf2 in white fat or muscle precursor cells cooperates with a mixture of adipogenic inducers to drive a brown-fat cell-selective differentiation program (18). We wondered whether Ebf2 reprograms precursor cell identity before adipogenic induction. To assess this question, we expressed Ebf2 or a control retroviral vector in C2C12 myoblasts and measured the expression of brown preadipose signature genes. Ebf2 expression was sufficient to activate the expression levels of many brown preadipose selective genes, including a 28-fold increase in Meox1 and 6- to 10-fold increases in Fam129a, Sfrp4, and Adamts15 (Fig. 5D). Ebf2 expression also led to reduced levels of muscle-specific genes such as MyoD and MyoG in myoblasts but had no effect on Myf5 mRNA levels (Fig. 5D). These data suggest that Ebf2 expression reprograms myoblasts into a brown-preadipose–like state.
Discussion
Brown and beige fat cells dissipate chemical energy to protect animals against obesity and insulin resistance. We discovered that Ebf2 is a selective marker of brown and beige preadipose cells—the lineage-committed cells that mediate brown and beige fat expansion. The ability to detect and isolate these precursor cells will open up new opportunities to identify the mechanisms that control brown/beige preadipose commitment, proliferation, and differentiation.
Brown adipocytes, muscle, and dorsal dermis arise from Pax7- and En1-expressing cells in the central dermomyotome (15, 16). Our analyses indicate that Myf5Cre labels these same three differentiated cell types in late-stage embryos. Within this Myf5Cre-marked lineage, Ebf2 was only expressed in brown adipogenic precursor cells and was absent from both muscle and dermal cells. Indeed, embryonic brown adipogenic cells could be purified by their selective expression of Ebf2-GFP. Ebf2 was expressed as early as E11.5 in Myf5Cre-lineage–marked cells and was not detected in MyoD+ muscle cells, suggesting that brown adipogenic cells are committed by the time that Ebf2 is activated.
The stable committed state of brown preadipose cells is well established (33). These cells can be isolated and propagated ex vivo without losing their brown-specific differentiation potential. However, whereas much is known about the factors that promote brown fat-selective transcription in mature adipocytes, no transcription factors that function at the preadipose stage had been identified. Our results indicate that Ebf2 acts at the precursor cell stage to instruct and/or maintain preadipose cell fate. Future studies will examine the function of Ebf2-regulated preadipose genes in brown fat lineage commitment and differentiation. It will also be important to identify the cues that activate Ebf2 during brown fat induction. BMP signaling, which is critically required for early BAT development (22), is one such candidate for regulating Ebf2 expression/activity.
Ebf2-deficient mice develop dysmorphic fatty tissue devoid of brown-fat–specific characteristics in place of BAT (18). Thus, whereas Ebf2 is required to induce the brown-fat cell-selective gene program, it is dispensable for adipogenesis per se. We speculate that other members of the Ebf family act to support adipogenesis in the absence of Ebf2. In line with this speculation, Ebf1 and Ebf3 can stimulate adipocyte differentiation when expressed in fibroblasts (34). It is also interesting to note that Ebf3 was identified here (Fig. 5) and previously as a brown-adipose–enriched factor (14).
Beige adipocytes in WAT express many brown-fat–selective genes and seem to function much like classical brown fat cells (35). The induction of beige adipocytes is strongly associated with a lean and healthy phenotype in mice (2). In iWAT, beige adipocytes are thought to arise in situ from the de novo differentiation of resident precursor cells (14, 24, 29, 36). We found that Ebf2 is specifically expressed by adipogenic cells in WAT that are competent to activate a brown/beige fat-selective differentiation program. Elegant studies by the Granneman group showed that Pdgfrα-expressing cells in gonadal WAT can differentiate into both brown/beige and white adipocytes (24). The relationship between these bipotent Pdgfrα+ cells and the Ebf2+ cells identified here is unclear. One possibility is that the Pdgfrα+ cell population isolated by Lee et al. are upstream progenitor/stem cells that give rise to more committed populations of Ebf2+ beige and Ebf2− white preadipose cells in response to different cues. However, while our data strongly suggest that Ebf2+ cells are beige-specific precursors, we cannot exclude the possibility that these cells also give rise to white adipocytes in vivo under certain conditions. Finally, it is also possible that Ebf2+ cells are uniquely found in iWAT to provide that depot with a more robust capacity to undergo browning.
In conclusion, we report a critical role for Ebf2 in the control of brown preadipose cell fate and identify Ebf2 and other gene markers for committed brown/beige preadipose cells. This study opens up opportunities to examine the molecular pathways that control brown preadipose cell development and function. Gaining a better understanding of brown/beige committed precursors will be crucial for the design and implementation of brown and/or beige fat-targeted therapies.
Materials and Methods
Animals.
All procedures were approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee. The following mouse lines were obtained from The Jackson Laboratory: Myf5Cre (B6.129S4-Myf5tm3(cre)Sor/J), stock no. 010529; Pdgfrα-Cre (C57BL/6-Tg(Pdgfra-cre)1Clc/J), stock no. 013148; mTmG (B6.129(Cg)-Gt(ROSA)26Sortm4 (ACTB-tdTomato,-EGFP)Luo/J), stock no. 007676; Pparγflox (B6.129-Ppargtm2Rev/J), stock no. 004584; CAGGCreERT (B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J), stock no. 004682; and Rosa-dTomato (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J), stock no. 007909. Ebf2GFP knock-in mice and Ebf2-GFPTG BAC (bacterial artificial chromosome)-transgenic mice were described before (37, 38). Ebf2-GFPTG mice were developed by the Mutant Mouse Regional Resource Center.
Cell Preparation and Flow Cytometry.
For embryonic fibroblasts, excised tissue was digested in 6.1 mg-mL−1 collagenase (Roche; 11088882001) and 2.4 mg⋅mL−1 dispase (Roche; 04 942 078 001) in DMEM for 40 min at 37 °C in a shaking water bath. For SVF from iWAT, we performed two cycles of digestion. Digested tissue was successively filtered through 100-µm and 40-µm cell strainers (BD Biosciences). Cells were suspended in DMEM with 5% (vol/vol) FBS, and stained with antibodies for 30 min at 4 °C in the dark. Antibodies and secondary antibodies used for staining were: Pdgfrα APC 1:200 (Biolegend; 135907), Itga7-PE 1:200 (AbLab; University of British Columbia), CD31-PE 1:200 (BD Biosciences; 553373), CD45-PE 1:200 (BD Biosciences; 553081), Sca1-PE 1:200 (eBioscience; 12–5981-81), CD24-biotin 1:400 (eBioscience; 13–0242-81), CD34-biotin 1:400 (eBioscience; 13–0341-82), and streptavidin-PE 1:500 (Biolegend; 405204). Stained cells were analyzed by BD LSR II or sorted with BD FACS Aria. Debris and dead cells were excluded by forward scatter, side scatter, and DAPI gating. Data analysis was performed using FlowJo. Pparγ-knockout cells were obtained by treating primary SVF cells from the P1 BAT of CAGGCreERT; PparγFlox/Flox mice with 1 μM 4-hydroxytamoxifen (Sigma) for 3–5 d.
Cell Culture.
Freshly sorted cells were cultured on 8-well chamber slides (Ibidi; 80826) or collagen-coated 24-well plates (BD Biosciences; 354408) in DMEM with 10% (vol/vol) FBS, 10 ng/mL bFGF for 3 d. After confluence, cells were exposed to brown adipogenic induction medium [DMEM with 10% (vol/vol) FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 µM dexamethosone, 20 nM insulin, and 1 nM T3]. After 48 h, cells were switched to maintenance medium [DMEM with 10% (vol/vol) FBS, 1 nM T3, 20 nM insulin] for 4 d before harvesting. To induce adipogenesis in WAT-derived precursors, 0.5 μM rosiglitazone was included in the induction and maintenance medium.
RNA Extraction and Real-Time PCR.
Total RNA was extracted from cultured cells or 5 × 104 freshly sorted cells using the TRIzol method (Invitrogen) combined with Qiagen RNeasy Micro Column and then reverse transcribed into cDNA using an ABI high-capacity cDNA synthesis kit. Real-time PCR was performed on an ABI7900HT PCR machine using SYBR green fluorescent dye (ABI).
Cytochemistry, Immunohistochemistry, and Oil Red O Staining.
Freshly sorted cells were collected on chamber slides and fixed with 4% (wt/vol) paraformaldehyde (PFA) for 10 min. Primary and secondary antibodies used for staining are: Pparγ 1:250 (Thermo Scientific; MA5-14889), Ebf2 5 ng⋅mL−1 (R&D; AF7006), Prdm16 (in house) 1:200, Alexa Fluor 647 chicken anti-rabbit IgG 1:500 (Invitrogen), Alexa Fluor 647 donkey anti-sheep IgG 1:500 (Invitrogen). Tissues were fixed in 2% (wt/vol) PFA overnight. Paraffin-embedded sections were subjected to citrate-based antigen retrieval. Primary antibodies used for immunohistochemistry are: Pparγ 1:500 (Thermo Scientific; MA5-14889), perilipin 1:200 (Cell Signaling; 3470), Pdgfrα 1:50 (R&D; AF1062), GFP 1:500 (Abcam; AB6673), Ebf2 15 ng⋅mL−1 (R&D; AF7006), and MyoD 1:20 (Novocastra; NCL-MyoD). To stain lipid, cells were fixed in 10% (vol/vol) formalin, rinsed with PBS and 60% (vol/vol) isopropanol, incubated with Oil Red O in 60% isopropanol, and then rinsed in PBS.
Whole-Mount Confocal Microscopy.
Whole-mount confocal microscopy was performed as previously described (23). Briefly, 4-wk-old male mice were perfused transcardially with 1% PFA for 10 min. The 5-mm3 pieces of adipose depots were dissected and fixed in 1% PFA for another 20 min. Images were captured on a Leica TCS SP8 confocal microscope.
RNA-Seq and cDNA Microarray.
RNA-seq libraries were generated with the Illumina Truseq RNA Sample Preparation kit. High-thoughput sequencing was performed using a HiSeq 2000. Expression values of mRNA were called by TopHat/Cufflink pipeline. For microarray analysis, total RNA was purified from three biological replicates per cell type. Array hybridization and scanning were performed by the Penn Molecular Profiling Facility using Affymetrix GeneChip Mouse Genome 2.0 arrays.
Statistical Analysis.
Error bars show SD. Statistical significance was assessed by the Student t test, *P < 0.05, **P < 0.01.
Supplementary Material
Acknowledgments
We thank Min Min Lu and the Histology and Gene Expression Core of the Penn Cardiovasular Institute for immunohistochemistry; Randall R. Reed for the Ebf2GFP knockin mice; Hong Qian and Giacomo Consalez for providing the Ebf2-GFPTG transgenic mice; and the Functional Genomics Core of the Penn Diabetes and Endocrinology Research Center (DK19525) for high-throughput sequencing and data analyses. This work was supported by National Institute of General Medical Sciences/National Institutes of Health Award DP2OD007288 and a Searle Scholars Award (to P.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE60443).
See Commentary on page 14318.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412685111/-/DCSupplemental.
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Harms M, Seale P. Brown and beige fat: Development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 3.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J Clin Invest. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowell BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366(6457):740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 5.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106(5):563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 6.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopecky J, Clarke G, Enerbäck S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96(6):2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9(2):203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes. 2009;58(7):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19(5):631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 11.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013;19(5):635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013;17(5):798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE. 2012;7(11):e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atit R, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296(1):164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 16.Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48(7):424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17(4):562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Gurmaches J, et al. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16(3):348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JH, et al. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev Cell. 2013;26(4):393–404. doi: 10.1016/j.devcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495(7441):379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15(3):302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135(2):240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 26.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12(2):143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 27.Blanco-Bose WE, Yao CC, Kramer RH, Blau HM. Purification of mouse primary myoblasts based on alpha 7 integrin expression. Exp Cell Res. 2001;265(2):212–220. doi: 10.1006/excr.2001.5191. [DOI] [PubMed] [Google Scholar]
- 28.Joe AW, Yi L, Even Y, Vogl AW, Rossi FM. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 2009;27(10):2563–2570. doi: 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- 29.Schulz TJ, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci USA. 2011;108(1):143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464(7288):619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322(5901):583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Cserjesi P, Olson EN. Dermo-1: A novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172(1):280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 33.Klaus S, Ely M, Encke D, Heldmaier G. Functional assessment of white and brown adipocyte development and energy metabolism in cell culture. Dissociation of terminal differentiation and thermogenesis in brown adipocytes. J Cell Sci. 1995;108(Pt 10):3171–3180. doi: 10.1242/jcs.108.10.3171. [DOI] [PubMed] [Google Scholar]
- 34.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27(2):743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabalina IG, et al. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Reports. 2013;5(5):1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 36.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131(6):1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 38.Qian H, et al. Molecular characterization of prospectively isolated multipotent mesenchymal progenitors provides new insight into the cellular identity of mesenchymal stem cells in mouse bone marrow. Mol Cell Biol. 2013;33(4):661–677. doi: 10.1128/MCB.01287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.