Significance
We describe the smallest (220 nt) replicating covalently closed circular (CCC) RNA (nanogenome) that contains numerous unique and novel properties of coding and translation. It is the smallest of all viroids and virusoids and the only one to encode proteins. No noncoding sequences could be identified. Initiation and termination codons are combined in UGAUGA, and read-throughs generate larger proteins. This is the only CCC RNA to be directly translated by eukaryotic ribosomes. It also contains an internal ribosome entry site and it defies the classical Kozak scanning model for initiation of translation. Is it a living fossil or is it a novel trend in genomic evolution?
Keywords: hammerhead ribozyme, sobemovirus, circular RNA translation, leaky termination codons
Abstract
The highly structured (64% GC) covalently closed circular (CCC) RNA (220 nt) of the virusoid associated with rice yellow mottle virus codes for a 16-kDa highly basic protein using novel modalities for coding, translation, and gene expression. This CCC RNA is the smallest among all known viroids and virusoids and the only one that codes proteins. Its sequence possesses an internal ribosome entry site and is directly translated through two (or three) completely overlapping ORFs (shifting to a new reading frame at the end of each round). The initiation and termination codons overlap UGAUGA (underline highlights the initiation codon AUG within the combined initiation-termination sequence). Termination codons can be ignored to obtain larger read-through proteins. This circular RNA with no noncoding sequences is a unique natural supercompact “nanogenome.”
Viroids and virusoids (viroid-like satellite RNAs) are typically small (220–450 nt) covalently closed circular (CCC) RNAs with no coding capacity (i.e., no genetic information) (1–3) and are the smallest replicating circular RNA pathogens (3, 4). Because of their circular nature, they usually replicate through a rolling circle model to produce larger concatemers (4, 5) which are then processed into monomeric forms with a self-splicing hammerhead ribozyme (virusoids and viroids in the Avsunviroidae family) (6, 7) or by cellular enzymes (8). We have previously reported (9) the characterization and nucleotide sequence of the smallest circular virusoid (220 nt), that of the rice yellow mottle virus (sobemovirus) (RYMV). Like other known virusoids, the small circular satellite of RYMV (scRYMV) depends on a helper virus RYMV for replication and packaging (9, 10).
In silico translation of scRYMV revealed the presence of an unusual ORF capable of initiating translation from the AUG in the sequence UGAUGA of the 220-nt circular RNA by internal ribosome binding site (IRBS). As 220 is not an integer multiple of 3, after the first round of translation, the same circular sequence (or possibly the linear head-to-tail concatemers generated in vivo during rolling-circle replication) would be read in a different frame register. After the second round of translation, termination at the same initiation–termination sequence UGAUGA would result in the production of a highly basic 16-kDa protein with the N- and C-terminal halves of the protein encoded by the same 220-nt sequence but read in two distinct, totally overlapping frames. This virusoid could also suppress the leaky tandem termination codons (UGAUGA) to read the same sequence in a third frame and produce a new (18 kDa) read-through protein with an 18-aa C-terminal extension ended by a UAG codon. However, even the latter UAG termination codon may occasionally be ignored to generate longer proteins.
In this report, we present evidence demonstrating that the putative ORF(s) deduced from the 220-nt circular RNA sequence as described above are indeed operational.
Although this scRYMV RNA is classified as a virusoid, we report here that this virusoid is the only one so far found to encode for a protein. Modalities of initiation of translation (e.g., overlapped initiation and termination codons), the distinct N- and C-terminal halves of the 16-kDa protein translated from the same 220-nt circular sequence and the generation of read-through proteins were examined. We discuss the evolutionary implications of the genetic information and biological functions densely packed into a 220-nt nanogenome.
Results
Expression in Escherichia coli of a Head-to-Tail Trimer Copy of a scRYMV cDNA Clone.
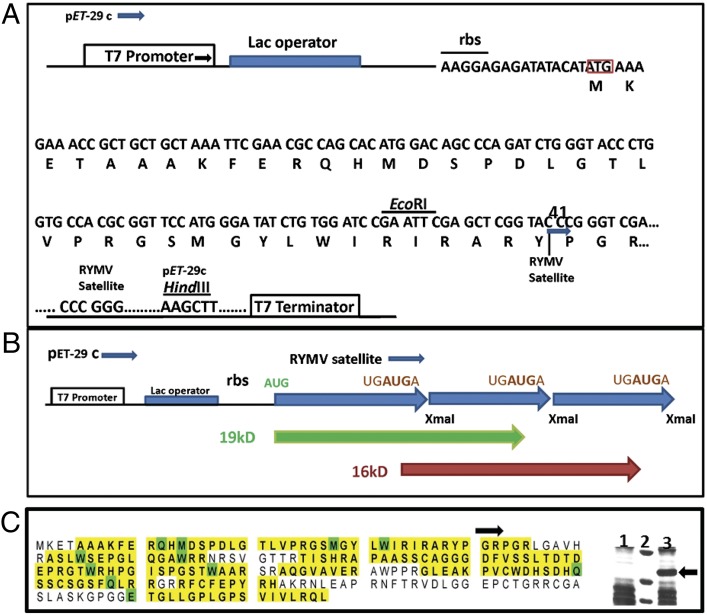
The original isolation, cloning, and several other properties of scRYMV RNA are described elsewhere (9, 11). The putative ORF generated from a linear trimeric head-to-tail cDNA copy of the 220 nt of scRYMV RNA (representing a three-frame overlapped ORF present in the monomeric form of circular RNA) was constructed as a fusion protein with S-Tag/thrombin under the T7 transcriptional and translational controls in plasmid expression tag (pET)29(c) vector. The resulting clone was designated pET52 (Fig. 1). Thirteen amino acids at the N terminus of the putative scRYMV protein were replaced by 39 aa from thrombin to generate a fusion protein. Upon induction of pET52 in E. coli cells, a major protein species of 19 kDa was detectable by SDS/PAGE and Coomassie brilliant blue R-250 staining (Fig. 1C, Right).
Fig. 1.
pET52 is constructed by the insertion of a head-to-tail trimer of the scRYMV satellite sequence into the empty pET29(C) plasmid at the XmaI site. (A) Detailed diagram depicting part of the nucleotide and amino acid sequences for 19-kDa scRYMV/thrombin fusion protein. Ribosome binding site (rbs) with Shine/Dalgarno sequence, and T7 promoter and terminator are also depicted. (B) Schematic diagram showing the translation of 19 kDa (starting at AUG) (green) and 16 kDa [starting at UGAUGA (brown) protein]. Thick blue arrowheads indicate the start of each monomeric form of the scRYMV head-to-tail trimer sequence with (XmaI site) at nucleotide 41 in scRYMV sequence (Fig. 6 includes numbering). (C) Identification of the scRYMV/thrombin fusion 19-kDa protein. The prominent band of 19-kDa protein purified from SDS/PAGE of total E. coli proteins containing pET52, was subjected to MS analysis. LC-MS/MS recovered peptides are shown in yellow. Modified amino acids are shown in green (Materials and Methods and Table S1). The 163/227 aa (72% coverage) are shown. Arrow indicates the start of scRYMV sequence in the 19-kDa fusion protein. (Right) Identification of the 19-kDa scRYMV/thrombin fusion protein (arrow). Coomassie brilliant blue-R250 staining of total E. coli expressed proteins from pET29 (empty plasmid control, lane 1) and pET52 (19-kDa protein, lane 3). Lane 2: molecular size markers from top to bottom: 23, 18, and 14 kDa, respectively.
MS Identification of the 19-kDa and 16-kDa Polypeptides in E. coli Overexpressing the pET52 Clone.
The region of the gel containing the major 19-kDa fusion polypeptide band obtained as described earlier was subjected to liquid chromatography (LC)/tandem MS (LC-MS/MS). Comparison of the theoretical amino acid sequence of the scRYMV ORF-derived protein (in silico translation) with the peptides resulting from the LC-MS/MS identified 17 peptides (Fig. 1C and Table S1) with 72% overall sequence coverage over the scRYMV-derived 19-kDa E. coli fusion protein of 172 aa. These peptides corresponded exactly to the expected sequence of the 19-kDa protein. Table S1 shows peptides detected along with their abundance and other essential parameters. In particular, six peptides were found with the sequence (T)AAAKFERQHMDSPDLGTLVPR(G), which occurs close to the N terminus, typical of the 19-kDa fusion protein. This major recombinant 19-kDa protein was subsequently used to raise specific polyclonal antibodies in rabbit (scRYMV antiserum).
Further analysis of the LC-MS/MS profile (Table S1) revealed five peptides of the sequence MSQEELGGTQEFHPGRPGRLGA(V) typical of the start of the 16-kDa protein. We also observed 77 peptides ending with the sequence (R)RFCFEPYRH, characteristic of both C-termini of the 19 kDa and the 16 kDa that terminated at the tandem stop sequence UGAUGA (see Fig. 6). The 16 kDa could only have arisen from internal initiation of translation at the first combined initiation–termination sequence UGAUGA in the pET52 sequence and terminated at the third initiation–termination sequence (Fig. 1 A and B). Analysis of equivalent protein bands from E. coli expressing the empty vector pET29 did not reveal any peptides corresponding to the scRYMV-derived proteins. However, both samples revealed the presence of E. coli-specific proteins when compared with peptides available in data banks.
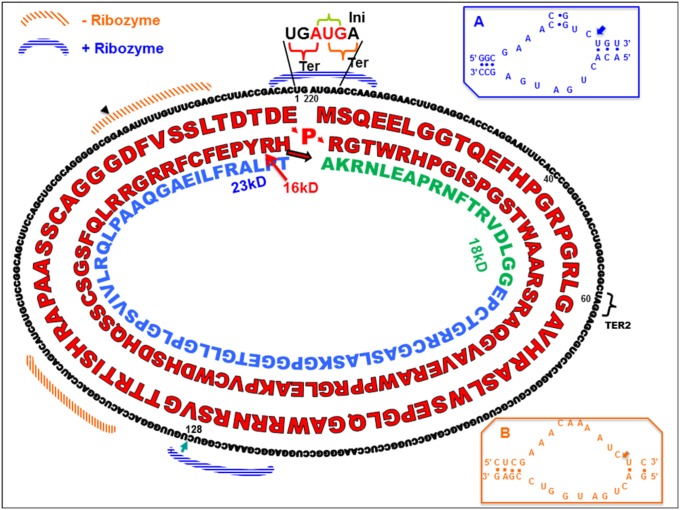
Fig. 6.
Schematic diagram showing the nucleotide and amino acid sequences of the encoded proteins and many other functions of the CCC RNA. Positions of initiation AUG codon (INI) and termination codons UGA (TER for 16 kDa) and UAG (TER2 for 18 kDa) are indicated. Locations of (+) and (−) hammerhead ribozymes are indicated. Positive-sense hammerhead ribozyme is shown in Inset A and that of minus-sense ribozyme is shown in Inset B. Arrowheads indicate the splice site for both ribozymes [on circular RNA depicting secondary structure and cleavage sites for both ribozymes (Insets)]. The amino acid sequences of 16-, 18-, and 23-kDa proteins are indicated. Shaded areas indicate the nucleotide sequences involved in (−) and (+) ribozymes. Nucleotide numbering starts at (position 1) UGAUGA.
In Vitro Translation of Transcripts of pET52 and Natural scRYMV CCC RNA.
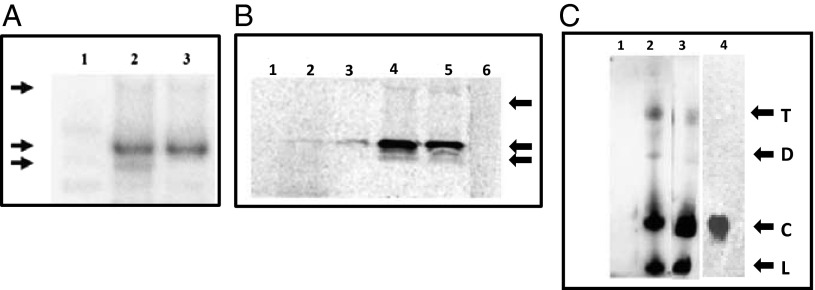
A coupled in vitro transcription/translation of linearized pET52 using a wheat germ lysate translation system followed by immunoprecipitation with the scRYMV antiserum revealed mainly two translated polypeptides of 16 and 19 kDa (Fig. 2A, lane 2). The 16-kDa protein was generated only by internal initiation (Fig. 1B). Indeed, when the trimer (pET52) was truncated (at the third XmaI site; Fig. 1B) into a dimer, the 16-kDa product, but not the 19-kDa fusion protein, was lost (Fig. 2A, lane 3). Translation of transcripts from pET29, as expected, did not produce any immunoprecipitation product with the scRYMV antiserum (Fig. 2A, lane 1).
Fig. 2.
In vitro transcription/translation and immunoprecipitation of translated products using antiserum raised against the scRYMV ORF-encoded protein. (A) Lane 2 shows 19-kDa and 16-kDa proteins from reaction using pET52 DNA, whereas lane 3 shows only the 19 kDa from reaction using the truncated pETdimer DNA. Lane 1: Negative control reaction using pET29 (empty vector). Arrows on left indicate molecular size of proteins, from top to bottom: 36 kDa, 19 kDa, and 16 kDa. (B) In vitro translation reaction depicting the 16-kDa protein from the scRYMV circular RNA (purified from denaturing polyacrylamide gels) is shown in lane 3, whereas that of the reaction from total RNA of RYMV-infected rice is shown in lane 2. Lane 5 demonstrates the 16-kDa product from reaction using total viral RNA extracted from RYMV virus particles, and lane 4 shows the enhanced 16-kDa signal from reaction using RYMV total viral RNA but supplemented with the same amount of gel-purified circular RNA as that used in the reaction of lane 3. Lanes 1 and 6 represent negative control reactions using healthy rice and the endogenous empty lysate, respectively. Arrows on right depict the position of molecular weight markers 25, 16, and 14 kDa from top to bottom, respectively. (C) Northern analysis to detect the nature of scRYMV RNA species. Denaturing 4–20% PAGE in presence of 8 M urea was carried out. Lanes 2 and 3 demonstrate presence of linear (marked as “L”), circular (“C”), dimer (“D”), and trimer (“T”) forms of the scRYMV RNA in total RNA preparations from RYMV-infected rice and RYMV particles, respectively. Lane 1 shows the negative control of total RNA from healthy rice. Lane 4: 7 M urea-PAGE stained with ethidium bromide and showing the purified circular RNA extracted from the band corresponding to circular RNA (shown in lane 3). This purified circular RNA is used for in vitro translation. Arrows indicate the positions of RNA size markers: 220 (marked as “L” or “C”), 440 (“D”), and 660 (“T”) nt.
Before in vitro translation of the scRYMV circular RNA obtained from RYMV-infected rice plants, we analyzed the types of scRYMV RNAs found in the virus particle and in infected plants and detected scRYMV circular RNA and its replicative intermediates (concatemers). Viroids and viroid-like circular RNAs are known to replicate through a “rolling circle” model (4, 5), which generates linear head-to-tail repeated genome-length concatemers. Northern analysis, using a specific 35S-labeled scRYMV probe, under denaturing conditions (7 M urea) of RNA species isolated from purified virus particles (Fig. 2C, lane 3), as well as from total cellular RNAs of RYMV-infected rice plants (Fig. 2C, lane 2), revealed the presence of several scRYMV RNA forms. We identified an intense fastest-moving linear form (220 nt), followed by a more intense slower-migrating circular form (also 220 nt) and two minor species of higher molecular weight forms of 440 and 660 nt (Fig. 2C, lanes 2 and 3). The identities and sizes of these concatemers were confirmed to be derived from scRYMV by hybridization to 35S-labeled scRYMV specific probes and by RT-PCR and sequencing.
When total viral RNA extracted from purified RYMV particles was used for the in vitro wheat germ transcription/translation system, a major protein with a molecular weight of 16 kDa was immunoprecipitated with the specific scRYMV antiserum (Fig. 2B, lane 5). Native CCC satellite RNA (i.e., scRYMV), purified from contaminating linear RNA and concatemers by PAGE in the presence of 7 M urea, translated into a typical 16-kDa protein (Fig. 2B, lane 3). Further, when total viral RNA (genomic and CCC RNA) was supplemented with purified circular RNA, a much-enhanced signal of 16 kDa was also observed (Fig. 2B, lane 4; compare lanes 4 and 5). Smaller bands were observed below the 16 kDa (lanes 4 and 5) and probably result from incomplete translation of the 16-kDa protein. Total RNA extracted from RYMV-infected plants, when translated, produced the 16-kDa protein (Fig. 2B, lane 2). Total RNA extracted from healthy rice plants and empty wheat germ lysate did not produce any immunoprecipitation products (Fig. 2B, lanes 1 and 6). It seems likely that most of the translation is directly related to amounts of CCC RNA.
Identification of scRYMV-Derived 16 kDa in RYMV-Infected Rice by Western and Tandem LC-MS/MS Analyses.
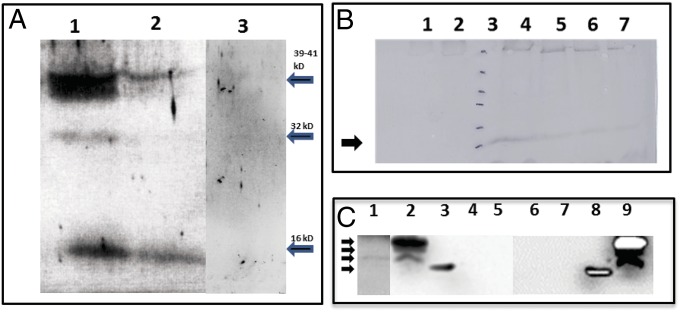
Total proteins extracted from RYMV-infected and healthy (control) rice plants were subjected to Western analysis using the scRYMV antiserum. The 16-kDa protein and higher molecular weight proteins (32 and 39–41 kDa), derived from the translation of scRYMV circular RNA (see Fig. 6) were seen only in RYMV-infected plants (Fig. 3A, lane 1). Total protein extracted from healthy uninfected rice plants did not react with the antiserum (Fig. 3A, lane 3). On the contrary, a fainter 16-kDa band is also seen in extracts from purified (i.e., RYMV) virus particles (Fig. 3A, lane 2), revealing that the protein is packaged in the virus particles. The scRYMV antiserum did not cross-react with the capsid protein of RYMV. Western analysis also revealed the temporal expression of the 16 kDa, which was not detectable in the first 3–4 d postinoculation (Fig. 3B, lane 2), but subsequently appeared and increased to peak at 7–10 d postinoculation (Fig. 3B, lane 4), followed by a steady decline at 10–28 d postinoculation (Fig. 3B, lanes 5–7).
Fig. 3.
SDS/PAGE, Western detection, and time course of expression of the scRYMV-encoded proteins. (A) Western analysis using antibodies specific to the scRYMV ORF-encoded protein to detect the 16-kDa proteins among total plant proteins from RYMV infected (lane 1), healthy uninfected plant (lane 3), and purified RYMV virus (lane 2). Arrows on right indicate the positions of the 39-, 32-, and 16-kDa proteins (from top to bottom). (B) Western analysis time course of the appearance of the scRYMV 16 kDa during infection in rice plants. Lane 1 contains total proteins extracted from noninfected rice plants (negative control). Lane 2 contains total proteins from RYMV-infected plants at 4 d postinoculation. Lanes 4–7 contain total rice proteins extracted from RYMV-infected plants at 10, 14, 21, and 28 d postinoculation, respectively. Lane 3 and arrow (16 kDa) depict molecular weight size markers 116, 66, 45, 35, 25, and 18 kDa, from top to bottom, respectively. (C) North-Western analysis showing the RNA-binding activity of scRYMV-encoded proteins. Lane 1 shows the presence of the 16-kDa RNA-binding protein in infected rice, whereas lanes 2 and 9 demonstrate the detection of the 16-kDa, 18-kDa, and 19-kDa RNA-binding proteins in protein extracts from E. coli carrying pET52. Lanes 4 and 5 are the negative controls from healthy rice and pET29 protein extracts, respectively. Lanes 6 and 7 are negative control from E. coli alone and E. coli carrying pET29, respectively. Lanes 3 and 8 contain the molecular weight markers including the 14-kDa positively charged lysozyme. The 35S-labeled scRYMV and PVX probes were used in lanes 1–5 and 6–9, respectively. Arrows on left indicate the positions of molecular size markers (from top to bottom, 19, 18, 16, and 14 kDa, respectively).
Tandem LC-MS/MS analyses of total rice proteins corresponding to the 16-kDa band from RYMV-infected rice (at 10 d postinoculation) presented a sequence coverage of 94% (Fig. 4). Nine peptides were obtained with the sequence MSQEELGGTQEFHPGRPGRL mapping at the N terminus of the scRYMV 16-kDa protein, demonstrating that internal initiation of translation takes place at the unique AUG sequence within the UGAUGA initiation–termination sequence (Table S2). Approximately 63 peptides were obtained with the sequence ending at (R)RFCFEPYRH, demonstrating that the 16 kDa is likely the major protein species synthesized by the scRYMV in RYMV-infected rice. Analysis of the corresponding bands from healthy rice plants did not show any peptides related to scRYMV. When peptides (from healthy or RYMV-infected plants) were compared with protein databases, a large number of peptides were identified as expected to be rice-derived proteins. However, none of the peptides related to 16-kDa proteins were found in any of the available databases, reflecting the uniqueness of this protein.
Fig. 4.

Mapping of the scRYMV-derived LC-MS/MS peptides from RYMV-infected rice on the scRYMV protein sequence generated from translation (in all three reading frames) of scRYMV RNA. Matching sequences are highlighted. LC-MS/MS recovered peptides are shown in yellow. Modified amino acids are shown in green. Table S2 provides raw data and other details regarding peptide detection.
RNA Binding and Other Properties of the 16 kDa-Protein: North-Western Analysis.
Amino acid analysis of the 16-kDa protein (146 aa) revealed that this protein is unusual in many respects. It does not have any significant sequence homology with any known protein in GenBank or Swiss-Prot. This protein is very rich in basic amino acids, including 22 arginines (13%), seven histidines (4%), and two lysine residues. A cluster of five arginines is also found toward the C terminus. The isoelectric point is calculated to be approximately 10.8. The high positive charge property of the 16-kDa protein was used to determine the capacity of this protein to bind homologous and heterologous RNAs. North-Western blots indicated that the 16-kDa protein is expressed in only RYMV-infected rice plants, and it binds to homologous (i.e., scRYMV) (Fig. 3C, lane 1) as well to heterologous [i.e., potato virus X (PVX)] RNAs. The 16-, 18-, and 19-kDa proteins expressed in E. coli also bind to scRYMV (i.e., homologous) and PVX (i.e., heterologous) labeled RNAs (Fig. 3, lanes 2 and 9). Lysozyme (used here as a molecular weight size marker) is also positively charged and nonspecifically binds RNAs (Fig. 3C, lanes 3 and 8). No other plant, E. coli, or size marker proteins reacted with labeled RNAs (Fig. 3, lanes 4–7).
Tandem LC-MS/MS Identification of scRYMV-Derived Read-Through Proteins.
In E. coli overexpressing the pET52 clone, evidence for the 18 kDa (read-through protein), which terminated at the downstream UAG (amber) codon is derived from the detection of the peptide AKRNLEAPRNFTRVDLGG, as shown in Table S1. This 18-kDa protein is also seen in results of North-Western analysis of E. coli expressing pET52 (Fig. 3C, lanes 2 and 9). Interestingly, we also observed LC-MS/MS peptides showing perfect matches with the sequence in the third reading frame beyond that of the 18-kDa read-through protein (see Fig. 6).
From LC-MS/MS analysis of RYMV-infected rice proteins, we also observed peptides translated in the third reading frame around the circular RNA. However, unlike in the E. coli system, we did not observe any peptide ending at the UAG codon, and therefore no 18-kDa polypeptide, also corroborated by our North-Western results (Fig. 3C, lane 1). Interestingly, Western blots (Fig. 3A), showed, in addition to the major 16-kDa protein, a 32-kDa polypeptide, which may be a dimer of the 16 kDa as well as a broad band (39–41-kDa polypeptides). Analysis of the LC-MS/MS profile in the SDS/PAGE gel region of the 39 kDa showed (at very low frequency) peptide RALPLTMSQEELGGTQEFHPGRPGRLGA(V) connecting the third reading frame back to the first reading frame, implying continued protein synthesis around the circular RNA (Table S2). It is possible that the 39-kDa protein could result from a five-round translation of the circular RNA, by ignoring once the tandem UGAUGA termination codons and then by recognizing them (after a second round of translation).
Mutational Analysis.
To determine the role of AUG to initiate translation, the sequence UGAUGA in the pBlueScript (pBS)-trimer clone was modified to UGAAUA and used in a coupled in vitro transcription/translation reaction followed by immunoprecipitation. As shown in Fig. 5A (lane 3), the synthesis of the 16-kDa protein was completely abolished, whereas the WT pBS-trimer with intact UGAUGA translated the 16-kDa protein (Fig. 5A, lane 5). This clearly demonstrated the role of the AUG within the UGAUGA sequence in the 16-kDa translation.
Fig. 5.
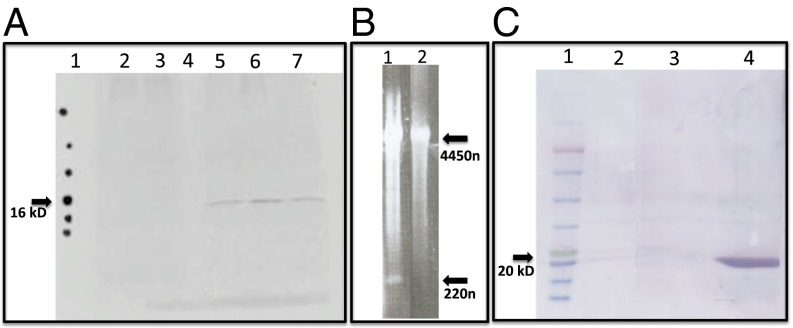
Mutational analysis of scRYMV. (A) In vitro transcription/translation and immunoprecipitation profile of scRYMV AUG to AUU (in the sequence UGAUGA) mutant shows abolition of 16-kDa translation. Lane 1: radiolabeled dots indicating the positions of protein molecular weight size markers (top to bottom, 35, 25, 20, 17, 11, and 5 kDa). Lane 2: in vitro reaction (with scRYMV 19-kDa antibody) with purified RYMV genomic RNA alone (without scRYMV) used as template shows no 16-kDa protein and was used as infectious transcript in plants. Lane 3: same reaction as in lane 2 except for addition of mutated scRYMV construct (AUG to AAU mutant pBS-trimer). Lane 4: empty lane. Lane 5: same reaction as in lane 2 except for the addition of pBS-trimer (intact construct) demonstrating the synthesis of a 16-kDa protein. Lane 6: reaction containing total RYMV viral RNA obtained from purified virus containing scRYMV. Lane 7: reaction containing purified WT scRYMV RNA alone (obtained from virus particles). (B) Replication of AUG to AAU scRYMV mutant is abolished in plants. Lane 1: RYMV viral RNA profile from purified virus particles of plants infected with infectious RYMV RNA along with T7 polymerase-generated in vitro transcripts from WT pBS-trimer scRYMV clone. Lane 2: RYMV infectious RNA plus in vitro transcripts from AUG to AAU mutant pBS-trimer clone showing complete absence of the scRYMV RNA (no replication). Upper arrow indicates the position of genomic RYMV RNA (4450 nt) and the bottom arrow indicates the position of circular scRYMV RNA (220 nt). (C) UGAUGA codons influence translation termination: Western blotting analysis of total E. coli proteins using pET dimer and anti–His-tag antibody for intact and a UGAUGA-to-CUCGAG mutant construct. Lane 1: protein molecular weight ladder (top to bottom, 135, 100, 75, 63, 48, 35, 25, 20, 17, and 11 kDa). Lane 2: E. coli containing empty pET29 plasmid. Lane 3: E. coli containing intact construct with UGAUGA terminator upstream of a His-tag. No protein product is detected. Lane 4: E. coli containing UGAUGA to CUCGAG mutant construct. A 20-kDa protein is detected by using anti–His-tag antibody. This is in accord with a translation read-through over the mutated UGAUGA-to-CUCGAG region producing a His-tagged fusion protein.
Transcripts of the intact pBS-trimer and AUG-to-AAU mutant clone were inoculated into rice plants along with RYMV infectious transcripts. At 2–3 wk postinoculation, RYMV was isolated from the rice leaves and the RNA profile of the virus particles was examined. Results indicated that mutation of AUG to AAU resulted in the abolition of replication of scRYMV as indicated by the total absence of scRYMV RNA (Fig. 5B, lane 2) in comparison with WT scRYMV (Fig. 5B, lane 1). This demonstrated that the AUG sequence plays a crucial role in the scRYMV replication and life cycle.
When the tandem termination codons UGAUGA were mutated to CUCGAG at the end of the 19-kDa ORF in the pETdimer, the 19 kDa is now extended by a His-tag sequence to produce a 20-kDa protein in E. coli. Results in Fig. 5C show that Western blotting analysis using His-tag antibody can only detect a product when the termination codons UGAUGA upstream of the His-tag are mutated (Fig. 5C, lanes 3 and 4). This clearly indicates that the UGAUGA was used for termination of translation.
Discussion
Identification of scRYMV Protein.
In this report, we have presented several lines of evidence for the presence of the 16-kDa protein from scRYMV. In silico translation revealed the presence of the 16-kDa protein not only in our isolate, but also in all 37 other known natural RYMV isolates (from Africa) (12). Expression in E. coli, supported by MS, confirmed the translatability of such a protein. In vitro translation and immunoprecipitation of purified CCC satellite RNA and RNAs extracted from purified virus particles also confirmed the synthesis of the 16-kDa protein (Fig. 2). Western analysis using specific antiserum against 16-kDa protein confirmed the presence of the protein only in RYMV-infected plants and in purified RYMV particles (Fig. 3A). North-Western experiments showed the presence of the positively charged protein not only in RYMV-infected rice plants but also in E. coli expressing the trimer clone pET52 (Fig. 3C). Tandem LC-MS/MS analysis of the 16-kDa polypeptides from RYMV-infected rice plants also confirmed the novel translation of the scRYMV RNA (Fig. 4). scRYMV is the smallest and the only known natural viroid-like CCC RNA to code for a translatable ORF (with an overlap comprising the entire sequence). Partial overlap was reported for viruses and eukaryotic genes (13–15), whereas small linear satellite RNA of cucumber mosaic virus has been shown to have ORFs translatable in vitro, but the ORFs are very short (16). The hepatitis delta virus is the only other known circular RNA with a coding capacity. However, in this case, translation is carried out from a linear subgenomic RNA (17).
Novel Characteristics of Circular RNA Translation.
We presented evidence for translation from CCC RNA using the initiation–termination sequence UGAUGA to initiate translation. The genome is read in two (or three) totally overlapped reading frame registers (off by a single nucleotide in each round), leading to new amino acid sequences. Then, the same UGAUGA sequence turns into two consecutive opal termination UGA codons (see Fig. 6). This arrangement results in the initiation and termination codons that are not only adjacent but also share the same sequence. This appears novel and unusual compared with millions of linear mRNAs in which the AUG is at the 5′ end and the termination at the 3′ end of mRNA. The N- and C-terminal halves of the 16-kDa protein are coded for by the same 220-nt sequence but read in two distinct frames. Another interesting feature of this RNA molecule is that it contains no noncoding sequences before the AUG or after the termination codon(s). As proposed in the model in Fig. 6, it is predicted that the termination codon UGA (opal) may be read through to give rise to the 18 kDa, which then terminates at the downstream amber termination codon UAG. Data described in Table S2 indicated that the tandem UGAs and UAG codons can be occasionally skipped to produce longer read-through peptides covering the entire third frame register of the CCC RNA. Furthermore, peptides connecting the third translational frame to the first one are seen in Table S2. Larger polypeptides produced from a continuous five-round translation of CCC RNA and termination at the same tandem UGAUGA will have a molecular weight of 39 kDa. Western blots from infected rice plants (Fig. 3A) show broad bands at 39–41 kDa, in concordance with the MS results.
It is interesting to determine the mechanisms of translation of this RNA, not only because it is circular, but also because of its high degree of secondary structure (64% CG content) (9), which probably hinders translation (18). The in vitro wheat germ translation of circular RNA seems to be more efficient when translated with the helper RYMV RNA (Fig. 2, lane 4) than the circular RNA alone (Fig. 2, lane 3). We postulate that a translation product of RYMV may destabilize the secondary structure of circular RNA, thereby enabling more efficient translation. However, the in vivo situation may be quite different, possibly because of many host proteins interacting with the scRYMV RNA that could help destabilization of the circular RNA secondary structure, facilitating not only translation, but also self-cleavage by the hammerhead ribozymes required for the replication of this RNA.
Eukaryotic ribosomes generally initiate translation through the Kozak cap-dependent scanning model (19). However, many viral RNAs (e.g., Picornaviridae) contain internal ribosome entry sites (IRESs). In addition, some multicistronic viral RNAs are also capable of cap-independent translation through internal initiation (20). Interestingly, a survey by Kozak (21) indicated that most of the IRES sequences seem to harbor cryptic splice sites, resulting in the formation of a monocistronic 3′ gene from a dicistronic mRNA. An IRBS is a sine qua non condition for translation of scRYMV CCC RNA. The structure of this IRBS must be quite different from that described for IRES. The entire 220-nt sequence of scRYMV RNA is smaller than the IRES. Translation from circular RNA defies the principles of the scanning model of translation. Indeed, the early proof that eukaryotic ribosomes lack the capacity to initiate translation internally was demonstrated by the circularization of mRNA (22). The scRYMV is an excellent model to study internal initiation. It is a natural CCC RNA that does not seem to have any cryptic splicing sites. However, if we assume that there is a cryptic site (other than the ribozyme) to linearize the circular 220-nt molecule before translation, a maximum of 73-aa polypeptide may be theoretically obtained provided that the AUG is at the 5′ end. In such a case, there will not be a termination codon except that of the amber UAG codon located 58 nt downstream of the AUG or at the end of the 220-nt long linear RNA (Fig. 6). No such small peptides were detectable from in vivo or in vitro translation. Consequently, linearization of the circular RNA will abolish its translation because the 16-kDa protein requires the reading of the same sequence two or even three times.
Possible Biological Function of scRYMV and Its Protein.
The 16-kDa protein is unique, does not have any sequence homology to any known protein in the databases, and is highly basic [theoretical iso-electric point (pI) of 10.51]. Because this protein is found not only in infected plants but also in purified RYMV, it is hypothesized that it would likely interact with its own CCC RNA and that of the helper RYMV RNA, and consequently controls its own translation. North-Western experiments (Fig. 3C) showed that the positively charged 16-kDa protein binds to its own as well as to foreign RNAs. The exact function(s) of the 16-kDa protein for the biology of the scRYMV or helper RYMV is/are mostly speculative. The 16-kDa protein level reached a peak at 10 d postinoculation (Fig. 3B) early on in the RYMV infection process, and may play a role in CCC RNA replication, although there is no sequence homology to any movement proteins, RNA-dependent RNA polymerases (GDD motif), or helicases (GXXXGSK motif) encoded by plant viruses. As the positively charged 16-kDa protein is packaged inside the RYMV capsid, an RNA neutralization function (like histones) is likely not only for the CCC RNA but also to that of RYMV helper virus.
The essential characteristic of scRYMV is genetic economy, which enables a reduction in genome size without compromising the ability to translate. From our previous studies, multiple functions attributed to the CCC RNA are reported. Besides the generation of proteins via the two or three overlapping frames (described here), the self-splicing processing of both positive (+) and negative (−) sense RNAs by hammerhead ribozyme sequences are also described (9). Other predictable functions are expected, including a ribosome entry site for translation, a packaging signal allowing the circular/linear scRYMV RNA to be packaged into RYMV capsid protein, and recognition signals for the RNA-dependent RNA polymerase (supplied by the RYMV helper) for replication of both (+) and (−) sense CCC RNAs replication. A recent finding by Stergachis et al. described codons with dual function (i.e., duons) that can bind to transcriptional factors involved in gene expression besides executing their classical genetic code function (23). In this report, we describe that each nucleotide of CCC RNA is involved not only in three different codon functions but also in all of the aforementioned functions. Condensing many functions in a small nucleotide sequence reveals a fascinating evolutionary result to accomplish numerous required functions with a minimal size genome. The miniaturization may offer advantages in terms of copy number, movement, transport, and even protection from nucleases and other harmful enzymes. However, the drawback is certainly in the limitation for mutation and the resulting consequences.
The 37 natural isolates of scRYMV are mainly classified into two groups, group A (11 isolates), with a 100% identity to our isolate, and group B (25 isolates), which differs from group A by six mutations and B′ (one isolate, seven mutations) (12). Mutations are mainly compensatory substitutions (G to C and A to U) clustered in one area of the scRYMV molecule and totally preserved the secondary structure as well as the ORF. The conservation of secondary structure is critical for the replication of scRYMV by the helper virus RNA-dependent RNA polymerase. These results are similar to those described earlier by our group for the 322-nt CCC RNA from lucerne transient streak virus (24). Preservation of the ORF in all natural isolates indicates the essential role of 16-kDa protein for the CCC RNA. There is no single mutation near the UGAUGA (start/termination sequence) in any of the natural isolates. This sequence is also critical for the functioning of the ribozyme (Fig. 6). Mutation of AUG into AAU (in UGAUGA) resulted in the total loss of replication of CCC RNA in rice plants. However, in vitro evidence using the same mutation resulted in total loss of translation of the 16-kDa protein.
Is the scRYMV a living fossil of primitive entities with RNA genome and self-splicing ribozymes, which were replaced by the present organisms with genomic DNA containing intervening, noncoding, and intron sequences, or is it a new attempt at a novel trend in genomic evolution and translation?
Materials and Methods
Isolation of RYMV and Cloning into pET29(c).
RYMV virus particles and total viral nucleic acids were purified and examined for integrity as described previously (9, 25). A full-size cDNA copy of the scRYMV RNA was cloned in pBS (+) (9).
Overexpression of the scRYMV-Generated Polypeptide in E. coli and Generation of Antisera.
Head-to-tail trimer copy of scRYMV (pET52) is expressed in E. coli, and the 19-kDa protein is used to raise antibodies (SI Materials and Methods).
In Vitro Translation and Immunoprecipitation Reactions.
Purified scRYMV was translated in vitro, followed by immunoprecipitation (26). SI Materials and Methods includes further details.
Northern Blot Analysis for Detection of scRYMV Concatemers.
Linear, circular, and concatemeric forms of scRYMV RNA were detected by Northern blot analysis (SI Materials and Methods).
Protein Extractions and Western and North-Western Analyses.
scRYMV proteins expressed in E. coli and in rice plants and prepared from virus particles were analyzed by North-Western analysis as described previously (27) (SI Materials and Methods).
Mutation of Initiation AUG to AAU and Tandem Termination Codons (UGAUGA to CUCGAG).
Initiation and termination codons were mutated and their effect was determined by in vitro translation or by infectivity as well as by the expression of a His-tag constructs (SI Materials and Methods).
LC-MS/MS Analysis of scRYMV-Specific Proteins from pET52-Expressing E. coli and RYMV-Infected Rice.
scRYMV proteins were purified, digested with trypsin, and treated for LC-MS/MS analysis, followed by database search and validation by Scaffold 3.6 (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Jacquelyn Jhingree for her excellent work related to LC-MS/MS at the Center for Analysis of Genome Evolution and Function facility and Dr. Kathleen Hefferon for reading the final version of this manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402814111/-/DCSupplemental.
References
- 1.Diener TO. Viroids. Adv Virus Res. 1983;28:241–283. doi: 10.1016/s0065-3527(08)60725-3. [DOI] [PubMed] [Google Scholar]
- 2.Symons RH, Randles JW. Encapsidated circular viroid-like satellite RNAs (virusoids) of plants. Curr Top Microbiol Immunol. 1999;239:81–105. doi: 10.1007/978-3-662-09796-0_5. [DOI] [PubMed] [Google Scholar]
- 3.Taliansky ME, Palukaitis PF. Satellite RNAs and satellite viruses. In: Granoff A, Webster RG, editors. Encyclopedia of Virology. 2nd Ed. Vol 3. Academic; San Diego, CA: 1999. pp. 1607–1615. [Google Scholar]
- 4.Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNA’s. Science. 1984;223(4635):450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 5.Bruening G, Passmore BK, van Tol H, Buzayan JM, Feldstein PA. Replication of a plant virus satellite RNA: Evidence favors transcription of circular templates of both polarities. Mol Plant Microbe Interact. 1991;4(3):219–225. doi: 10.1094/mpmi-4-219. [DOI] [PubMed] [Google Scholar]
- 6.Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 7.Symons RH. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 8.Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. doi: 10.1146/annurev.phyto.43.040204.140243. [DOI] [PubMed] [Google Scholar]
- 9.Collins RF, Gellatly DL, Sehgal OP, Abouhaidar MG. Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology. 1998;241(2):269–275. doi: 10.1006/viro.1997.8962. [DOI] [PubMed] [Google Scholar]
- 10.AbouHaidar MG, Paliwal YC. Comparison of the nucleotide sequences of the viroid- like satellite RNA of the Canadian and Australasian strains of lucerne transient streak virus. J Gen Virol. 1988;69:2369–2373. [Google Scholar]
- 11.Sehgal OP, AbouHaidar MG, Gellatly DL, Ivanov I, Thottapilly G. An associated small RNA in rice yellow mottle sobemovirus homologous to the satellite RNA of lucerne transient streak sobemovirus. Phytopathology. 1993;83:1309–1311. [Google Scholar]
- 12.Pinel A, Abubakar Z, Traoré O, Konaté G, Fargette D. Molecular epidemiology of the RNA satellite of Rice yellow mottle virus in Africa. Arch Virol. 2003;148(9):1721–1733. doi: 10.1007/s00705-003-0138-1. [DOI] [PubMed] [Google Scholar]
- 13.Barrell BG, Air GM, Hutchison CA., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs A, Keese PK. In: Molecular Basis of Virus Evolution. Gibbs A, Calisher CH, García-Arenal F, editors. Cambridge Univ Press; Cambridge, UK: 1995. [Google Scholar]
- 15.Makalowska I, Lin CF, Makalowski W. Overlapping genes in vertebrate genomes. Comput Biol Chem. 2005;29(1):1–12. doi: 10.1016/j.compbiolchem.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Hidaka S, Hanada K, Ishikawa K. In vitro messenger properties of a satellite RNA of cucumber mosaic virus. J Gen Virol. 1990;71(pt 2):439–442. doi: 10.1099/0022-1317-71-2-439. [DOI] [PubMed] [Google Scholar]
- 17.Taylor JM. Virology of hepatitis D virus. Semin Liver Dis. 2012;32(3):195–200. doi: 10.1055/s-0032-1323623. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. New ways of initiating translation in eukaryotes? Mol Cell Biol. 2001;21(6):1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 20.Hefferon KL, Khalilian H, Xu H, AbouHaidar MG. Expression of the coat protein of potato virus X from a dicistronic mRNA in transgenic potato plants. J Gen Virol. 1997;78(pt 11):3051–3059. doi: 10.1099/0022-1317-78-11-3051. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33(20):6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280(5717):82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 23.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342(6164):1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gellatly D, Mirhadi K, Venkataraman S, AbouHaidar MG. Structural and sequence integrity are essential for the replication of the viroid-like satellite RNA of lucerne transient streak virus. J Gen Virol. 2011;92(Pt 6):1475–1481. doi: 10.1099/vir.0.029801-0. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 26.Philipson L, et al. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- 27.Tsai M-S, Hsu Y-H, Lin N-S. Bamboo mosaic potexvirus satellite RNA (satBaMV RNA)-encoded P20 protein preferentially binds to satBaMV RNA. J Virol. 1999;73(4):3032–3039. doi: 10.1128/jvi.73.4.3032-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.