Significance
Imprinted genes are normally selectively expressed, depending on the parent from whom the gene is inherited. These genes play important roles in control of growth and metabolism. We find that the loss of insulin and insulin-like growth factor 1 (IGF1) receptors in cells induces a major decrease in expression of multiple imprinted genes, regardless of whether they would be maternally or paternally expressed. This deregulation is associated with changes in methylation of DNA, but is not due to classical imprinting control. Our results demonstrate a previously unrecognized function of insulin and IGF1 receptors independent of these hormones and provide a new pathway by which insulin and IGF1 receptors may regulate growth and metabolism during early development and thereafter.
Keywords: transcriptional control, diabetes, developmental control
Abstract
In addition to signaling through the classical tyrosine kinase pathway, recent studies indicate that insulin receptors (IRs) and insulin-like growth factor 1 (IGF1) receptors (IGF1Rs) can emit signals in the unoccupied state through some yet-to-be-defined noncanonical pathways. Here we show that cells lacking both IRs and IGF1Rs exhibit a major decrease in expression of multiple imprinted genes and microRNAs, which is partially mimicked by inactivation of IR alone in mouse embryonic fibroblasts or in vivo in brown fat in mice. This down-regulation is accompanied by changes in DNA methylation of differentially methylated regions related to these loci. Different from a loss of imprinting pattern, loss of IR and IGF1R causes down-regulated expression of both maternally and paternally expressed imprinted genes and microRNAs, including neighboring reciprocally imprinted genes. Thus, the unoccupied IR and IGF1R generate previously unidentified signals that control expression of imprinted genes and miRNAs through transcriptional mechanisms that are distinct from classical imprinting control.
Insulin and insulin-like growth factor 1 (IGF1) control many biological processes such as cellular metabolism, proliferation, differentiation, and apoptosis. These effects are mediated through ligand activation of the tyrosine kinase activity intrinsic to their receptors (1, 2). We have recently shown that preadipocytes lacking both insulin receptors (IRs) and IGF1 receptors (IGF1Rs)—i.e., double-knockout (DKO) cells, which lack any evidence of classical insulin/IGF1 signaling as measured by Akt and MAP kinase activation—are resistant to apoptosis (3, 4). This resistance is due to posttranscriptional regulation of key proapoptotic and antiapoptotic proteins and is not only independent of ligand binding, but is also independent of the kinase activity of the receptor, indicating that IR and IGF1R serve as both tyrosine kinase and dependence receptors (4, 5).
Genomic imprinting is a key modulator of developmental and physiological processes in mammals (6). Imprinting results in parental origin-specific gene expression and controls expression of a variety of genes often located in clusters, including protein-coding genes, retrotransposon-derived genes, and long noncoding RNAs (lncRNAs), as well as small-regulatory RNAs, such as microRNAs (miRNAs) and C/D-box small nucleolar RNAs (C/D snoRNAs) (7–9). Imprinting in mouse is regulated by germ-line–derived differential DNA methylation acting at specific CpG-rich DNA sequences. These differentially methylated regions (DMRs) or imprinting control regions (ICRs) can regulate monoallelic expression of upstream and downstream alleles through cis-acting mechanisms (10, 11).
Imprinted genes play an important role in the control of growth and other processes, are widely expressed during prenatal development, and tend to be down-regulated after birth (6, 12). The mechanisms through which imprinted genes control growth remain to be fully elucidated, although perturbations in imprinted gene dosage have recently been shown to alter the IGF1 signaling pathway (13, 14). In humans, altered imprinted gene dosage plays a role in a number of well-studied disorders, including Beckwith–Wiedemann, Silver–Russell, Angelman, and Prader–Willi syndromes (15–17).
In the present study, we have further investigated the effects of the unoccupied IR and IGF1R by analysis of miRNA and mRNA gene expression in cells lacking both IR and IGF1R—i.e., DKO cells. We find that DKO cells show a major decrease in expression of multiple paternally and maternally expressed imprinted miRNAs and mRNAs. This decrease is accompanied by increased methylation at some imprinted loci. In addition, we observed that mouse embryonic fibroblasts (MEFs) with homozygous or heterozygous inactivation of the IR gene show a similar and gene-dose–dependent down-regulation of imprinted miRNAs and mRNAs. A decrease in expression of imprinted genes is also observed in brown adipose tissue of mice with fat-specific IR deletion. Thus, loss of IRs and IGF1Rs in somatic cells induces a coordinated regulation of an imprinted gene network associated with changes in the epigenome, indicating a previously unrecognized effect of unoccupied IRs and IGF1Rs on the regulation of expression of imprinted genes and miRNAs.
Results and Discussion
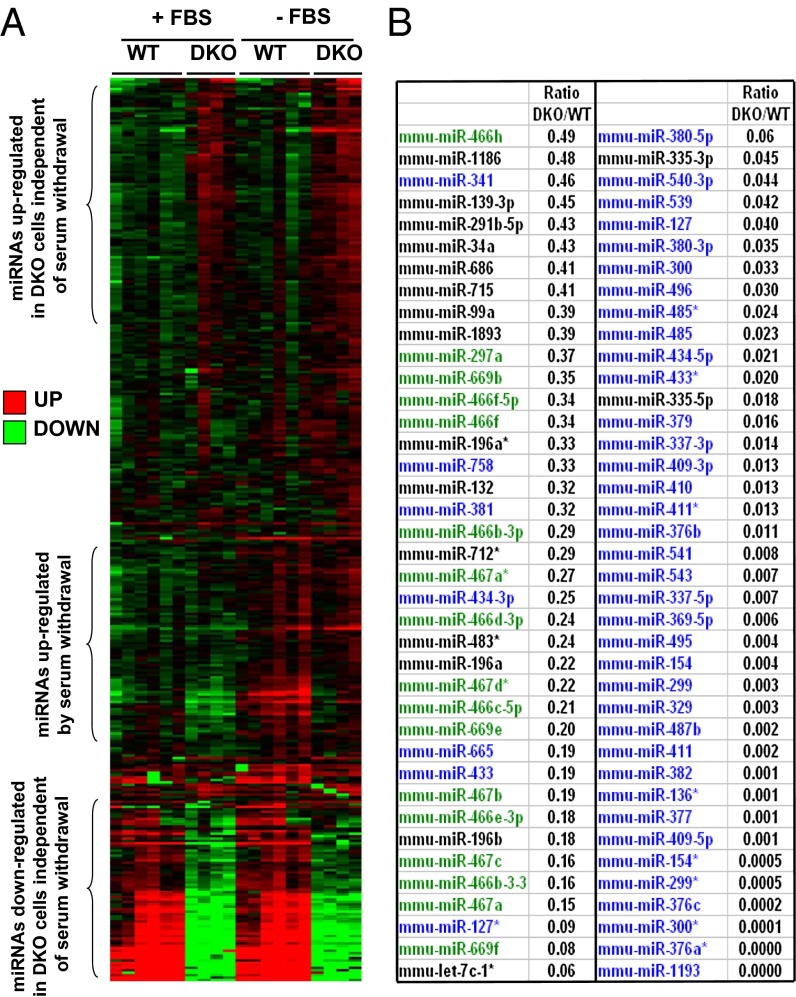
To explore the role of unoccupied IRs and IGF1Rs on cellular function, we performed global mRNA and miRNA expression profiling in confluent control (WT) and IR/IGF1R DKO brown preadipocyte cells in the presence or absence of FBS—i.e., under normal or proapoptotic conditions. Among 632 miRNA assessed by quantitative real-time PCR (real-time qPCR), 371 were detectable in WT cells, and the abundance of 134 of these was significantly different between WT and DKO cells. Among these, 45 miRNAs were significantly up-regulated between 1.4- and 4.4-fold in DKO cells compared with WT cells, whereas 89 miRNAs were significantly down-regulated in DKO cells (Fig. 1A). This down-regulation of miRNAs in the DKO cells was observed in both the serum-fed and -starved conditions and was very striking, with differences in expression of 100-fold or more (Fig. 1 A and B). The decrease in miRNA expression in DKO cells occurred without changes in enzymes or proteins playing a role in miRNA formation or maturation—including Dicer, Drosha, DGCR8, Exp-5, Ago2, TRBP, PACT, and DDX5—suggesting a decrease in expression of the primary miRNA transcripts rather than a change in miRNA processing (Fig. S1). Genomic analysis revealed that, whereas up-regulated miRNAs in the DKO cells were randomly distributed across the genome, 61 of the 89 down-regulated miRNAs resided in clusters on two chromosomes: 44 resided in a single cluster on mouse chromosome 12, and another 17 were in a cluster on chromosome 2 (Fig. 1B). Both of these regions have been shown to be imprinted (18–21).
Fig. 1.
miRNA expression profiling in WT and IR/IGF1R DKO cells. Large-scale miRNA expression profiling was performed by qPCR in confluent WT and DKO cells under normal (in the presence of serum: +FBS) or apoptotic (in the absence of serum for 6 h: −FBS) conditions. (A) Heat map representing the miRNA expression profile in WT and DKO cells in normal or apoptotic conditions. Red represents high expression and green low expression. (B) List of miRNAs significantly down-regulated by more than twofold in DKO cells compared with WT cells in the presence of serum. miRNAs in blue share the same chromosomal location in a cluster in region 12qF1; miRNAs in green share the same chromosomal location in a cluster in region 2qA1.
Microarray analysis of WT and DKO cells was also performed by using Affymetrix microarrays. Of the 20,530 probe sets detectable either in WT or DKO cells, 699 (3.4%) were significantly up-regulated and 798 (3.9%) were significantly down-regulated more than twofold in DKO cells. When these data were subjected to gene set enrichment analysis (GSEA), 16 gene sets of 474 were significantly altered [false discovery rate (FDR) < 25%]. One of the highest rankings of these altered gene sets was the set of imprinted genes, which was significantly enriched in WT compared with DKO cells (nominal P < 0.001; FDR 0.225). Among the list of known imprinted genes represented on the microarray, 28 were detectable in either WT or DKO cells. Of these, only one, Zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 or Zrsr1, was significantly up-regulated in DKO cells (1.7-fold increase compared with WT cells), whereas 20 were down-regulated by 30% or more in DKO cells (Table S1). Several of these (Necdin, Igf2, Cdkn1c, Dlk1, and H19) were down-regulated by >98%. Although some of those genes are normally imprinted in a tissue-specific fashion, their coregulation in brown preadipocytes lacking IRs and IGF1Rs is indicative of a common mechanism of regulatory control, which is different from traditional imprinting control.
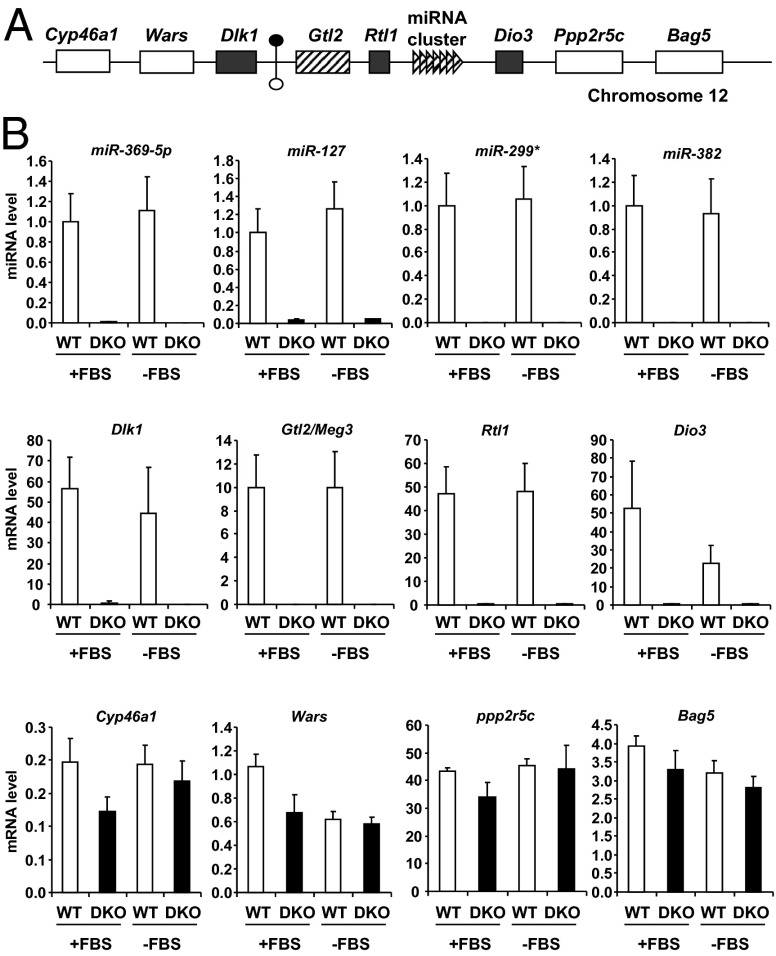
The decrease in expression of imprinted genes and miRNAs in the DKO cells was striking and involved genes on multiple chromosomes. The miRNA cluster on chromosome 12 is the largest known miRNA cluster in mammals and is part of a larger imprinted locus that includes the paternally expressed (maternally repressed) genes Dlk1, Rtl1, and Dio3 and the maternally expressed (paternally repressed) gene Meg3/Gtl2 and associated maternally expressed noncoding RNAs including the miRNAs (18, 19). All imprinted genes in this locus were detectable by real-time qPCR to varying degrees in WT cells, but all were either markedly down-regulated (>100-fold) or undetectable in DKO cells, regardless of the presence or absence of serum (Fig. 2B). Furthermore, down-regulation of genes in this locus occurred whether the genes were normally expressed from the paternal allele or the maternal allele (Fig. 2A). By contrast, genes directly upstream (Cyp46a1 and Wars) or downstream (Ppp2r5c and Bag5) of this imprinted locus showed no difference in expression between WT and DKO cells.
Fig. 2.
Expression of miRNAs and mRNAs in imprinted Dlk1–Dio3 locus on chromosome 12 in WT and DKO cells. (A) Graphical representation of mouse imprinted Dlk1–Dio3 locus on chromosome 12qF1. Genes are shown as colored: white, biallelic expression; hatched, maternal expression; dark gray, paternal expression. An IG-DMR located between Dlk1 and Gtl2 genes is represented by circles (filled, hypermethylated; open, hypomethylated). The genomic region is not drawn to scale. (B) Expression of miRNAs and mRNAs was measured by real-time PCR in confluent WT and DKO cells in the presence (+FBS) or absence (−FBS) of serum for 6 h. Results represent the average ± SEM of six independent experiments.
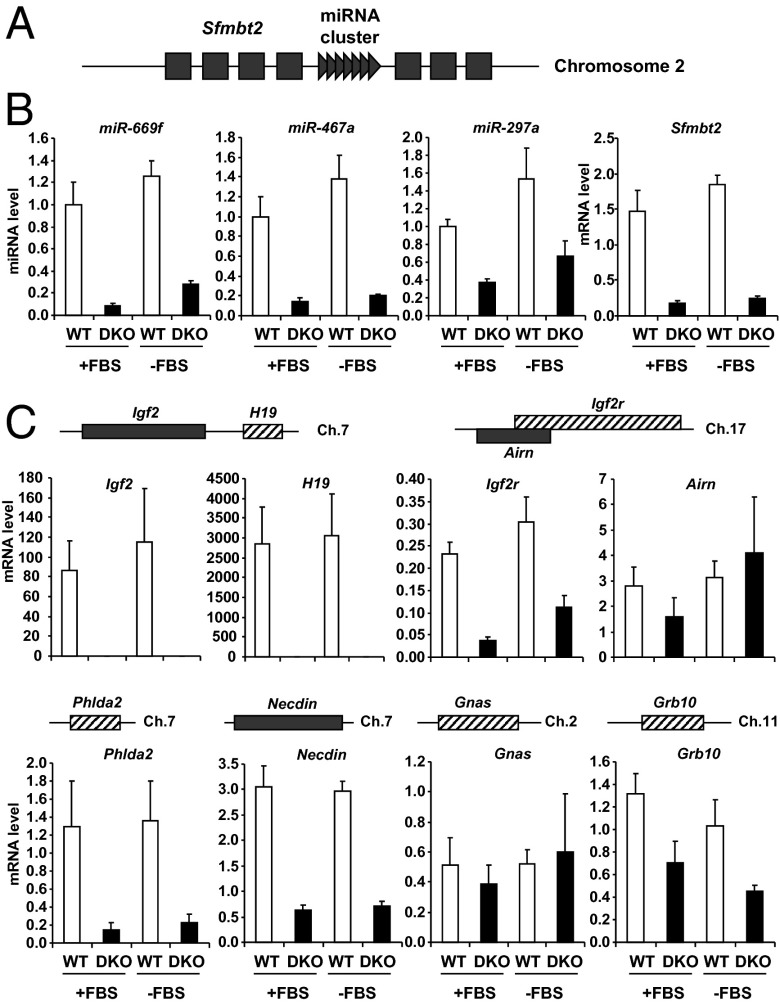
The miRNA cluster on mouse chromosome 2 is part of an intron of the imprinted gene Sfmbt2 (20, 21) (Fig. 3A). Again, expression of all miRNAs in this cluster and Sfmbt2 mRNA were decreased in DKO cells—in this case by 70–80% compared with WT cells (Fig. 3B). Likewise, the paternally expressed Igf2 gene and the maternally expressed lncRNA H19 on mouse chromosome 7 were easily detected in control cells, but virtually undetectable in DKO cells (Fig. 3C). Expression of the imprinted genes Phlda2 on chromosome 7, Necdin on chromosome 7, Igf2r on chromosome 17, and Grb10 on chromosome 11 were also decreased between twofold and eightfold in DKO cells compared with WT cells. Despite these marked changes in many imprinted genes, not all imprinted genes were altered in DKO cells, with no significant differences in mRNA between WT and DKO cells for Airn (a noncoding RNA which regulates imprinting of Igf2r on chromosome 17) and Gnas (chromosome 2) (Fig. 3C). Thus, DKO cells displayed decreased expression of multiple maternally and paternally expressed imprinted genes and miRNAs on several different chromosomes, indicating a broad-ranging dysregulation of imprinted gene expression in these cells.
Fig. 3.
Expression of other imprinted genes in WT and DKO cells. (A) Graphical representation of mouse imprinted Sfmbt2 gene in chromosomal region 2qA1. Dark gray boxes represent exons of the paternally expressed Sfmbt2 gene; dark gray triangles represent the miRNA cluster in an intron of Sfmbt2. (B and C) Expressions of Sfmbt2 and miRNAs from the cluster on chromosome 2qA1 (B) and of other imprinted genes (C) were measured by real-time PCR in confluent WT and DKO cells in the presence (+FBS) or absence (−FBS) of serum for 6 h. Results represent the average ± SEM of six independent experiments.
To determine whether the change in expression of these imprinted genes was related to a loss of insulin or IGF1 signaling, WT brown preadipocytes and differentiated adipocytes derived from them were treated with 100 nM insulin or IGF1 for 30 min or 6 h. All imprinted genes tested that were decreased in the DKO cells were either unchanged by insulin/IGF1 stimulation or modestly decreased by agonist stimulation of WT cells. For example, Dio3 expression was decreased by 50% in preadipocytes or adipocytes treated with insulin or IGF1 for 6 h, whereas expression of Dlk1/Pref1, Igf2, and H19 was not regulated by insulin or IGF1 treatment at either time point (Fig. S2). Thus, the down-regulation of expression of imprinted genes observed in DKO cells was not simply due to a lack of positive regulation of these genes by insulin and IGF1. Furthermore, the changes observed in imprinted gene expression were not reversed in DKO cells reexpressing either the IR or IGF1R or both (Fig. S3), suggesting a stable and heritable maintenance of the effect specifically at imprinted genes in DKO cells that was initiated by the absence of the receptors in the DKO cells.
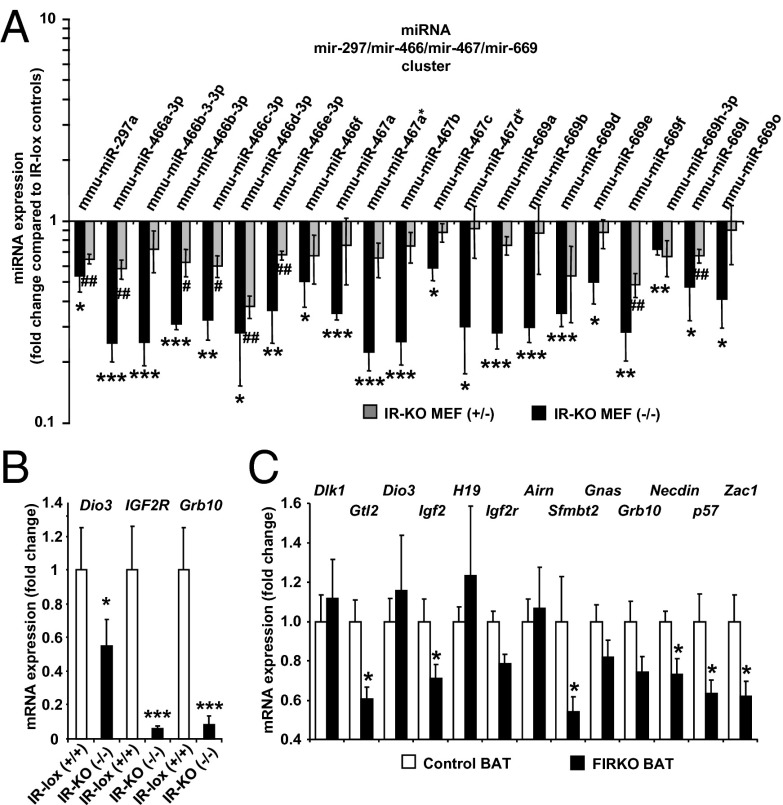
To determine whether this regulation of expression of imprinted genes was cell-type–specific, we used mouse embryonic fibroblasts (MEFs) from IRlox/CreERT2 mice in which the IR gene could be disrupted in vitro by treating the cells with tamoxifen. A similar qPCR-based miRNA expression profile in control MEFs or IR heterozygous or homozygous KO (IRKO MEF+/− or MEF−/−) cells was then performed. In this model, miRNAs from the imprinted cluster on chromosome 2 were expressed in control MEFs and were significantly decreased 50–80% in homozygous IRKO MEFs and by 25–50% in the heterozygous IRKO MEFs (Fig. 4A), suggesting a dose-dependent effect of the extent of disruption of the IR on the regulation of these imprinted miRNAs. Whereas the miRNAs from the imprinted cluster on chromosome 12 were not expressed at a detectable level in either control or IRKO MEFs, expression of Dio3, also on chromosome 12, was decreased by 45% (Fig. 4B). Likewise, expression of Grb10 on chromosome 11 and Igf2r on chromosome 17 was decreased in homozygous IRKO MEFs by 90% and 95%, respectively, compared with control cells. Thus, MEFs lacking the IR show similar, but somewhat less marked, changes in expression of imprinted genes as the DKO preadipocytes. A similar decrease in expression of imprinted genes was also observed in vivo in brown adipose tissue of mice with genetic inactivation of the IR gene [fat-specific IRKO (FIRKO) mice]. Expression of Gtl2, Igf2, Sfmbt2, Necdin, p57, and Zac1 was significantly decreased between 30% and 50% in FIRKO brown fat (Fig. 4C).
Fig. 4.
Imprinted gene expression in IR-KO MEFs and FIRKO brown fat. (A) miRNA expression profiling was performed by qPCR in control, heterozygous KO (IR-KO MEF+/−), and homozygous KO (IR-KO MEF−/−). Expression of imprinted miRNAs from cluster in intron of Smbt2 gene in chromosomal region 2qA1 is shown. Results are expressed as fold change compared with expression in control cells normalized to 1. Logarithmic scale. (B) Expression of imprinted genes Dio3, IGF2R, and Grb10 in confluent control and IR-KO MEF−/− was measured by real-time PCR. Results represent the average ± SEM of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with control cells). (C) Expression of imprinted mRNAs was measured by real-time PCR in brown adipose tissue of 6-mo-old male control and FIRKO mice. Results represent the average ± SEM of five mice. *P < 0.05 (compared with control mice).
Imprinted genes are regulated at two levels: first, like all other genes, through the direct effects of transcription factors; and second, through imprinting control that confers monoallelic expression in a parental origin-specific manner. Factors mediating imprinting control include expression of regulatory lncRNAs that impact the expression of protein-coding genes in cis or blocking of enhancer elements by CCCTC-binding factor (CTCF) insulators (22). Because both maternally and paternally expressed imprinted genes within the same cluster showed coordinated rather than reciprocal changes in gene expression, we postulated that the deregulation was not associated with perturbed imprinting control.
In some instances, imprinted lncRNAs serve as host transcripts for antisense transacting short RNAs such as siRNAs, miRNAs, or snoRNAs (13, 23–25). Three lncRNAs were measured in this study—namely, Gtl2, H19, and Airn. Both Gtl2 and H19 were expressed in WT cells, but undetectable in DKO cells, making it unlikely that these noncoding RNAs have a role in the regulation of potential trans targets. Airn lncRNA, with expression required for repression of Igf2r on the paternal chromosome (26), was unaffected in the DKO cells, despite Igf2r being down-regulated, again confirming repression in an imprinting-independent manner.
CTCF is a highly conserved zinc finger protein that plays a role in transcriptional activation/repression, insulation, imprinting, and X chromosome inactivation (27, 28). CTCF expression was not different between WT and DKO cells (Fig. S4). Ezh1 and Ezh2 are Polycomb-group family members, which form multimeric protein complexes involved in maintaining the transcriptional repressive state of genes. They act mainly to maintain gene silencing by trimethylating histone H3 at lysine 27 (29–31). Again, no change in expression of Ezh1 and Ezh2 was observed in DKO cells (Fig. S4). Zinc finger protein 57 homolog (ZFP57) and its cofactor KAP1 regulate imprints with loss of ZFP57 affecting the maintenance of their DNA methylation (32). ZFP57 expression was similar between WT and DKO cells, but KAP1 was down-regulated threefold in DKO cells (Fig. S4). This difference between KAP1 and ZFP57 expression is again consistent with dysregulation at the transcriptional level rather than at the level of imprinting.
The transcription factor Zac1 has been shown to regulate a large imprinted gene network, including Igf2, H19, Cdkn1c, and Dlk1. Zac1 directly regulates the Igf2/H19 locus through binding to a shared enhancer and transactivating Igf2 and H19 promoters (33). Zac1 inactivation in mice results in intrauterine growth restriction, altered bone formation, and neonatal lethality (33). Transient overexpression of Zac1 in neural crest-derived cells leads to the potent induction of several imprinted genes, whereas expression of these genes is decreased in livers from Zac1-deficient embryos. In DKO cells, Zac1 expression was decreased by 80% compared with WT cells (Fig. S5A). Furthermore, p21, a downstream target of Zac1 (34), was decreased fivefold at the mRNA level (Fig. S5A) and more than fivefold at the protein level in DKO cells compared with WT cells (Fig. S5B). However, reexpression of Zac1 in DKO cells only partially restored expression of some imprinted genes, with nonsignificant twofold to fivefold increases in expression of Gtl2, IGF2, and H19, and a significant 10-fold increase in Dlk1 (Fig. S5C). These findings indicate that the decrease in Zac1 is not the major cause of the general decrease in expression of imprinted genes observed in DKO cells.
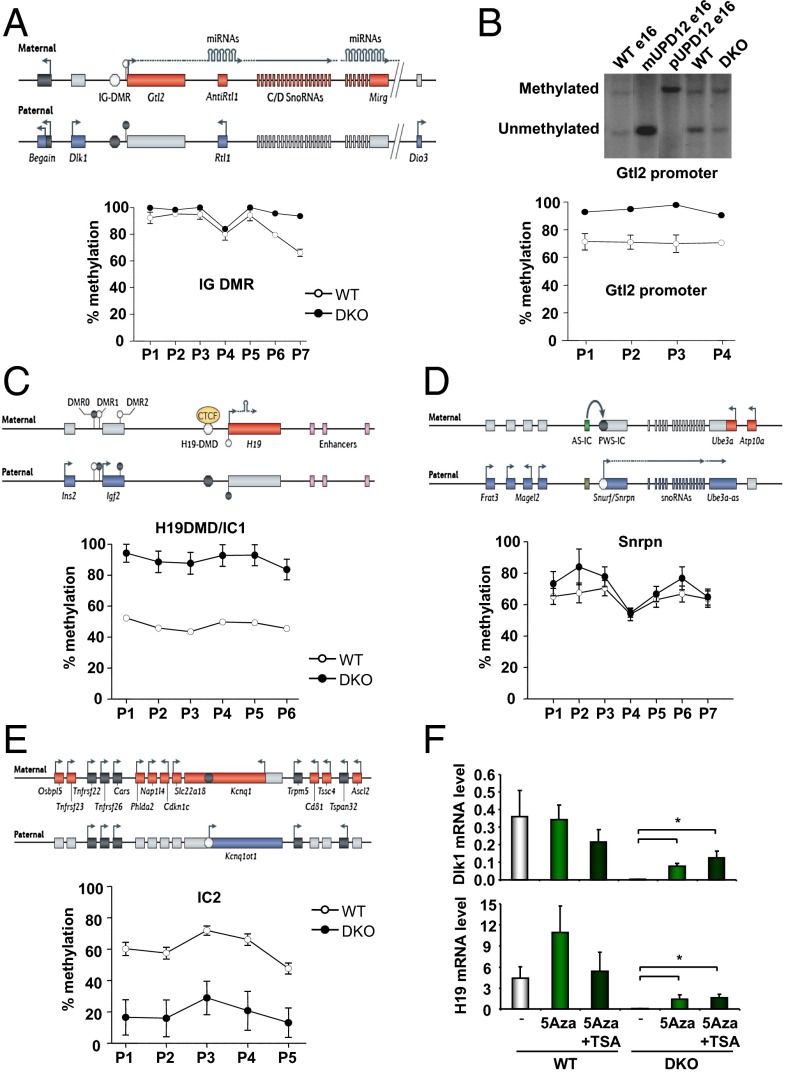
DNA methylation is an important regulator of imprinted genes (35). Imprinting in clusters is regulated by parental origin-specific germ-line DNA methylation at ICRs and sometimes at secondary DMRs acquired after fertilization in response to methylation at the ICR. DNA methylation at DMRs in the Dlk1–Dio3 locus includes the Gtl2 DMR that spans the Gtl2 promoter and its first exon and intron, the intergenic DMR (IG-DMR), which is located 13 kb upstream of the Gtl2 promoter, and the Dlk1 DMR which overlaps intron 4 and exon 5 of Dlk1 (36) (Fig. 5A). Quantitative pyrosequencing revealed a high level of methylation in both control and DKO cells at multiple sites in the IG-DMR, but those sites, which were not almost completely methylated in the WT cells (p6 and p7), showed increased methylation in DKO cells (Fig. 5A). Increased methylation was also observed at the Gtl2 promoter in DKO cells compared with WT cells, as measured by Southern blotting after digestion with methylation-sensitive restriction enzymes, and quantitative pyrosequencing (Fig. 5B). Similarly, methylation at the H19 differentially methylated domain (H19 DMD) or IC1, as measured by quantitative pyrosequencing, increased from 50% in WT cells to almost 100% in DKO cells at all sites tested (Fig. 5C). The Snrpn and Nnat promoters were also hypermethylated in DKO cells compared with control cells, consistent with the decreased expression of Necdin and Nnat, respectively (Fig. 5D and Fig. S6A). Interestingly, the Kcnq1ot1 promoter/IC2 and the Zrsr1 promoter were hypomethylated in DKO cells, consistent with the decreased expression of Cdkn1c and the increased expression of Zrsr1 (Fig. 5E, Fig. S6B, and Table S1). Thus, the marked decrease—or in some cases virtual absence—of expression of multiple imprinted mRNAs and miRNAs in DKO cells was associated with changes in regional DNA methylation. This decrease occurred without changes in expression of the de novo DNA methyltransferases Dnmt3a and Dnmt3b or the maintenance DNA methyl transferase Dnmt1 in DKO cells (Fig. S7). It is important to recall that methylation at the IG-DMR and the H19 DMD results in repression of Gtl2 and H19 and is normally associated with activation of the reciprocally imprinted Dlk1 and Igf2 loci (11). Similarly, decreased methylation of the Kcnq1ot1 promoter/IC2 is normally associated with an increase in Kcnq1ot1. The changes in methylation observed in the DKO cells, however, are associated with a generalized transcriptional repression rather than a classical canonical change in imprinting status, indicating that this repression is the result of a different and unique mechanism of action of transcriptional control.
Fig. 5.
DNA methylation in WT and DKO cells. (A) Schematic representation of the Dlk1–Dio3 locus and methylation analysis of the IG-DMR by pyrosequencing after bisulfite treatment of DNA from confluent WT and DKO cells. (B) Methylation analysis of the Gtl2 promoter: Southern blotting was performed on DNA from confluent WT and DKO cells digested with methylation-sensitive restriction enzymes and probed for the Gtl2 promoter. Controls using DNA from maternal and paternal uniparental disomy for chromosome 12 (UPD12) were added. Pyrosequencing was performed on DNA from WT and DKO cells after bisulfite treatment. (C–E) Schematic representation of imprinted loci and methylation analysis by pyrosequencing after bisulfite treatment of DNA from confluent WT and DKO cells. (F) Proliferating WT and DKO cells were treated with 0.2 μM 5-Azacytidine (5Aza) for 6 d, with or without 1 μM trichostatin A (TSA) for the last 18 h. Dlk1 and H19 expression were measured by real-time PCR in confluent cells. Results represent the average ± SEM of three independent experiments. *P < 0.05 (compared with control cells). Schematic representations were obtained from ref. 9.
Treatment of cells with the DNA methyltransferase inhibitor 5-azacytidine (5Aza), alone or in combination with histone deacetylase inhibitor trichostatin A (TSA), results in reactivation of epigenetically repressed genes (37). The 5Aza treatment of WT cells for 6 d induced a modest increase in expression of imprinted genes Dio3, Gtl2, and Sfmbt2 (Fig. S8). This increase was further enhanced by the addition of TSA for the last 18 h of 5Aza treatment. More importantly, treatment of DKO cells with 5Aza restored expression of Dlk1, H19, Dio3, Gtl2, and Sfmbt2 to levels comparable to those of WT cells, and treatment with TSA further increased these mRNA levels (Fig. 5F and Fig. S8), suggesting that repression of the imprinted genes in DKO cells is directly or indirectly related to increased DNA methylation, with or without some increase in histone deacetylation. However, the already-high level of methylation present in WT cells at some loci, and the relatively modest increase in methylation in DKO cells at other loci, suggest that additional mechanisms play a role in the decrease in expression of imprinted genes observed in DKO cells or that normal imprinted methylation may have been affected by cell culture conditions.
Together, our results show that loss of IRs and IGF1Rs in somatic cells leads to coordinated transcriptional down-regulation of multiple imprinted genes. This finding is distinct from canonical imprinting regulation in that perturbed imprinting control should lead to down-regulation of some imprinted genes with a reciprocal up-regulation of others. This result was not observed in our model. This previously unidentified pathway of regulation appears to be stably maintained, at least in part, by changes in DNA methylation. Imprinted genes are generally dosage-sensitive, and their appropriate expression is important for normal development and physiological responses, including those associated with growth and involving insulin and IGF1 signaling (13, 14, 38). Our observation points to a role of the unoccupied IRs and IGF1Rs in the coordinate control of multiple imprinted genes. Interestingly, it has recently been reported that pancreatic islets from patients with type 2 diabetes also have markedly reduced expression of the miRNAs in the Dlk1–Dio3 gene cluster (39). Clearly, further study of this unique role of IRs/IGF1Rs in the regulation of imprinted gene expression will be needed to fully elucidate its molecular mechanism and its relationship to normal physiology and disease.
Materials and Methods
Generation and Culture Conditions of Brown Preadipocytes.
DKO brown preadipocyte cell lines were generated from immortalized IR and IGF1R floxed brown preadipocytes infected with an adenovirus encoding Cre recombinase as described (3). Control cells used in the study were either immortalized WT or brown preadipocytes with floxed IR and IGF1R alleles. Results from the different control cell lines were pooled because no difference in miRNA and mRNA expression was noted and are referred to as WT. Cells were maintained in DMEM (high glucose) containing 10% (vol/vol) FCS at 37 °C in a 5% CO2 environment. For the gene expression study in adipocytes, control cells were differentiated as described (3).
Generation and Culture Conditions of MEFs.
The IRKO MEFs were obtained as described (40). Briefly, pregnant female mice from breeding pairs of mice heterozygous for the IRloxP mutation and heterozygous for the tamoxifen-inducible Cre recombinase were killed at day 16.5 postconception. Isolation and cultivation of the MEFs was carried out and the genotypes of the embryos were determined by PCR. For the induction of recombination, IR+/− or IR−/− fibroblasts were exposed to tamoxifen (Sigma-Aldrich) at a concentration of 1 μM for 7 d. All cells were maintained and analyzed in DMEM (Sigma-Aldrich) containing 10 mM d-glucose and 10% (vol/vol) FBS (Biochrom AG). MEFs were cultured at 37 °C in a humidified atmosphere of 5% CO2.
FIRKO Mice.
Homozygous FIRKO mice were generated as described by breeding IRlox/lox mice with mice expressing the Cre recombinase under the control of the adipose tissue-specific aP2 promoter (41). The mice were kept on a regular chow diet. All protocols were approved by the Institutional Animal Care and Use Committee of the Joslin Diabetes Center.
Zac1 Overexpression.
Stable overexpression of Zac1 was achieved by introducing murine Zac1 cDNA (Mm30411, GeneCopoeia) into the pBabe retrovirus vector. Plates (10 cm) of phoenix cells were transiently transfected with 10 μg of control (empty pBabe vector) or Zac1-containing retroviral expression vectors (SuperFect; Qiagen). At 48 h after transfection, virus-containing medium was collected and passed through a 0.45-mm pore-size filter. An equal volume of fresh growth medium and Polybrene (hexadimethrine bromide; 12 mg/mL) were added to the virus-loaded medium. This medium was then applied to proliferating (∼40% confluent) DKO cells. At 48 h after infection, cells were treated with trypsin and replated in medium supplemented with zeocin (Invitrogen) as a selection antibiotic.
Quantification of mRNA and miRNA Expression by qPCR.
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen). A total of 1 μg of RNA was reverse-transcribed with a high-capacity cDNA reverse-transcription kit (Applied Biosystems). Synthesis of stem–loop miRNA cDNAs was achieved by addition of specific miRNA hairpin reverse transcription primers to the reverse transcriptase reaction as described (42). Real-time PCR was performed starting with 12.5 ng of cDNA and both sense and antisense oligonucleotides (300 nM each) in a final volume of 10 μL with the iQ SYBR Green Supermix (Bio-Rad). Fluorescence was monitored and analyzed in a CFX384 Real-Time PCR Detection System (Bio-Rad). Amplification of specific transcripts was confirmed by analyzing melting curve profiles at the end of each PCR. Analysis of TATA box–binding protein expression was performed in parallel to normalize gene expression.
miRNA qPCR Array.
QuantiMir Genome-Wide PCR Arrays (System Biosciences) were used to assess the expression profile of 632 mouse miRNAs. The method is based on SYBR-Green real-time PCR quantitation, which provides a highly sensitive and quantitative approach for measuring the full repertoire of biologically validated miRNAs. Briefly, small RNAs in the sample of total RNA isolated from cultured cells were tagged at the 3′ end and extended to generate detectable cDNAs for qPCR. The quantitation step was based on the SYBR-Green real-time qPCR protocol as described above, using primers provided by the manufacturer. The miRNA was considered expressed when the Ct values of the samples was <30.
Microarray Analysis.
A total of 15 μg of cRNA from five WT and five DKO samples were hybridized to murine Affymetrix U74Av.2 arrays. Target preparation, hybridization, and scanning were performed in the Joslin Diabetes Center Genomics Core. Signal intensities were quantitated by using GeneChip Operating Software. Global scaling was used to standardize signal intensities. GSEA was performed by using GSEA software (www.broad.mit.edu/gsea) to identify differentially expressed gene sets.
Methylation Analysis.
For Southern blotting, 10 μg of genomic DNA was digested with StuI (IG-DMR), NheI (Gtl2 promoter), or PstI (Dlk1 DMR) with or without MspI and HpaII/HhaI and probed as described (43, 44). For bisulfite conversion, genomic DNA (1 μg) was treated by using the Imprint DNA Modification Kit (Sigma) in accordance with the manufacturer’s instructions for the two-step conversion, and eluted in 20 μL. Pyrosequencing was carried out according to ref. 45 and was performed on the PSQ HS96 System by using PyroGold Q96 SQA Reagents (Qiagen). The degree of methylation at CpG sites (without distinguishing between maternal and paternal alleles) was determined by Pyro-Q CpG software.
Statistical Analyses.
All data are presented as mean ± SEM and analyzed by the unpaired two-tailed Student t test. A P value of <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Mary Elizabeth Patti for helpful discussions and Katie Hughes for assistance with the microarray analysis. This work was supported by National Institutes of Health Grants DK31036 and DK82659; Diabetes Endocrinology Research Center Grant DK34834, an American Diabetes Association mentor-based award, and the Mary K. Iacocca Professorship. A.C.F.-S. and M.C. are supported by FP7 EpigeneSys and grants from the Medical Research Council and Wellcome Trust. K.Z. was supported by an Albert Renold Fellowship of the European Foundation for the Study of Diabetes.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415475111/-/DCSupplemental.
References
- 1.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: Insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 2.Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6(1):409. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher J, Tseng YH, Kahn CR. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J Biol Chem. 2010;285(22):17235–17245. doi: 10.1074/jbc.M110.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher J, et al. A kinase-independent role for unoccupied insulin and IGF-1 receptors in the control of apoptosis. Sci Signal. 2010;3(151):ra87. doi: 10.1126/scisignal.2001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehlen P. Dependence receptors: The trophic theory revisited. Sci Signal. 2010;3(151):pe47. doi: 10.1126/scisignal.3151pe47. [DOI] [PubMed] [Google Scholar]
- 6.Radford EJ, Ferrón SR, Ferguson-Smith AC. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 2011;585(13):2059–2066. doi: 10.1016/j.febslet.2011.05.063. [DOI] [PubMed] [Google Scholar]
- 7.DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345(6270):78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 8.Charalambous M, et al. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci USA. 2003;100(14):8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson-Smith AC. Genomic imprinting: The emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12(8):565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 10.Hudson QJ, Kulinski TM, Huetter SP, Barlow DP. Genomic imprinting mechanisms in embryonic and extraembryonic mouse tissues. Heredity (Edinb) 2010;105(1):45–56. doi: 10.1038/hdy.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19(3):281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3(7):a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charalambous M, da Rocha ST, Hernandez A, Ferguson-Smith AC. Perturbations to the IGF1 growth pathway and adult energy homeostasis following disruption of mouse chromosome 12 imprinting. Acta Physiol (Oxf) 2014;210(1):174–187. doi: 10.1111/apha.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapunzina P, Monk D. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol Cell. 2011;103(7):303–317. doi: 10.1042/BC20110013. [DOI] [PubMed] [Google Scholar]
- 16.Butler MG. Prader-Willi syndrome: Obesity due to genomic imprinting. Curr Genomics. 2011;12(3):204–215. doi: 10.2174/138920211795677877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomizawa S, Sasaki H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J Hum Genet. 2012;57(2):84–91. doi: 10.1038/jhg.2011.151. [DOI] [PubMed] [Google Scholar]
- 18.Seitz H, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34(3):261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 19.Seitz H, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14(9):1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzmin A, et al. The PcG gene Sfmbt2 is paternally expressed in extraembryonic tissues. Gene Expr Patterns. 2008;8(2):107–116. doi: 10.1016/j.modgep.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, et al. Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genomics. 2011;12:204. doi: 10.1186/1471-2164-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: Noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 23.Royo H, Cavaillé J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100(3):149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 24.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, Hu H, Lai M. Non-coding RNAs and their epigenetic regulatory mechanisms. Biol Cell. 2010;102(12):645–655. doi: 10.1042/BC20100029. [DOI] [PubMed] [Google Scholar]
- 26.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 27.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 28.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 30.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varrault A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Liu PY, et al. Modulation of the cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa Cells. Mol Cancer Res. 2008;6(7):1204–1214. doi: 10.1158/1541-7786.MCR-08-0123. [DOI] [PubMed] [Google Scholar]
- 35.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 36.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 38.Charalambous M, et al. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15(2):209–221. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kameswaran V, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19(1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarse K, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15(4):451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blüher M, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3(1):25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 42.Clancy JL, et al. Methods to analyze microRNA-mediated control of mRNA translation. Methods Enzymol. 2007;431:83–111. doi: 10.1016/S0076-6879(07)31006-9. [DOI] [PubMed] [Google Scholar]
- 43.Takada S, et al. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10(18):1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- 44.Takada S, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: Implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet. 2002;11(1):77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 45.Sun B, et al. Status of genomic imprinting in epigenetically distinct pluripotent stem cells. Stem Cells. 2012;30(2):161–168. doi: 10.1002/stem.793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.