Significance
RNAi-mediated antiviral immunity directs specific virus resistance by virus-derived siRNAs in contrast to broad-spectrum resistance triggered in innate immunity by host pattern recognition receptors. Here we show that induction of antiviral RNAi in Arabidopsis is associated with production of a genetically distinct class of virus-activated siRNAs (vasiRNAs) by RNA-dependent RNA polymerase-1 to target hundreds of host genes for RNA silencing by Argonaute-2. Production of vasiRNAs is induced by viruses from two different supergroups of RNA virus families, targeted for inhibition by Cucumber mosaic virus, and correlated with virus resistance independently of viral siRNAs. We propose that antiviral RNAi activates broad-spectrum antiviral activity via widespread silencing of host genes directed by vasiRNAs in addition to specific antiviral defense by viral siRNAs.
Keywords: RNA silencing, microbiology, plant biology, viral suppressor of RNAi
Abstract
Antiviral immunity controlled by RNA interference (RNAi) in plants and animals is thought to specifically target only viral RNAs by the virus-derived small interfering RNAs (siRNAs). Here we show that activation of antiviral RNAi in Arabidopsis plants is accompanied by the production of an abundant class of endogenous siRNAs mapped to the exon regions of more than 1,000 host genes and rRNA. These virus-activated siRNAs (vasiRNAs) are predominantly 21 nucleotides long with an approximately equal ratio of sense and antisense strands. Genetically, vasiRNAs are distinct from the known plant endogenous siRNAs characterized to date and instead resemble viral siRNAs by requiring Dicer-like 4 and RNA-dependent RNA polymerase 1 (RDR1) for biogenesis. However, loss of EXORIBONUCLEASE4/THYLENE-INSENSITIVE5 enhances vasiRNA biogenesis and virus resistance without altering the biogenesis of viral siRNAs. We show that vasiRNAs are active in directing widespread silencing of the target host genes and that Argonaute-2 binds to and is essential for the silencing activity of vasiRNAs. Production of vasiRNAs is readily detectable in Arabidopsis after infection by viruses from two distinct supergroups of plant RNA virus families and is targeted for inhibition by the silencing suppressor protein 2b of Cucumber mosaic virus. These findings reveal RDR1 production of Arabidopsis endogenous siRNAs and identify production of vasiRNAs to direct widespread silencing of host genes as a conserved response of plants to infection by diverse viruses. A possible function for vasiRNAs to confer broad-spectrum antiviral activity distinct to the virus-specific antiviral RNAi by viral siRNAs is discussed.
RNA silencing, also referred as RNA interference (RNAi), regulates gene expression in eukaryotes by Argonaute protein complexes loaded with small interfering RNAs (siRNAs) or microRNAs (1, 2). RNA silencing is highly specific because the targeted genes are selected by the base pairing between the small RNA in an Argonaute complex and its target RNA. Studies from the last two decades have shown that RNA silencing acts as a major natural antiviral defense mechanism in plants and invertebrates (1, 3, 4). Recent reports have further provided evidence for a similar antiviral function of RNAi in mammals (5, 6). A unifying feature of antiviral silencing in all of these host organisms is the production of virus-derived siRNAs processed from virus-specific double-stranded RNA (dsRNA) by a Dicer endoribonuclease (1, 3–6). As a result, antiviral silencing is considered to be highly specific and target only the virus that initially triggers the immune response (1, 3, 4). Silencing of specific host genes occurs when there is near-perfect complementarity between a viral siRNA and the cellular mRNA (7, 8).
Genetic studies in the model plant Arabidopsis thaliana have identified both hierarchical and overlapping pathways for the production and amplification of viral siRNAs (3, 4). Viral siRNAs targeting RNA viruses are primarily made by Dicer-like 4 (DCL4) and are 21 nucleotides (nt) in length. The 22-nt viral siRNAs made by DCL2 are much less abundant, but are sufficient to confer protective immunity in the absence of 21-nt viral siRNAs processed by DCL4 (9−12). The majority of viral siRNAs produced during antiviral silencing are “secondary” viral siRNAs amplified by RNA-dependent RNA polymerase 1 (RDR1) or RDR6 of Arabidopsis (13, 14). Among the 10 Argonaute proteins in Arabidopsis, Argonaute 1 (AGO1) and AGO2 have been found to load two different sets of viral siRNAs and mediate antiviral silencing in a cooperative manner (15–18).
RNAi-mediated antiviral immunity shares key features with the animal and plant innate immunity initiated following recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) (19). For example, both types of immunity mechanisms become active immediately after infection and antiviral silencing also depends on the recognition of viral dsRNA as a broadly conserved PAMP by the host Dicer complex. However, the PRR-activated innate immunity mechanisms, including PAMP-triggered and effector-triggered immunities (PTI and ETI) in plants, involve activation of transcriptional signaling cascades and confer broad-spectrum pathogen resistance (20–25). Moreover, degradation of single-stranded RNA by RNaseL and repression of protein translation by protein kinase R target both viral and host genes and are an integral part of the vertebrate innate antiviral immunity regulated by type 1 interferons (26). By contrast, recognition of viral dsRNA by Dicer in antiviral silencing produces viral siRNAs to direct virus-specific resistance. The impact of active antiviral silencing on either the global host gene expression or the broad-spectrum local and systemic acquired virus resistance is unknown.
In this study, we show that activation of antiviral silencing in Arabidopsis is accompanied by RDR1-dependent production of abundant siRNAs encoded by the host genome to target hundreds of host genes for silencing. Genetic analyses reveal that these virus-activated Arabidopsis siRNAs resemble viral siRNAs in the biogenesis but distinct from all of the Arabidopsis siRNAs characterized to date (27–33). Our findings reveal that RDR1-dependent production of the endogenous siRNAs is a conserved response of Arabidopsis to infection by either Cucumber mosaic virus (CMV) or Turnip mosaic virus (TuMV) and is targeted for inhibition by the viral suppressor of RNAi (VSR) encoded by CMV, but not by the VSR of TuMV. We propose that production of the endogenous siRNAs confers a broad-spectrum antiviral activity complementary to the virus-specific resistance directed by viral siRNAs.
Results
Detection of a Distinct Class of Arabidopsis siRNAs Induced by Virus Infection.
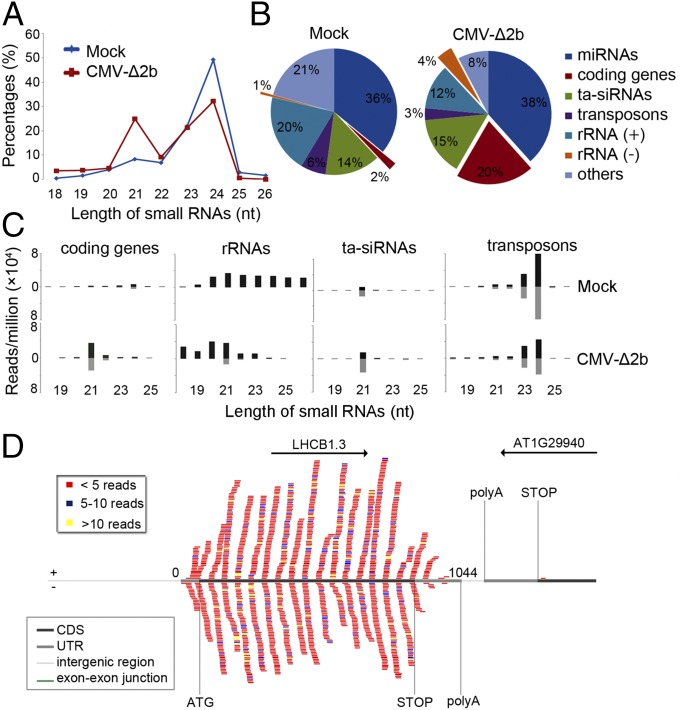
Viral siRNAs are amplified by both RDR1 and RDR6 in Arabidopsis during CMV infection when its VSR protein 2b is rendered nonexpressing (13). Therefore, we searched for novel Arabidopsis small RNAs activated by virus infection in wild type (WT) plants challenged by the VSR-deficient mutant of CMV, CMV-∆2b (13). Comparative analysis of the populations of small RNAs with 100% sequence identity to the Arabidopsis genome revealed that CMV-∆2b infection induced a markedly enhanced population of 21-nt RNAs in contrast to the remaining size classes of endogenous small RNAs (Fig. 1A). To identify the genomic origins of these virus-activated small RNAs, the total sequenced Arabidopsis genome-specific 21-nt RNAs were divided into microRNAs (miRNAs), trans-acting (ta)siRNAs, and small RNAs mapped to protein-coding genes, transposons, sense and antisense strands of rRNAs, and other loci. As shown in Fig. 1B, CMV-∆2b infection caused greatly increased accumulation of 21-nt RNAs mapped to the protein-coding genes and the antisense strand of rRNAs by 10 and 4 folds, respectively. The total small RNAs mapped to these protein-coding genes exhibited an overwhelming size preference for 21-nt (71.2%) with a minor peak of 22-nt species (Fig. 1C). Notably, both 21-nt and 22-nt RNAs from these target genes were divided approximately equally into sense and antisense strands (Fig. 1C) and were mapped only to the mature mRNA of the target genes without spreading into introns or neighboring genes (Fig. 1D and Fig. S1A). All of the three rRNAs (5.8S, 18S, and 25S) were targeted in the infected plants by antisense small RNAs (Fig. S2A), which also were predominantly 21 nucleotides long in contrast to the random size distribution of sense small RNAs of rRNAs (Fig. 1C). These properties of the dominant 21-nt RNAs derived from the protein-coding genes and antisense strand rRNAs suggest that they are novel Arabidopsis siRNAs, designated as virus-activated siRNAs (vasiRNAs). In total, 21-nt vasiRNAs corresponding to the protein-coding genes and antisense strand rRNAs represented 20% and 4%, respectively, of the total endogenous 21-nt RNAs sequenced from the virus-infected plants (Fig. 1B).
Fig. 1.
Properties of virus-activated Arabidopsis siRNAs. Relative abundance of unique Arabidopsis small RNAs according to their lengths (A) and of the total 21-nt RNAs from different sequence groups (B) in plants without (mock) or with infection by CMV-∆2b. (C) Length distribution (in nucleotides) and abundance (reads per million of total reads) of the total Arabidopsis small RNAs derived from protein-coding genes, rRNAs, tasiRNAs, and transposons from WT plants after mock or CMV-∆2b infection. (D) Distribution pattern of sense (top) and antisense (bottom) vasiRNAs specific to one representative RDR1 target gene, photosystem II light harvesting complex gene 1.3 (LHCB1.3). Various regions of the target gene and the neighboring gene(s) are indicated by colored lines. CDS, the protein-coding region; UTR, untranslated region.
Unique Genetic Requirements for the Biogenesis of vasiRNAs.
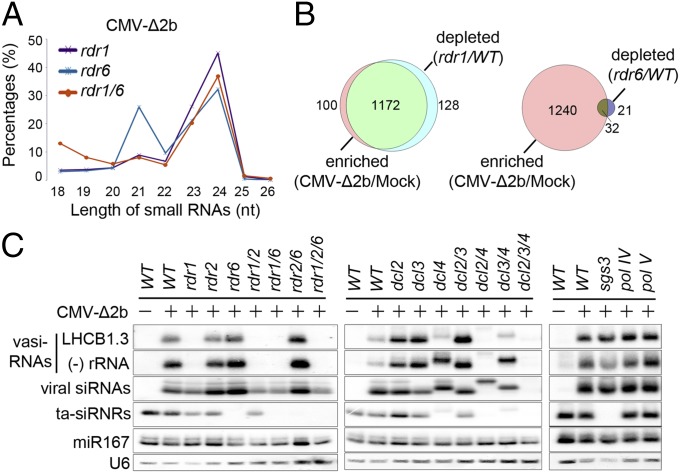
Arabidopsis siRNAs characterized to date are dependent on either RDR2 or RDR6 although it is curious that RDR1 as the first characterized RDR has no known endogenous siRNAs so far (27). To investigate the genetic pathway for the biogenesis of vasiRNAs, we examined total small RNAs sequenced from CMV-∆2b-infected Arabidopsis mutants carrying a loss-of-function allele for RDR1, RDR6, or both. As expected (34, 35), the RDR6-dependent 21-nt tasiRNAs became undetectable in both rdr6 and rdr1 rdr6 mutants whereas the RDR2-dependent 24-nt heterochromatic (het)siRNAs accumulated to high levels in all of the three mutants (Fig. 2A and Fig. S1 C and D). The population of 21-nt vasiRNAs remained highly abundant in rdr6, but disappeared in both rdr1 and rdr1 rdr6 mutants (Fig. 2A and Fig. S1 C and D), indicating that vasiRNA production requires RDR1. Similar to WT plants, the small RNAs mapped to the protein-coding genes and antisense rRNAs in rdr6 plants infected with CMV-∆2b were predominantly 21-nt with a small population of 22-nt RNAs (Fig. S1 C and D). In contrast to WT and rdr6 plants, however, both rdr1 and rdr1 rdr6 mutants failed to produce these vasiRNAs after CMV-∆2b infection (Fig. S1 C and D).
Fig. 2.
Genetic requirements for vasiRNA biogenesis. (A) Relative abundance of unique Arabidopsis small RNAs according to their lengths induced by CMV-∆2b in mutant plants defective for RDR1, RDR6, or both. (B) Venn diagram depicting the proportion of loci that posses a twofold or greater enrichment of vasiRNAs in WT plants after CMV-∆2b infection and are also depleted twofold or more in rdr1 plants (Left) or in rdr6 plants (Right) compared with WT plants after CMV-∆2b infection. (C) Northern blot detection of vasiRNAs derived from LHCB1.3 and the antisense strand of 25S rRNA in WT plants and 17 different mutants after CMV-∆2b infection (+) as well as in WT plants after mock infection (−). The same set of RNA samples were also probed for CMV-∆2b siRNAs, tasiRNA ASRP255, microRNA 167 (miR167), and U6 RNA.
We focused on the 1,708 protein-coding genes with at least 10 reads per million of the total sequenced Arabidopsis small RNAs in small RNA libraries from mock and CMV-∆2b infected WT plants. We found that 1,272 from the 1,708 genes were targeted by vasiRNAs with a twofold or greater increase in read count in WT plants after CMV-∆2b infection compared with mock infection (Fig. 2B and Fig. S1B). Notably, vasiRNAs targeting 1,172 of these 1,272 genes exhibited a twofold or greater decrease in read count in rdr1 plants compared with WT plants after CMV-∆2b infection. In contrast, vasiRNAs targeting only 32 of these 1,272 genes were depleted twofold or greater in rdr6 plants. These 1,172 were thus defined as the genes targeted specifically by RDR1-dependent vasiRNAs in subsequent analyses (Dataset S1).
We found that vasiRNAs specific to the RDR1 target genes and 25S rRNA were abundant and readily detectable by Northern blot hybridization in WT plants after CMV-∆2b infection, but not after mock inoculation (Fig. 2C). Gel blot hybridization also verified the accumulation of vasiRNAs in rdr2, rdr6, and rdr2 rdr6 mutants, but not in any of the single, double, or triple mutants that contained the rdr1 allele after CMV-∆2b infection (Fig. 2C). The accumulation patterns of ta-siRNAs and hetsiRNAs in these mutants (Fig. 2C) were as expected (34, 35). These results illustrate that the biogenesis of vasiRNAs requires RDR1 but neither RDR2 nor RDR6, thus identifying vasiRNAs as the first endogenous siRNAs of RDR1. Because many vasiRNAs map to exon–exon junctions (Fig. 1D and Fig. S1A), it is likely that mature mRNAs are used as templates for their synthesis by RDR1.
To further characterize the biogenesis pathway of vasiRNAs, we infected an expanded panel of Arabidopsis mutants with CMV-∆2b (Fig. 2C). Our analysis of the set of dcl mutants showed that the dominant 21-nt vasiRNAs were produced by DCL4 as predicted by the dominant size of vasiRNAs, because they were undetectable in the dcl4-containing single, double, or triple mutant plants (Fig. 2C). We noted that the disappearance of the 21-nt vasiRNAs in dcl4 and dcl3/4 mutant plants was accompanied with the accumulation of a major 22-nt species and that neither 21-nt nor 22-nt vasiRNAs were detectable in the dcl2/3/4 triple mutant plants (Fig. 2C). No obvious defect was detected for the biogenesis of vasiRNAs in mutant plants carrying a loss of function allele in SGS3, Pol IV, or Pol V (Fig. 2C), known to be essential for the biogenesis of various types of siRNAs of Arabidopsis (27). Together, our findings suggest that vasiRNAs are genetically distinct from the known plant endogenous siRNAs characterized to date, but instead resemble viral siRNAs by requiring both DCL4 and RDR1 for biogenesis (3, 27).
Detection of vasiRNAs in AGO1 and AGO2 Complexes.
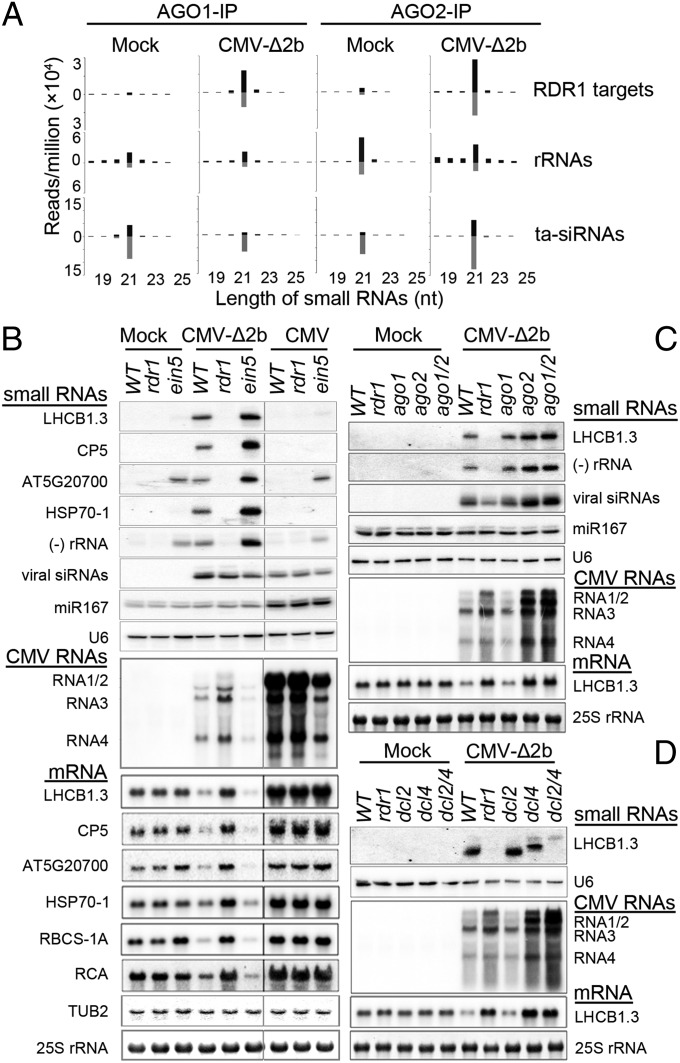
Viral siRNAs have been previously shown to load into AGO1 and AGO2 among the 10 AGOs of Arabidopsis (15–17). We found that the small RNAs mapped to the RDR1 target genes and rRNAs were detectable and predominantly 21 nucleotides in length with approximately equal strand ratios in both AGO1 and AGO2 complexes coimmunoprecipitated from healthy WT Arabidopsis plants (Fig. 3A). By comparison, vasiRNAs targeting rRNAs were more abundant than those derived from the RDR1 target genes and both types of vasiRNAs were more abundant in AGO2 complex than in AGO1 complex (Fig. 3A). CMV-∆2b infection triggered strong enrichment of the vasiRNAs specific for the RDR1 target genes in both AGO1 and AGO2 complexes, but such enrichment was not observed for rRNA-specific vasiRNAs (Fig. 3A) even through these vasiRNAs were strongly induced (Fig. 2C). Specifically, vasiRNAs were enriched twofold or greater for 86.7% and 79.9% of the 1,172 RDR1 target genes in AGO1 and AGO2 complexes, respectively, or for 71.9% of the RDR1 target genes in both AGO1 and AGO2 complexes (Fig. S3A). In total, 21-nt vasiRNAs from the infected Arabidopsis represented 5.7% and 10.9% of the total endogenous 21-nt small RNAs found in AGO1 and AGO2 complexes, respectively. Similar to viral siRNAs and Arabidopsis siRNAs and miRNAs (15–17), vasiRNAs loaded into AGO1 and AGO2 complexes exhibited strong bias for U and A at the 5′-termini, respectively (Fig. S3B).
Fig. 3.
VasiRNAs are biologically active. (A) Length distribution and abundance (reads per million of total reads) of the small RNAs derived from the 1,172 RDR1 target genes, rRNAs, and tasiRNAs found in AGO1 and AGO2 complexes coimmunoprecipitated (IP) from WT plants after mock or CMV-∆2b infection. (B−D) Northern blot analyses of the accumulation of the large and small RNAs in WT and mutant plants after inoculation with buffer (mock), CMV, and/or CMV-∆2b. LHCB1.3 was examined as the RDR1 target gene in C and D whereas five additional target genes (membrane related protein CP5; AT5G20700; HSP70-1, heat shock protein 70–1; RBCS-1A, Rubisco small subunit 1A; RCA, Rubisco activase) were analyzed in B. Methylene blue staining of 25S rRNA and Northern blot detection of Tubulin beta 2 (TUB2) mRNA, viral siRNAs, miR167, and U6 RNA were shown as controls. All of the mutant alleles (dcl2-1, dcl4-2, and ago2-1) used were null alleles except for ago1-27, which is hypomorphic because AGO1 is indispensable for development.
Widespread Silencing of Host Genes Directed by vasiRNAs.
Detection of vasiRNAs in the in vivo Argonaute complexes suggests that they are biologically active during infection. To investigate whether vasiRNAs direct silencing of the corresponding host genes, we compared the mRNA levels of six RDR1 target genes in WT and rdr1 plants after infection with CMV-∆2b or CMV using mock-inoculated WT plants as controls (Fig. 3B). Northern blotting analyses detected consistently reduced accumulation of the transcripts from all of the six RDR1 target genes in WT plants after CMV-∆2b infection compared with mock infection (Fig. 3B, Bottom). In contrast, reduced expression of these RDR1 target genes was not detected either in rdr1 plants after CMV-∆2b infection or in WT plants after CMV infection, in both of which production of vasiRNAs was undetectable (Fig. 3B). These results suggest that infection triggers widespread silencing of host genes by the RDR1-dependent vasiRNAs. However, the accumulation of rRNAs in the infected plants was not altered in an RDR1-dependent manner (Fig. S4), suggesting that rRNA-specific vasiRNAs may not direct RNA silencing during infection.
Arabidopsis EXORIBONUCLEASE4 (XRN4)/ETHYLENE-INSENSITIVE5 (EIN5) encodes a cytoplasmic exoribonuclease that degrades RNA intermediates from mRNA decay and Argonaute slicing, which in turn inhibits RDR6-dependent production of siRNAs targeting transgene transcripts (36–38). The ein5 mutant also accumulates 21-nt small RNAs from an unknown biogenesis pathway to target ∼130 endogenous mRNAs in immature flower bud tissues (38, 39), 19 of which were identified here as RDR1 target genes in leaf tissues. Northern blotting analyses detected a greatly increased accumulation of vasiRNAs in CMV-∆2b-infected ein5 plants compared with either mock-inoculated ein5 plants or CMV-∆2b-infected WT plants (Fig. 3B). We found that CMV-∆2b infection also induced stronger silencing of the six RDR1 target genes in ein5 plants than in WT plants (Fig. 3B), consistent with the increased levels of vasiRNAs. Moreover, no obvious changes in the accumulation of vasiRNAs were observed in ein5 plants with or without infection by CMV that expresses the VSR 2b protein, and the silencing of the RDR1 target transcripts was not detected in CMV-infected ein5 plants (Fig. 3B). These results further support a role of vasiRNAs in the widespread silencing of host genes induced by virus infection.
CMV accumulated to lower levels in ein5 plants than in WT plants (Fig. 3B, Middle), similar to that described previously in a study based on the infection of a different xrn4/ein5 mutant by a different CMV isolate (40). We found that ein5 plants were also more resistant to CMV-∆2b than WT plants and that CMV-∆2b accumulated to the highest level in rdr1 plants (Fig. 3B). In contrast to the dramatic effects of ein5 and rdr1 alleles on the accumulation of vasiRNAs, the accumulation of the viral siRNAs exhibited no obvious differences among WT, ein5, and rdr1 plants infected by either CMV-∆2b or CMV (Fig. 3B, Top). CMV-∆2b replicated to lower levels in rdr1 plants than CMV did in WT plants because the 2b protein suppresses the amplification of viral siRNAs to target CMV-∆2b by both RDR1 and RDR6 (11, 13). Therefore, the increased virus resistance in ein5 plants was correlated with an increased accumulation of vasiRNAs whereas loss of vasiRNA production in rdr1 plants was associated with the highest accumulation of CMV-∆2b. These results strongly suggest that the RDR1-dependent widespread silencing of host genes directed by vasiRNAs plays a role in the virus resistance activated during antiviral silencing in a manner independent of the antiviral activity of viral siRNAs.
DCL4 and AGO2 Are Essential for RNA Silencing by vasiRNAs.
We next examined vasiRNA silencing in mutant plants defective in those DCL and AGO genes involved in the biogenesis and loading of vasiRNAs (Figs. 2 and 3A). CMV-∆2b replicated to higher levels in dcl4 and ago2 mutant plants than in dcl2, ago1, or WT plants (Fig. 3 C and D). A strong decrease in LHCB1.3 mRNA accumulation was detected in both dcl2 and ago1 plants as found in WT plants following the infection of CMV-∆2b (Fig. 3 C and D). By contrast, LHCB1.3 mRNA levels were nearly as high in dcl4 and ago2 mutant plants as they were in rdr1 plants after CMV-∆2b infection (Fig. 3 C and D). Moreover, LHCB1.3 mRNA did not accumulate to higher levels in dcl2/dcl4 and ago1/ago2 double mutant plants than in dcl4 and ago2 single mutant plants (Fig. 3 C and D). These results show that both DCL4 and AGO2 are essential for RNA silencing by vasiRNAs. These findings also indicate that the 22-nt vasiRNAs produced by DCL2 in absence of DCL4 are inactive in RNA silencing and that AGO1 does not independently mediate vasiRNA silencing in ago2 plants. We noted that production of vasiRNAs was efficiently induced by CMV-∆2b in both ago1 and ago2 plants (Fig. 3C). Thus, targeting the mature mRNA for efficient vasiRNA biogenesis by DCL4 and RDR1 in ago2 plants is insufficient to ensure vasiRNA silencing, suggesting that RNA silencing of the target genes requires the AGO2-mediated silencing activity of vasiRNAs.
Production of vasiRNAs in Arabidopsis Infected by Distinct Viruses.
We further investigated the production of vasiRNAs in response to infection by Q strain of CMV (Q-CMV) related distantly to Fny-CMV used above and TuMV, which is classified into a supergroup of plant RNA virus families that does not include CMV. Production of vasiRNAs was also induced by Q-CMV-∆2b in WT and ein5 plants, but not in rdr1 plants, and vasiRNAs were undetectable in all of the Arabidopsis lines infected by Q-CMV (Fig. S5) that expresses the 2b protein with 53.5% sequence identity to Fny-CMV 2b protein (41). Notably, infection by a recombinant green fluorescent protein-expressing isolate of TuMV (TuMV-GFP) strongly induced production of vasiRNAs targeting the gene LHCB1.3 and 25S rRNA in WT, rdr6, and ein5 plants, but not in rdr1 plants (Fig. 4A). Similar to the infections by VSR-deficient mutant of either CMV strain, ein5 plants produced the highest vasiRNA levels in response to TuMV-GFP, which expresses its VSR helper component proteinase (HC-Pro). These results suggest that the biogenesis of vasiRNAs is inhibited by the VSRs of CMV strains, but not by the VSR of TuMV. Our findings together identify the RDR1-dependent production of vasiRNAs as a conserved host response to infection by diverse RNA viruses.
Fig. 4.
Production and properties of vasiRNAs induced by TuMV-GFP. (A) Northern blot detection of vasiRNAs specific to LHCB1.3 and the antisense 25S rRNA in WT and mutant plants after inoculation with buffer (mock) or TuMV-GFP. Viral genomic RNA and siRNAs as well as miR167 and U6 RNA were also probed. (B) Relative abundance of unique Arabidopsis small RNAs according to their lengths in WT and mutant plants after TuMV-GFP infection. (C) Length distribution and abundance of the total Arabidopsis small RNAs derived from protein-coding genes, rRNAs, tasiRNAs, and transposons from WT and mutant plants after TuMV-GFP infection. (D) Venn diagram depicting the proportion of loci that posses a twofold or greater enrichment of vasiRNAs in WT plants after CMV-∆2b infection and are also enriched twofold or more after TuMV-GFP infection. (E) Distribution pattern of sense (top) and antisense (bottom) vasiRNAs specific to LHCB1.3.
We next sequenced the small RNA populations from WT, rdr1, and rdr6 plants infected with TuMV-GFP. TuMV-GFP infection in both WT and rdr6 plants, but not in rdr1 plants, induced a population of 21-nt RNAs (Fig. 4B) mapped predominantly to the protein-coding genes and rRNAs of the Arabidopsis genome (Fig. 4C and Fig. S2B). The total small RNAs specific to these protein-coding genes and rRNAs were predominantly 21-nt with a minor peak of 22-nt species, were divided approximately equally into sense and antisense strands (Fig. 4C), and targeted only the mature mRNA of the target genes without spreading into introns or neighboring genes (Fig. 4E and Fig. S6B). Thus, these RDR1-dependent 21-nt Arabidopsis small RNAs induced by TuMV-GFP infection, representing 17% of the total sequenced endogenous 21-nt RNAs, are genetically identical to the vasiRNAs characterized above in CMV-∆2b infections (Fig. S6A). A total of 1,068 protein-coding genes were defined as the RDR1 target genes in response to TuMV-GFP infection (Dataset S1) because vasiRNAs targeting these genes were enriched twofold or greater in WT plants after TuMV-GFP infection, but were depleted twofold or more in rdr1 plants compared with WT plants after TuMV-GFP infection. Notably, we found that a substantially overlapping set of host genes was targeted for silencing by the RDR1-dependent vasiRNAs as part of the Arabidopsis antiviral silencing response to either CMV or TuMV (Fig. 4D and Dataset S1). Gene ontology (GO) analysis revealed that genes responsive to biotic and abiotic stimuli were significantly enriched in the RDR1 target genes induced by CMV-∆2b, TuMV-GFP, or both (Fig. S7 A−C). These findings suggest that vasiRNAs act to modulate host responses to virus infection.
Discussion
In this study, we examined the population of the total host-specific small RNAs in Arabidopsis plants after induction of antiviral silencing by WT or mutant viruses that do not suppress the amplification of viral siRNAs. This led to the discovery of vasiRNAs as an abundant class of 21-nt Arabidopsis siRNAs that are mapped to the exon regions of more than 1,000 genes and are genetically distinct to all of the Arabidopsis siRNAs characterized to date, including hetsiRNAs, tasiRNAs, natural antisense siRNAs, epigenetically activated siRNAs, and DNA double-strand break-induced small RNAs (27–33). These vasiRNAs are produced by DCL4 and RDR1, and loss of XRN4/EIN5 enhances vasiRNA biogenesis. Northern blot analysis indicates that vasiRNAs are active in directing the widespread silencing of the target genes. Our results further reveal that AGO2 binds to vasiRNAs in vivo and is essential for vasiRNA silencing although it is dispensable for vasiRNA biogenesis. However, the vasiRNAs induced by virus infection to target rRNA do not appear to direct RNA silencing in the infected plants and are not specifically loaded in AGO1 or AGO2. Notably, vasiRNAs are readily detectable only after infection with TuMV-GFP or the VSR-deficient mutants of the two distantly related CMV strains. This requirement for vasiRNA biogenesis explains why the endogenous siRNAs produced by RDR1 remain elusive until now even through RDR1 was the first cellular RDR cloned in 1998 (42). The discovery of vasiRNAs expands the repertoire of siRNAs and suggests that the siRNA-processing activity of Dicer proteins may play a more important role in the regulation of plant and animal gene expression than what is currently known (5, 6, 27).
The hypersensitive response triggered by plant virus infection is associated with both local and systemic acquired resistance (LAR and SAR) effective against broad-spectrum viruses (43). Much is known about the mechanisms of PTI, ETI, and SAR that control the plant resistance to bacterial and fungal pathogens regulated by the hormone salicylic acid (21, 25, 44). However, the genetic pathway and the effector molecules that control LAR and SAR against viral pathogens are poorly characterized. RNAi-mediated antiviral immunity clearly acts as the major virus resistance mechanism in plants and exhibits features of PTI including Dicer detection of the viral dsRNA and induction of ETI to specifically recognize VSRs (19, 45, 46). However, the properties of antiviral silencing described to date are not consistent with the broad-spectrum antiviral activity as defined in LAR and SAR. Several observations made in this study led us to propose that induction of antiviral silencing confers broad-spectrum antiviral activity as a result of widespread silencing of host genes directed by vasiRNAs in addition to specific antiviral defense by viral siRNAs. First, the relative abundance of vasiRNAs in WT and mutant plants is positively correlated to the host resistance to infection by either CMV or CMV-∆2b under conditions in which viral siRNAs accumulated to similar levels. Second, rRNA and an overlapping set of host genes, particularly genes responsive to biotic and abiotic stimuli, are targeted by vasiRNAs in plants after immune challenge by viruses from distinct supergroups of RNA viruses. Third, the DCL4-RDR1-AGO2 genetic pathway for the biogenesis and activity of vasiRNAs is identical to the pathway controlling the production of one of the viral siRNA populations, which is analogous to the broad-spectrum targeting of viral and host genes by the same innate immunity components in vertebrates (26). Fourth, production of vasiRNAs is targeted for viral inhibition as found in plants infected by CMV strains. Although TuMV does not inhibit vasiRNA production, it remains possible that the silencing activity of vasiRNAs is susceptible to suppression by the potyviral VSR HC-Pro (47, 48).
Materials and Methods
Plant Materials, Viruses, and Infection Assays.
Use of WT Arabidopsis thaliana ecotype Columbia, dcl, rdr, or ago single, double, and/or triple mutants, sgs3-1, and transgenic lines expressing HA-tagged AGO2 in the WT background and FLAG-tagged AGO1 in the ago1-36 mutant background driven by their native promoters were described previously (11, 13, 15). Mutants ein5-6, polIV-3, and polV-11 were described by others (49–51). The plant growth room was set with 10 h in light and 14 h in dark at 24 °C. CMV-∆2b from both Fny and Q strains contained a deletion of the 2b coding sequence and triggered amplification of viral siRNAs by both RDR1 and RDR6 as described previously (11, 13). Unlike previous studies, the inocula for WT and mutant CMV isolates used in this study contained purified virions at 5 μg/mL The recombinant TuMV isolate used here, TuMV-GFP, expresses green fluorescent protein and its propagation and infection followed the method described (14).
Northern Blot Hybridizations.
Both high and low molecular weight RNAs were extracted from the upper uninoculated leaves of Arabidopsis seedlings 14 d after virus inoculation and analyzed by Northern blot hybridizations as described previously (15). High molecular weight RNA gel blots were probed with 32P-labeled DNAs corresponding to the conserved 3′ terminal sequence of RNA2 of Fny-CMV or Q-CMV, the coat protein-coding region of TuMV, to the protein-coding region of the six RDR1 target genes photosystem II light harvesting complex gene 1.3 (LHCB1.3), membrane related protein CP5 (CP5), AT5G20700, heat shock protein 70-1 (HSP70-1), Rubisco small subunit 1A (RBCS-1A), and Rubisco activase (RCA). Tubulin beta 2 (TUB2) mRNA and 25S rRNA were probed as controls. A mixture of seven DNA oligonucleotides (Table S1) corresponding to the (+)-strand of CMV RNA3 was used for detecting the negative-strand siRNAs specific to the Fny and Q strains (11, 13). A PCR fragment of the cylindrical inclusion protein-coding region was used to synthesize the 32P-labeled probe for TuMV siRNAs by random priming as describe previously (14). A mixture of DNA oligonucleotides was designed and synthesized according to the deep sequencing profiles to hybridize to the antisense hot spot vasiRNAs of 25S rRNA or to the sense and antisense hot spot vasiRNAs of the individual RDR1 target genes, LHCB1.3, CP5, AT5G20700, and HSP70-1. Transacting siRNA ASRP255, microRNA 167, and U6 RNA were detected by 32P-labeled DNA oligonucleotide probes. The blot signals were detected by phosphor imager and multiple film exposures.
Small RNA Sequencing and Bioinformatics Analysis.
We previously reported the analysis of viral siRNAs in duplicate total small RNA libraries sequenced from WT and rdr1 plants 14 d after infection with CMV-∆2b of the Fny strain (13). This study constructed and sequenced two independent small RNA libraries from the upper uninoculated leaves of (i) WT Arabidopsis plants 14 d after mock inoculation and of (ii) rdr6 and (iii) rdr1 rdr6 plants 14 d after infection with Fny CMV-∆2b, and one library each from the upper uninoculated leaves of (iv) WT, (v) rdr1, and (vi) rdr6 plants 14 d after infection with TuMV-GFP as well as of (vii) the FLAG-AGO1/ago1-36 plants and (viii) the HA-AGO2 plants 14 d after mock inoculation. Coimmunoprecipitation with FLAG- and HA-specific antibodies was used to obtain (ix) AGO1 and (x) AGO2 complexes, respectively, from the FLAG-AGO1/ago1-36 and HA-AGO2 plants 14 d after infection with Fny CMV-∆2b for extracting total loaded small RNAs for the construction and sequencing of small RNA libraries as described (52). Bioinformatic analysis was carried out as described previously (13, 15, 53). Briefly, small RNAs in the size range of 18–26 nt were used for further analysis after removing the adaptor sequences. Bowtie software was used for alignment analysis (54). Small RNAs with perfect matches to the Arabidopsis genome were used for further analysis and normalization. The small RNAs were annotated with reference to the following databases: TAIR database (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR9_genome_release/) for Arabidopsis genome, miRBase (http://microrna.sanger.ac.uk/sequences) for miRNA sequences, Rfam (www.sanger.ac.uk/Software/Rfam/) for noncoding RNA sequences (rRNAs, tRNAs, snoRNAs, and snRNAs), and Repbase (www.girinst.org) for transposons and repeats. WebLogo was used for analyzing of relative frequencies of nucleotides at each position of the small RNAs. Other analysis was performed by in-house PERL scripts. GO enrichment analysis was performed by BiNGO software (www.psb.ugent.be/cbd/papers/BiNGO/Home.html) with default parameters and the corrected P values calculated using the whole A. thaliana genome annotation as a reference set.
Supplementary Material
Acknowledgments
We thank Jinfeng Lu and Jinfeng Chen for technical assistance. This project was supported by grants from US–Israel Binational Agricultural Research and Development Fund (BARD-IS-4513-12), USIsrael Binational Science Foundation (BSF-2011302), and US Department of Agriculture Research Service (6659-22000-025) and by Agricultural Experimental Station of the University of California, Riverside (to S.-W.D.) This work was also supported in part by grants from National Basic Research Program 973 (2014CB138402) and the Natural Science Foundation of China (to Y.L.). Y.-Q.Y. and Y.-H.Q. were supported by China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSM1493819–GSM1493827).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407131111/-/DCSupplemental.
References
- 1.Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding SW & Lu R (2011) Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr Opin Virol 1(6):533–544. [DOI] [PMC free article] [PubMed]
- 4.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11(11):745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342(6155):231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342(6155):235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7(5):e1002022. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimura H, et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011;7(5):e1002021. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313(5783):68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 10.Fusaro AF, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7(11):1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19(6):2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25(14):3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XB, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(1):484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Ruiz H, et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22(2):481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XB, et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011;23(4):1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbonell A, et al. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24(9):3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey JJ, et al. An antiviral defense role of AGO2 in plants. PLoS ONE. 2011;6(1):e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA. 2008;105(38):14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227(1):176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12(2):89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 21.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant−pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 22.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 23.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki A, Medzhitov R. Innate responses to viral infections. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Vol I. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 189–213. [Google Scholar]
- 27.Martínez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: A diversity of pathways. Biochim Biophys Acta. 2013;1829(12):1300–1308. doi: 10.1016/j.bbagrm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21(17):4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18(19):2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123(7):1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creasey KM, et al. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508(7496):411–415. doi: 10.1038/nature13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunoyer P, et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29(10):1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wei W, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149(1):101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38(6):721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2(5):E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15(2):173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306(5698):1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 38.Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim Biophys Acta. 2013;1829(6-7):590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Gy I, et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19(11):3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding SW, Anderson BJ, Haase HR, Symons RH. New overlapping gene encoded by the cucumber mosaic virus genome. Virology. 1994;198(2):593–601. doi: 10.1006/viro.1994.1071. [DOI] [PubMed] [Google Scholar]
- 42.Schiebel W, et al. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell. 1998;10(12):2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross AF. Systemic effects of local lesion formation. In: Beemster ABR, Dijkstra J, editors. Viruses of Plants. North-Holland; Amsterdam: 1966. pp. 127–150. [Google Scholar]
- 44.Fu ZQ, Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 45.Li HW, et al. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 1999;18(10):2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sansregret R, et al. Extreme resistance as a host counter-counter defense against viral suppression of RNA silencing. PLoS Pathog. 2013;9(6):e1003435. doi: 10.1371/journal.ppat.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Mallory AC, et al. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell. 2001;13(3):571–583. doi: 10.1105/tpc.13.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endres MW, et al. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 2010;6(1):e1000729. doi: 10.1371/journal.ppat.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olmedo G, et al. ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA. 2006;103(36):13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308(5718):118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 51.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19(17):2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102(33):11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du P, et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7(8):e1002176. doi: 10.1371/journal.ppat.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.